Abstract
The use of cryo-EM and three-dimensional image reconstruction is becoming increasingly common. Our vision for this technique is to provide a straightforward manner in which users can proceed from raw data to a reliable 3D reconstruction through a pipeline that both facilitates management of the processing steps and makes the results at each step more transparent. Tightly integrated with a relational SQL database, Appion is a modular and transparent pipeline that extends existing software applications and procedures. The user manages and controls the software modules via web-based forms, and all results are similarly available using web-based viewers directly linked to the underlying database, enabling even naive users to quickly deduce the quality of their results. The Appion API was designed with the principle that applications should be compatible with a broad range of specimens and that libraries and routines are modular and extensible. Presented here is a description of the design and architecture of the working Appion pipeline prototype and some results of its use.
Introduction
The use of cryo-electron microscopy (cryoEM) and three-dimensional image reconstruction to study and verify macro molecular complexes is becoming ubiquitous. These techniques provide researchers with a powerful means by which to investigate stable molecules at subnanometer resolutions and explore the dynamics of macromolecules. Recent advances in instrumentation and processing capabilities, combined with automated data collection software, facilitate routine collection of large single particle datasets containing hundreds of thousands of individual particle images. It is now fairly common for these reconstructions to achieve subnanometeror near-atomic resolutions, yet it remains unclear what parameters in the numerous processing and refinement steps have the most influence on achieving the best possible resolution (Bottcher et al., 1997; Conway et al., 1997; Jiang et al., 2008; Ludtke et al., 2008; Matadeen et al., 1999; Stagg et al., 2008; Yu et al., 2008; Zhang et al., 2008; Zhou, 2008). There are a large number of routines and associated parameters that potentially affect the outcome of a reconstruction, and it is challenging to systematically explore this parameter space using standard input and output files and flat file management systems.
In conjunction with increasing dataset sizes, there are also increasingly numerous software packages available for data processing (Smith and Carragher, 2008). Most researchers use a variety of packages to achieve the various steps required to proceed from raw micrographs to a 3D reconstructed density map. Variations in file formats, user interfaces, Euler angle conventions and symmetry axes definitions, however, makes transfer of information from one package to another a tedious and error prone process. Attempts to unify some of these packages have been made, combining libraries and infrastructure to streamline the transfer of data between routines under a single processing environment but unifying packages are currently not in wide-spread use (Hohn et al., 2007).
The Leginon software system for automated data acquisition, developed in our lab (Suloway et al., 2005), tracks and records all microscope and environmental parameters associated with every acquired image into a MySQL database (http://www.mysql.com). This provides a straightforward method for comparing and contrasting datasets acquired during different microscope sessions and also provides assurance that acquisition parameters required for further data processing are always available and accurate. This permanent record of the dataset survives changes in lab personnel and is available to anyone authorized to access the data. Part of the rationale for developing Appion was to provide a similar system for tracking and recording the subsequent processing steps that result in a 3D density map. A secondary goal was to facilitate the steps required for 3D reconstruction and make these as transparent as possible. Appion is tightly integrated with the Leginon acquisition system so that preliminary processing and analysis procedures (e.g. particle picking and contrast transfer function (CTF) estimation) can be launched to run concurrently with data collection, and subsequent steps (e.g. creating image particle stacks, classification, 3D reconstruction) can begin as soon as data collection is completed. All parameters associated with these processes, as well as the results arising from them, are stored in the processing database.
The Appion processing pipeline provides users with the ability to process EM data in an intuitive and guided fashion. The user is able to inspect the processed data during every step toward a reconstruction; data outputs are presented via web pages in a standardized format to the user for assessment. While an experienced user of EM software packages will understand how to inspect the output data of the processing steps, this data can often be distributed across a large number of files in a variety of formats and require specialized programs for viewing. For new users, single particle processing can appear as an arcane art form, involving a series of “black box” methods that obscure the data from the end users. Providing web-based reporting tools that present the output data in a standardized format makes it more difficult for a user to ignore problems that might not be obvious in the final outputs but would show up as anomalies in intermediate data or in the overall statistical evaluation of the dataset. The standardized output also makes it easy to compare results of one reconstruction to another. With the growing popularity of EM as a tool for structural assessment, the Appion processing pipeline makes the associated software tools accessible to novice electron microscopists, allowing transparent data processing and thereby reducing the possibility of incorrect or misinterpreted reconstructions.
