Abstract
In the Brassicaceae glucosinolates influence feeding, reproduction and development of many insect herbivores. Glucosinolate production and effects on herbivore feeding have been extensively studied in the model species Arabidopsis thaliana and Brassica crops, both of which constitutively produce leaf glucosinolates mostly derived from the amino acid methionine. Much less is known about the regulation or role in defense of glucosinolates derived from other aliphatic amino acids such as the branched-chain amino acids (BCAA) valine and isoleucine. We have identified a glucosinolate polymorphism in Boechera stricta controlling the allocation to BCAA- vs. methionine-derived glucosinolates in both leaves and seeds. Boechera stricta is a perennial species that grows in mostly undisturbed habitats of western North America. We have measured glucosinolate profiles and concentrations in 192 F2 lines that have previously been used for genetic map construction. We also performed herbivory assays on six F3 replicates per F2 line using the generalist lepidopteran Trichoplusia ni. Quantitative Trait Locus (QTL) analysis identified a single locus controlling both glucosinolate profile and levels of herbivory, the Branched Chain-Methionine Allocation or BCMA QTL. We have delimited this QTL to a small genomic region with a 1.0 LOD confidence interval just 1.9 cM wide, which in A. thaliana contains ∼100 genes. We also found that methionine-derived glucosinolates provided significantly greater defense than the BCAA-derived glucosinolates against feeding by this generalist insect herbivore. The future positional cloning of this locus will allow for testing various adaptive explanations.
Keywords: Boechera, glucosinolates, Quantitative Trait Loci, ecological genomics, herbivory
INTRODUCTION
The emerging field of ecological genomics strives to uncover the genetic and molecular mechanisms influencing organisms’ responses and adaptations to their natural environments (Feder and Mitchell-Olds, 2003; Ouborg and Vriezen, 2007; Ungerer et al, 2007; van Straalen and Roelofs, 2006). To achieve this aim requires species with a well-developed genomic tool-kit and a good understanding of ecologically important phenotypes. Rarely can both aims be met simultaneously. Molecular biological and genomic tools have been developed primarily for traditional model and agricultural organisms representing a narrow spectrum of phenotypes, whereas many organisms that are the focus of ecological research have had limited genomic resources devoted to them and/or are genetically intractable. Two solutions for uniting genomic and ecological approaches are to explore the natural variation found within model or crop species, or alternatively to turn to close relatives for which a broader range of adaptive traits can be investigated. These approaches have been successfully applied to understanding plant chemical defenses against insect herbivores by using the model species Arabidopsis thaliana and Brassica crops (both members of the family Brassicaceae) as well as their near relatives (Alonso-Blanco et al, 2005; Clauss and Koch, 2006; Koornneef et al, 2004; Padilla et al, 2007; Schranz et al, 2007a). In this report, we examine chemical defenses and their effects on insect herbivory in Boechera stricta, a close-relative of A. thaliana, by Quantitative Trait Locus (QTL) analysis for a set of glucosinolates not present in Arabidopsis or Brassica (Windsor et al, 2005).
The major chemical defense in the Brassicaceae and related families is the two-component glucosinolate-myrosinase complex (Rodman et al, 1998; Windsor et al, 2005) that can influence levels of feeding damage and oviposition by many insect herbivores (particularly Lepidoptera) (Beekwilder et al, 2008; Burow et al, 2006). Generalist insects are sensitive to glucosinolate-based plant defenses, whereas specialists can cope with these compounds (Grubb and Abel, 2006; Halkier and Gershenzon, 2006; Kliebenstein et al, 2005; Windsor et al, 2005). Levels of herbivory generally decrease the fitness of the host plants and it has been demonstrated that herbivores can act as agents of natural selection on plant chemical defenses (Mauricio and Rausher, 1997). When variable secondary compounds are found within a plant species, it is tempting to assume that there will also be differences in defense towards herbivores. However, this must be experimentally validated particularly when the defensive compound(s) being tested are not well studied, such as glucosinolates derived from branched-chain amino acids (BCAA).
In Arabidopsis, naturally occurring genetic polymorphisms and loss-of-function mutations have allowed identification of genes encoding glucosinolate biosynthetic enzymes (recent reviews by Grubb and Abel, 2006; Halkier and Gershenzon, 2006). The biosynthesis of glucosinolates occurs in three steps: chain-elongation of the amino acid (for Met and Phe only), formation of the core structure, and side chain modifications. Glucosinolates are synthesized from one of eight amino acid precursors. Chemicals derived from Met, Val, Ile, Leu, or Ala are classified as aliphatic glucosinolates; those derived from Trp are called indole glucosinolates; and those derived from Phe or Tyr are referred to as aromatic glucosinolates. Two consecutive reactions in the core pathway, catalyzed by the specific cytochrome P450s (CYP79 and CYP83 gene families), convert the amino acid to a S-alkylthiohydroximate (Halkier and Gershenzon, 2006; Kliebenstein et al, 2005). While structural variation among glucosinolates is derived from the precursor amino acids, modifications of the R-groups also contribute enormous diversity of this class of compounds. The carbon chain-elongation of methionine-derived glucosinolates in particular gives rise to much of the diversity of glucosinolates found in the Brassicaceae. Chain elongation is controlled by the GS-Elong locus, which encodes several methythioalkylmalate synthases (MAM). The final step in glucosinolate biosynthesis is possible side-chain modification, e.g. acylation, alkenylation, esterifcation, hydroxylation and oxidation.