The database that supports Appion has the added benefit of providing long-term provenance for the data and the reconstructions that result from it. Stored processing parameters and results enable new users to follow precisely how raw data was processed in order to arrive at a given reconstruction. This infrastructure provides insurance against the loss of knowledge upon the departure of a lab member and provides a record of all processing parameters and location of files pertaining to a reconstruction from years past.
A fundamental aspect of the Appion processing pipeline architecture is its ability to incorporate and integrate new routines, methods, and software. There are currently a myriad of data processing and reconstruction software packages available to the EM community, and new ones will undoubtedly continue to appear and evolve (Smith and Carragher, 2008). It is unlikely that a solitary monolithic software package will dominate in the foreseeable future, so that it is essential that any processing pipeline have the flexibility to easily incorporate new functions. To this end, the uniformity of the underlying MySQL database acts as a translator between the many different file formats and parameter definitions extant in the community, allowing for the modular transfer of data between packages. Such integration of routines from different packages also allows for side-by-side comparisons of competing functions maintained by the packages, providing a quantitative means to assess the strengths and weaknesses of different reconstruction software.
We present here a working prototype of the Appion pipeline, describing its design and infrastructure, and some results of its use in our own laboratory. We begin with a brief overview of the entire process and then go on to describe some of the details of the supporting infrastructure. A much more detailed description of the process of using the pipeline is provided in the supplementary material.
Overview of Appion Infrastructure
The organization of Appion is divided into 3 layers: projects, experiments, and processing runs, and is summarized in Table 1. The first step in the pipeline is to define a project by filling out a short web based form. This registers the new project by creating a new unique entry in the project database, maintaining a detailed summary that can be edited or updated by authorized users. All data acquisition experiments that use Leginon are linked to a project, so that users can access and view all the data associated with any given project using a web based summary page that queries the database for all associated experiments (Supplementary Figure 1), and subsequent analyses. Users can browse through the collected micrograph images or monitor the image statistics as the experiment progresses using a set of web based viewers. Summary pages provide concise graphical reports on information such as the drift rate, image statistics, contamination level, environmental conditions, etc (Fellmann et al., 2002).
Table 1.
Appion Organization
| Project | Description of the biological structure of study and related information, such as background information, preliminary data, user’s names, funding, biohazards, etc. |
| Experiments | A set of data acquired from Leginon microscopy sessions. The user provides a brief description for the experiment, and the database stores all acquisition parameters. All experiments are associated with a project. |
| Processing Runs | Individual attempts at processing and manipulation of the raw data collected during an experiment, e.g. particle picking, CTF estimation, reference-free classification, reconstruction. |
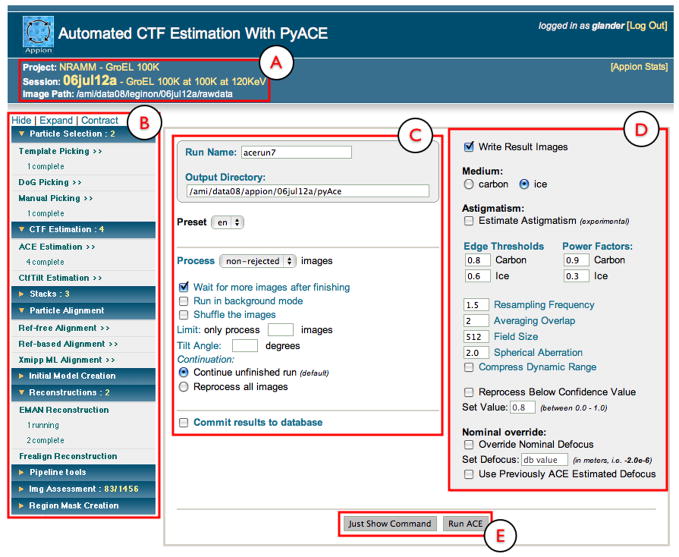
The Appion pipeline allows preliminary processing of the acquired images (e.g. particle picking and CTF estimation) to occur concurrently with data collection. Access to the Appion processing web pages is provided by a link from the web-based image viewing pages (Supplementary Figure 1D). The main page (Figure 1) provides a menu bar listing processing functions that are currently available to the experiment based on the current status of the analysis. Initially, the only data associated with a given experiment will be raw micrographs, so the functions available to the user are limited to particle picking, CTF estimation, and visual micrograph assessment. When these functions are launched, they process the micrograph data as soon as it is registered into the database, and keep pace with the rate of data collection. The menu of processing functions indicates the number of processes currently running or completed (Figure 1). As soon as the results from these initial processing routines are registered in the database, further options become available in the menu. For example, particle stack creation becomes available after particle picking, and classification and reconstruction procedures appear after stack creation. At each step users are able to view the results of a processing routine via a web browser that presents the outputs in a variety of formats, including output parameters, graphs, images, volume rendering. Our goal is to provide a transparent and comprehensive presentation of intermediate results that encourages and enables critical data assessment and identification of anomalies that mayarise during data processing.
Figure 1. Appion web-based interface.
In the upper left hand section, the project, experiment name, description and raw data path is displayed (A). On the left side of all Appion Web pages, there is a navigable menu that displays all available reconstructions steps, along with the number of processing jobs that are running or completed (B). Each section of this menu can be expanded or contracted to show or hide the subsections. To the right of the menu, a web form pertaining to the currently selected reconstruction step is shown. The left portion of the form (C) is termed the appion Loop form, and is the same for all pages that will perform a function on all the images in the database. The right portion of the form shows the parameters that are specific to the current reconstruction step (D). Default values are provided for the parameters. The user has the option of either directly launching the job to a processing cluster or of requesting the text version of the command so that it can be manually launched from a Unix terminal (E).
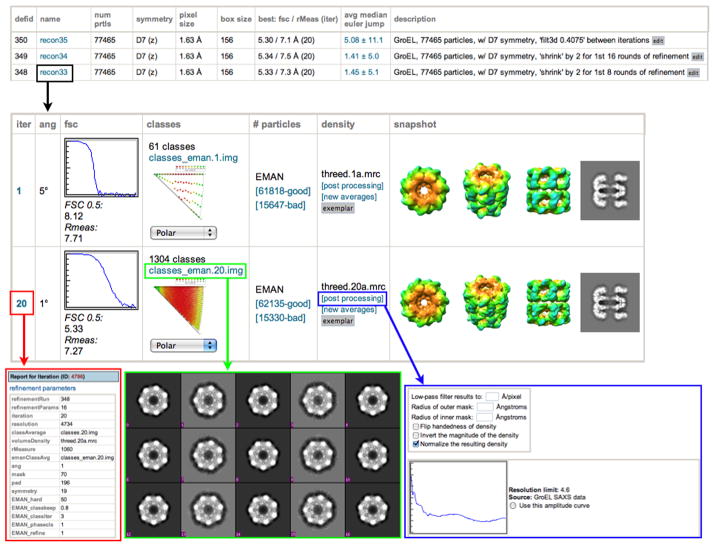
Once a final 3D reconstruction has been completed, the parameters specified during the reconstruction, as well as the results of all particle classifications, class averages, and three-dimensional models from each iteration are stored to the database. The resolution of the reconstruction is calculated using both the conventional Fourier shell correlation (FSC) method (Harauz and Van Heel, 1986), and the Fourier neighbor correlation method (which we here refer to as Rmeasure) (Figure 2)(Sousa and Grigorieff, 2007). The user then has the option of specifying a 3D reconstructed density map as an “exemplar”, which generates a web page that provides a detailed summary of all of the data collection and processing steps for that particular reconstruction. The exemplar page also provides a paragraph of text describing the methods in a format suitable for inclusion in a journal article (Supplementary Figure 4).
Figure 2. Reconstruction report page.
Overall statistics for every reconstruction stored in the database are displayed on the reconstruction summary page. Selecting one of these reconstructions links the user to a page displaying the results of each iteration (black box). For every iteration, the parameters for that iteration are available by clicking on the iteration number (red box). Plots showing the Euler angle distribution and the FSC curve. These plots, as well as most other images and graphs displayed, are thumbnails that can be displayed at full resolution by clicking on them. Class averages can be viewed by clicking on the “classes” link (green box) and particles that were used in the reconstruction vs. those that were rejected can be viewed by clicking on the “good” and “bad” links. The user has the option to do some post-processing on the 3D maps (blue box), e.g. applying a low-pass filter, masks, flipping the handedness, normalizing, and performing an amplitude adjustment.