Genomic and ecological approaches can by integrated through detailed investigations of natural variation displayed by model species. An example comes from the molecular evolutionary analysis of variation in Met glucosinolate chain-elongation among Arabidopsis ecotypes. Recent analysis has shown that glucosinolate chain-length variation, and resistance to feeding by the generalist herbivore Spodoptera exigua, is due to complex allelic and copy number variation of MAM genes at the GS-Elong locus (Kroymann et al, 2003). Comparative genomic studies have also implicated the MAM genes in natural genetic variation for glucosinolate chain elongation in the Brassica crops (Gao et al, 2006; Li and Quiros, 2002), and in Arabidopsis lyrata (Clauss et al, 2006; Heidel et al, 2006). In A. lyrata, genetic variation at GS-Elong determines biochemical variation in glucosinolate side chains both within and among populations, demonstrating widespread functional polymorphism at GS-Elong. While Met-derived glucosinolates are the dominant constitutive glucosinolates expressed in the leaves of Arabidopsis and Brassica species, we have little information about the possible ecological or adaptive role of aliphatic glucosinolates derived from other amino acids.
Transgenic methods can be employed to explore phenotypes beyond the natural range of variability (e.g. glucosinolates derived from other, non-methionine, amino acids). Transgenic over-expression of CYP79s, such as those coming from the cyanogenic pathway of non-glucosinolate producing species is one means of generating novel glucosinolates. Introduction of CYP79D2 from the biosynthetic pathway of the cyanogenic glucosides from Cassava (Manihot esculenta) (Mikkelsen and Halkier, 2003) led to the accumulation of glucosinolates derived from the branched-chain amino acids (BCAA) valine and isoleucine; namely, 1-methyl ethyl (isopropyl) and 1-methylpropyl, which are normally absent or present in trace amounts in Arabidopsis (Brown et al, 2003). When these transgenic plants were assayed with pathogenic fungi and bacteria, it was found that the Cyp79D2 transgenic line provided enhanced resistance to some pathogens such as Erwinia carotovora (Brader et al, 2006). However, the transgenic Cyp79D2 plants still produced predominantly endogenous Met-derived glucosinolates (Mikkelsen and Halkier, 2003).
Another approach to studying the effects of non-Met glucosinolates on herbivore defense is to examine additional species having variation beyond what is available in Arabidopsis. In a recent survey of glucosinolate diversity among the relatives of Arabidopsis, several examples of dominant non-Met glucosinolates were found (Windsor et al, 2005). In particular, the genus Boechera was found to have some lines that predominantly produced methionine glucosinolates (especially 6-methylsulfinyl hexyl) and others that predominantly produced valine-derived glucosinolates (1-methyl ethyl) (also, see Supplemental Data file which demonstrates this variation both within and between populations).
The genus Boechera contains an array of morphologically and ecologically diverse taxa with highest diversity in western North America. The genus has a base chromosome number of x=7 and contains both diploid sexuals, as well as diploid, aneuploid and polyploidy apomictic lines (Kantama et al, 2007). Boechera stricta is one of the most morphologically and molecularly well-defined Boechera species; it is predominantly diploid, sexual, highly self-fertilizing and most accessions form a monophyletic group (Dobeš et al, 2004; Schranz et al, 2005), making it a good system for ecological genomic studies. To this end, we have also created a number of genomic resources including extensive comparative analyses with Arabidopsis. The B. stricta line SAD12 was the focus of a partial genomic sequencing project in which we analyzed over 39,000 paired end-sequences (Windsor et al., 2006). We found that genic and inter-genic regions are very similar to A. thaliana, and conserved microsynteny can be used for rapid identification and cloning of Boechera genes (Windsor et al, 2006). These results were used to generate 196 microsatellite and candidate gene molecular markers and to create an F2 genetic map of B. stricta based on 192 individuals (Schranz et al, 2007b). The genetic map also established the detailed syntenic relationships to Arabidopsis and to the n=8 species such as A. lyrata and Capsella rubella (Schranz et al, 2006b).
The parents used for our genetic cross came from two diverse habitats and differ in their glucosinolate profiles. The maternal SAD12 locality in Colorado is a sagebrush grassland in a river valley occurring at an elevation of 2,530 m. This population produces predominately Met-derived 6-methylsulfinyl hexyl glucosinolate (6MSOH). The paternal LTM population from Idaho grows in a subalpine meadow occurring at an elevation of 2,390 m. Val-derived 1-methyl ethyl glucosinolate (1ME) is the predominant glucosinolate produced by LTM. Hence, Boechera provides an excellent opportunity to evaluate the effects that differences in glucosinolate amino acid precursors have on insect herbivory and whether these differences are adaptive. In this study, we perform QTL analysis on ecologically relevant quantitative variation in glucosinolates between Boechera populations and assess the effects of this variation on feeding by the generalist lepidopteran herbivore Trichoplusia ni (Noctuidae). These efforts are an important step towards the cloning of an ecologically variable trait that will allow for future direct testing of ecological hypotheses.