Appion Infrastructure
Infrastructure Overview
Appion, similar to Leginon, has been implemented primarily in the Python scripting language (http://www.python.org), which is in increasingly wide use in a range of scientific disciplines. This powerful language provides cross-platform compatibility and can be readily learned even by non-programmers. In Appion the main function of the python scripts is to provide generic “wrappers” for a wide array of existing software packages in use by the EM community, thus providing for inter-package compatibility. For example, functions from SPIDER, EMAN, Frealign, Imagic, FindEM, Xmippand Matlab have all been incorporated into the Appion pipeline by Python wrappers (Frank et al., 1996; Grigorieff, 2007; Ludtke et al., 1999; Mallick et al., 2005; Roseman, 2003; Scheres et al., 2008; van Heel et al., 1996). The relative simplicity of the python language makes it accessible even to non-programmers, so that anyone with an interest can incorporate a new software package into the pipeline.
Fundamental to the Appion pipeline is the underlying database, which tracks all information and provides the links between disparate packages. The creation and maintenance of the Appion database is based on an automatically adaptive database structure that also supports the Leginon and Project databases. The Appion database is relationally linked to these other databases such that information can be traced back from every point of processing; for example, an individual particle contributing to a reconstruction can be tracked back to its location in a specific image and thus to every parameter associated with the data acquisition. The Project database stores global data associated with the overall biological project, the Leginon database primarily stores information related to the raw data as it is collected, and the Appion database stores all the parameters, inputs, and outputs from every processing step. This consortium of databases provides a complete record of the workflow from micrograph to reconstruction, and querying this information provides insight into the results of varying algorithms and methods used during data collection and processing.
Database Management by Sinedon
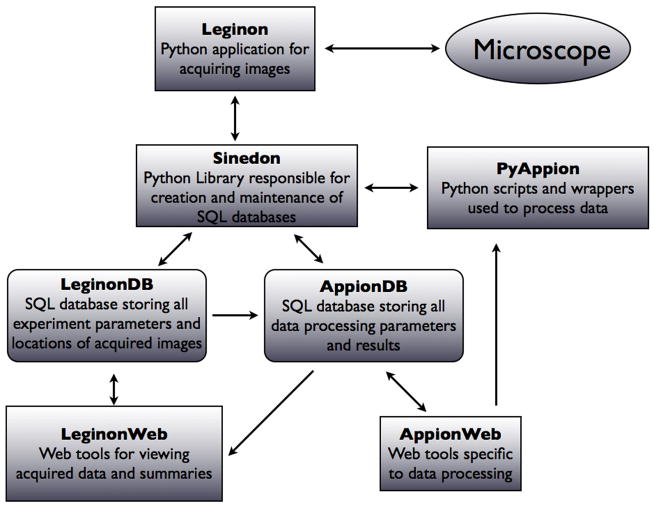
The structured query language (SQL) is a powerful open source relational database management system (RDMS) for the creation and administration of databases, and remains the standard-bearer for declarative database languages (http://www.mysql.com). Querying, updating and modifying an SQL database structure, however, requires knowledge of the SQL language, and manually updating the entries to support new procedures as they are added to the pipeline can be a tedious and confusing process, especially as the size of a database grows. One of the most critical components supporting the Appion infrastructure is the Sinedon software library, which was created to provide an interface between Appion’s Python scripts and the Appion SQL database (Figure 3) and to automatically maintain relational links between tables and columns.
Figure 3. Schematic representation of the Appion infrastructure.
The Leginon application controls the microscope and acquires data interacting with the Leginon database via the Sinedon library. A set of web based tools (Leginon Web), provide a view of the data stored in the Leginon database. Processing of the acquired data is controlled form the Appion Web pages, which launch pyAppion scripts on a cluster, and store the results into the Appion database via Sinedon.
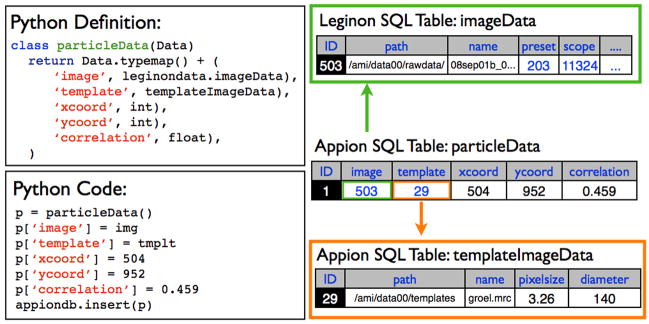
The Sinedon software is designed on the concept of object-relational mapping (ORM), which provides the ability to transfer data from object-oriented programming languages to a database system, with the added capability of creating new tables if they do not already exist. The goal of this library is to facilitate the creation and querying of database tables using a Python script, rather than through standard SQL commands. Utilizing the python module SQLDict, dictionary-like classes describing table structures are defined in Python within a single class named appion Data.py, and when an object of this class is created and filled with values in Python, the table corresponding to the class is created if it does not already exist, and the object information is entered as a new row in the table (Figure 4). All data are inserted as scalar values, and every row is unique through the use of primary keys. A timestamp is also stored for every element inserted into the database. An essential aspect of Sinedon is its ability to automatically create relational links between classes. If a class is created that contains within it an object of a different class, this information is stored in the SQL database using foreign keys (Figure 4). Additionally, altering the class definition in Python automatically performs updates of the table structure in the Appion database. Upon the next insertion of an object pertaining to this class, the changed structure will be mapped automatically to the SQL table. Through Sinedon, Appion developers are not required to write SQL queries, and the Appion database, which consists of nearly 100 tables (many containing dozens of columns) and which is constantly being modified and extended as new procedures are added to the pipeline, does not require any manual maintenance or specialized expertise.
Figure 4. The Sinedon library.
A python class is defined in appion Data.py, and instantiated in a python script. Parameters are assigned to the instance of the class, and entered into the database, using relational links, and creating new tables if necessary.
The PyAppion Library
The major functional component of Appion is a collection of scripts that control all the available analysis and reconstruction procedures. These scripts reference the classes defined in appion Data.py, defining how data is read from and written to the Appion database (Figure 4). Because many of these scripts operate on the acquired images as they are collected during an experiment, we have created a general module named appion Loop.py, which steps through all existing images in an experiment, and then pauses and waits for a new image to be entered into the database. Functions such as particle pickers or CTF estimators can be launched using this module to perform tasks during an experiment, so that processing of all newly collected images occurs concurrently with data collection. The Appion python scripts utilize Python’s “optparse” module for command line parsing, type assignment/conversion, and help document generation. The optparse module also provides for the assignment of default values in the case of an absent parameter. This supporting library provides a standardized method with which to develop and run procedures and to store parameters and results to the SQL database.
Launching processing jobs via Appion Web
While the Appion python scripts can always be launched from a command line setting, all of the major Python scripts required to perform a single particle reconstruction can also be accessed and launched from an intuitive web-based graphical user interface (Figure 1). This web-based user interface is generated using PHP, a server-side scripting language for the generation of dynamic web pages that is capable of reading and writing to the Appion SQL database (http://www.php.net). These web-based interfaces to the pipeline provide a graphical means by which to create and launch the Appion Python scripts but the user also always has the option to request the corresponding command-line text, which can be modified and manually submitted. This is especially useful when prototyping new options. The Appion web submission pages have been standardized, using a template designed specifically to the Appion structure. An “Appion Loop” template page emulates the Python appion Loop.py script, such that the user can set up the process to run through all existing images contained in an experiment. The user specifies the type of images upon which to perform the given function (e.g. close vs. far from focus), whether or not the processing is to run concurrently with data collection, and if the results should be stored to the database (Figure 1).
While the appion Loop section of the user interface remains consistent across similar procedures, parameters specific to the selected function appear in a separate section of the user interface. Whenever appropriate, default parameter values are automatically provided based on the experimental setup or previous processing results. Parameter values can also be recalled from earlier processing runs, which is particularly useful when it is desirable to process two separate datasets in the same way. Hovering the cursor over any of the parameter labels on the web page, pops up a documentation window describing the parameter, and may offer some advice on its usage (see Figure 5). Every processing run is associated with a unique job name tracked in the database. Default names are automatically assigned by the Appion scripts but the user is free to change the default name to make it more descriptive under the restriction that two jobs with the same name cannot be submitted for the same experiment. A series of other sanity checks are performed before the script is launched, ensuring that all necessary parameters have been assigned values, and that the values are appropriate for data that will be processed. For many of the procedures, the user is also offered the option of processing a single image from the dataset in order to test the suitability of entered parameters. A variety of applications for manual assessment and/or editing of automatically processed data are also available from the Appion web interface.
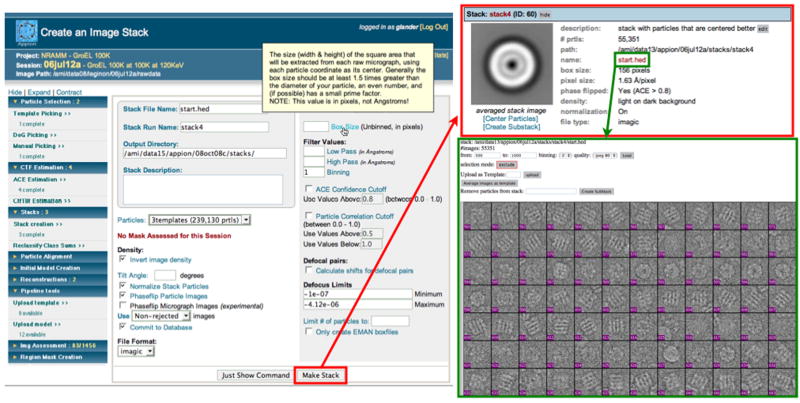
Figure 5. Creation of image stacks.
Users are able to create an image stack from the results of a particle selection routine, filtering the particles by the confidence of the CTF estimation, the particle correlation (if an automatic particle picker was used), or by defocus limits. Popup documentation is displayed describing the “box size” parameter. The resulting stack is summarized online, with an averaged image of all the particles comprising the stack. From this point the user may examine the individual particles within the stack, center the particles using EMAN’scenalignint, or create a subset of the stack.
Upon submission of the web form, the Appion Python command is sent to a queue on a local processing cluster, and an entry for monitoring the job’s status is entered into the database. Upon the commencement of the script, the job’s status is updated in the database to reflect this, and the user can track the progress of the job as it proceeds. In the event of undesired results or an analysis that is proceeding badly, the user can stop the processing job, at which point they can return to the web form and make any necessary changes and resubmit. Stopped jobs can be resubmitted to continue from the point at which they were previously stopped, and the number of jobs a user can submit is limited only by the implemented processing hardware and cluster queuing software.
Viewing job results via Appion Web
An extensive set of web tools have been developed to support the reporting of the processed results to the user. This is the most critical aspect of the Appion pipeline, as an early mistake in processing could otherwise go unnoticed, leading to confusing and incorrect results later on in the reconstruction process. The software that is required to read and parse data from the Appion database and display the results to web pages is primarily written in PHP. Results are displayed in a variety of ways, either as summaries of the entire process or as individual images, graphs or overlays onto existing images. Particle stacks are displayed using a PHP module capable of reading IMAGIC, SPIDER, and HDF5-formatted stack files, and which is modeled on the EMAN “v2” function (Figure 5C)(Ludtke et al., 1999). This same viewer is used to examine results of reference-free classification, class averages resulting from a reconstruction, slices of a three-dimensional model, or any other image stored into a stack file.
Results
The Appion prototype is currently in use on a day-to-day basis by multiple labs at our institute and by users with varying degrees of EM expertise, including visitors from external institutions. The pipeline has been used to perform two-dimensional assessment of structural variability and conformational heterogeneity, and to reconstruct dozens of three-dimensional structures ranging from 200 kilodalton complexes to 16 megadalton viruses (Lander et al., 2008; Nemecek et al., 2008; Stagg et al., 2008). To date, the database stores the locations of over 20 million selected particles, ~1000 particle stacks (with an average of roughly 15,000 particles per stack), and ~350 reconstructions corresponding to over 100 million particle classifications. Despite this accumulation of data, queries to the database are still very fast and the processing results from every experiment can be readily accessed and examined in detail through the standardized web reporting pages. Without the organizational structure maintained by the Appion database, these data would be difficult to track adequately.
The pipeline is under constant development by a team of developers with a range of expertise in computing. The codes are managed using a version control system (svn) (http://subversion.tigris.org) and daily builds of the committed codes takes place automatically. The sinedon library and existing examples makes it straightforward for almost any EM user with a facility for computers to add new functionality to the Appion pipeline by writing python scripts. A somewhat more challenging task is to build the web pages that provide the graphical user interface and report the results as this requires knowledge of the PHP language and the ability to understand direct SQL queries. There are, however, templates that can be used to generate new web pages and forms.
A detailed description of using the Appion pipeline to reconstruct a 3D map of GroELis provided in supplementary materials, and a summary of the currently available functions and dependencies are described in Table 2.
Table 2.
Major Functions available in Appion
| Function | Description | Package Requirement |
|---|---|---|
| Particle Selection | ||
| Template Picker | Automated template-based particle picker | FindEM |
| DoG Picker | Automated particle picker based on difference of gaussians | Appion |
| Manual Picker | Manual selection of particles, or editing of automated particle pickers | Appion |
| CTF Estimation | ||
| ACE | Automatic CTF estimation (Figure 1) | Matlab |
| CtfTilt | Automatic CTF estimation for tilted datasets (RCT/OTR/Tomography) | CtfTilt |
| Stack Creation | ||
| Makestack | Creation of boxed particle images, with ability to filter images based on the quality of the CTF estimation and particle filtering based on correlation value. Low/High pass filtering, binning, phase-flipping, normalization, inversion of density can be applied to the particles at this step. | EMAN |
| Particle Alignment | ||
| Reference-free classification | Particle alignment and classification by multivariate-statistical analysis of an image stack (Supplemental Figure 2) | SPIDER |
| Maximum- likelihood multireference alignment | Particle alignment and classification by maximum-likelihood approach | XMIPP |
| Reference-based classification | Particle alignment and classification using reference-based analysis | SPIDER |
| 3D Reconstruction (Chimera is used to generate density snapshots) | ||
| EMAN reconstruction | Projection-based matching particle classification for 3d reconstruction | EMAN, Rmeasure, Chimera |
| EMAN/SPIDER reconstruction | Projection-based matching particle classification with correspondence analysis for class averaging | EMAN, SPIDER, Rmeasure, Chimera |
| Frealign reconstruction | Refinement of particle orientations. SPIDER, EMAN, or Frealign can be used to determine initial euler angles for each particle | [EMAN, SPIDER], Frealign, Chimera |
Discussion
An important aspect regarding the architecture of Appion is that although it provides a directed manner in which an inexperienced user can proceed through a reconstruction, experienced users do not lose any flexibility. The Appion “pipeline” is not a stringent series of steps into which the user is locked, but rather a guide that provides a rational user interface to a wide variety of processing and analysis packages and routines. An experienced user, provided they are familiar with Python, can readily incorporate new methods into the pipeline and quickly make changes or add features to existing procedures. Using the Sinedon interface, new tables and columns automatically populate the database as the user runs the new scripts.
The value of the database underlying the Appion pipeline is its ability to both streamline the reconstruction process and integrate disparate software packages, as well as tracking and managing a staggering amount of metadata that can be used to assess the influence of various processing during the course of a reconstruction. We have begun to use this database infrastructure to probe some of the potential factors that limit the resolution of 3D maps. Questions addressing the influence of various factors during data collection, such as accelerating voltage, electron dose, ice thickness, and contamination have been explored using GroEL as a test-bed (Cheng et al., 2006; Stagg et al., 2008; Stagg et al., 2006). The pipeline makes is easy to ensure that each data set is analyzed in precisely the same manner, and variations within the data can be monitored throughout the process. We have also investigated the effect of particle number on classification and computation of a three dimensional map, and the methods implemented by various packages to exclude unacceptable particles or class averages from inclusion in the final map. Using the Appion pipeline it is straightforward to launch dozens of single particle refinements, each with specific reconstruction parameters varied, so as to understand their effect on the final quality of the density map.
While we have begun initial efforts toward displaying and analyzing the information that is stored in the database, this is the area of development that is likely to continue to change most rapidly. The results pages only provide a subset of the information in the database and this is often presented in a format that does not take full advantage of the available metadata. We also envision future versions of Appion where the reconstruction process could be actively driven based on the current results. For example, the parameters for the next iteration of the reconstruction could be modified based on the results of the current step; particles that are not assigned to stable orientations could be excluded, or assigned to alternative classification methods; and the reconstruction could be paused or halted if anomalous data are detected.
This pipeline, due to the simple web-based design of its interface, provides excellent potential as a teaching device. Although not currently implemented, it should be possible for students new to EM to follow tutorials that run in the web browsers alongside the Appion web interface. These tutorials could take a new student through each step of a reconstruction, demonstrating the results of properly processed data and problems that might arise.
The current implementation of Appion is tightly integrated with the Leginon database, but data collection by Leginon is not necessarily a mandatory requirement. Appion requires as input several microscopic parameters (such as KeV, pixel size, spherical aberration, nominal defocus) to be associated with every micrograph for processing. We have written a python module, “manualimageloader.py”, that allows for data acquired from sources other than Leginon to be uploaded into a database and from there it can be processed using Appion. In this manner data acquired utilizing other packages, e.g. JADAS (Zhang et al., 2009), could be processed using Appion. Development of a web-based version of this import module is currently underway.
Appion is freely available under the Apache Open Source License, Version 2.0. Software can be downloaded from http://www.appion.org.
Supplementary Material
Acknowledgments
We thank Dr. William Young for providing extensive computational support in the integration of the Appion pipeline into the Scripps Garibaldi cluster. We are also grateful to Dr. Arthur Horwichand Dr. Eli Chapman for providing us with the GroEL test specimen. This project was primarily funded by grants from the National Institutes of Health (NIH) through the National Center for Research Resources’ P41 program (grants RR17573, RR023093), and additionally by a fellowship from the ARCS foundation (to G.C.L.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Cheng A, Fellmann D, Pulokas J, Potter CS, Carragher B. Does contamination buildup limit throughput for automated cryoEM? J Struct Biol. 2006;154:303–11. doi: 10.1016/j.jsb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–4. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- Fellmann D, Pulokas J, Milligan RA, Carragher B, Potter CS. A relational database for cryoEM: experience at one year and 50 000 images. J Struct Biol. 2002;137:273–82. doi: 10.1016/s1047-8477(02)00002-3. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Harauz G, Van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon l5 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–4. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semi automated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–8. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Matadeen R, Patwardhan A, Gowen B, Orlova EV, Pape T, Cuff M, Mueller F, Brimacombe R, van Heel M. The Escherichia coli large ribosomal subunit at 7.5 A resolution. Structure. 1999;7:1575–83. doi: 10.1016/s0969-2126(00)88348-3. [DOI] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM. Particle finding in electron micrographs using a fast local correlation algorithm. Ultramicroscopy. 2003;94:225–36. doi: 10.1016/s0304-3991(02)00333-9. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–90. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Carragher B. Software tools for molecular microscopy. J Struct Biol. 2008;163:224–8. doi: 10.1016/j.jsb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157:201–10. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Lander GC, Quispe J, Voss NR, Cheng A, Bradlow H, Bradlow S, Carragher B, Potter CS. A test-bed for optimizing high-resolution single particle reconstructions. J Struct Biol. 2008;163:29–39. doi: 10.1016/j.jsb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, Lander GC, Pulokas J, Fellmann D, Cheng A, Quispe JD, Mallick SP, Avila RM, Carragher B, Potter CS. Automated cryoEM data acquisition and analysis of 284742 particles of GroEL. J Struct Biol. 2006;155:470–81. doi: 10.1016/j.jsb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–9. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, Jakana J, Marsh MP, Booth CR, Shinkawa T, Nakata M, Chiu W. JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J Struct Biol. 2009;165:1–9. doi: 10.1016/j.jsb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–72. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Towards atomic resolution structural determination by single-particle cryo-electron microscopy. Curr Opin Struct Biol. 2008;18:218–28. doi: 10.1016/j.sbi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.