MATERIALS AND METHODS
Plant materials and genetic map construction
As described previously (Schranz et al, 2007b), a genetic cross was made between two highly inbred lines of Boechera stricta (Graham) Al-Shehbaz. The maternal line “SAD12” was collected in Gunnison County, CO and the paternal line “LTM” was collected in Lemhi County, ID. Details about the plant populations, locations and the methods for genetic crossing were reported previously (Schranz et al, 2005).
A total of 192 F2 lines were grown, with seeds placed on moist filter paper in sealed Petri dishes and cold-treated at 4°C for three weeks in the dark. The Petri dishes were then transferred to a growth chamber until seed germination. The germinated seedlings were transferred to 96-well flats. Seedlings were grown for four weeks and then transplanted to pots (11×11×13 cm). The plants were grown in a controlled growth room at 21-23°C under long day conditions (16 hours light and 8 hours dark).
When plants were eight weeks old we sampled young leaves from each line directly into liquid nitrogen to minimize breakdown of glucosinolates. The two most recent fully expanded leaves (∼0.1 g) per line were first harvested to determine constitutive glucosinolate composition (see below). The collected tissue samples were freeze-dried and the dry weight of each sample was measured. Tissue was then harvested for DNA isolations from each of the F2 lines used for genetic map construction (Schranz et al. 2007).
The F2 lines were then allowed to grow for an additional four weeks, and were subsequently vernalized by transfer to 4°C under long day conditions (16 hours light and 8 hours dark) for four weeks. Post-vernalizaton, the plants were transferred back to 21-23°C. All lines flowered in approximately 3-5 weeks and were allowed to self-fertilize. The resulting F3 seeds were harvested from each line.
To investigate the genetic variation for insect resistance among lines of B. stricta, we used a randomized complete blocks design with 6 replicates for each line. The 192 F3 lines were divided equally among 6 blocks (each block was constituted by four 48-pot flats). In October 2006, ten F3 seeds per line were placed into Petri dishes and subjected to 4°C for cold stratification for four weeks. Once germinated, six seedlings per line (192 × 6 = 1152 plants in total) were individually grown in 5 × 5 cm pots filled with standard soil (Fafard® 4p mix; Massachusetts, USA). Pots were arranged randomly in flats (48 pots/flat). Plantlets were grown in the Duke University greenhouse and placed under controlled growth conditions. After ca. 4 weeks, plants were moved to a growth chamber and analyzed for insect herbivory (below).
Insect feeding, herbivory assessment and statistical analysis
An insect feeding trial was conducted using the generalist lepidopteran Trichoplusia ni (cabbage looper, Noctuidae). Second instar-larva were ordered from Benzon Research Inc. (Carlisle, PA, USA) and were available for experimentation 24 hours before the beginning of the feeding trial.
Without any previous starvation period, a single second-instar T. ni larva was placed on each plant. To avoid larvae roaming among plants and to guarantee an individual insect treatment for each plant during the whole experiment period (48 hours), each plant-larva pair was encapsulated using a cone-shaped pipe of plastic (5 cm diameter × 14 cm high). Preliminary observations determined that the use of these plastic pipes did not affect feeding behavior or survivorship of T. ni larvae even over longer periods (72 hours). After 48 hours of feeding, larvae were removed from the plants and herbivory levels for each plant recorded. We recorded both the proportion of leaves with some sign of insect herbivory and a visual estimation of the proportion of tissue removed per leaf (ranging from 1-100; see (Kliebenstein et al, 2002) for a similar procedure). The percentage of leaf area damaged was calculated as the product of the proportion of leaves harmed and the average proportion of tissue removed per leaf. In addition, all plants were investigated for the presence or absence of insect larvae at the end of the experiment; any plants lacking a larva were excluded from the analysis (8 plants out of 1152). Since plant size may influence both the type and the concentration of glucosinolates in Brassicaceous species (Brown et al, 2003; Petersen et al, 2002), rosette diameter was recorded 24 hours before the experiment as an estimate of plant size.
To obtain least square means for T. ni herbivory for each line, we fitted a generalized linear mixed model using the GLIMMIX procedure in SAS 9.1.2 (SAS Institute 2005). In this model, we considered the response variable (i.e., percent plant damage per plant) to have a binomial distribution and logit link function. “Line” was included in the model as fixed-effect factor, and flat-nested-within-block was treated as random-effect factor. Plant size (i.e., plant diameter, see above) was a covariate included in the model to control for developmental differences among plants (see (Kliebenstein et al, 2002). Finally, least square model-adjusted means for herbivory were obtained using the ILINK option of the LSMEANS statement in the GLIMMIX procedure (SAS Institute 2005).
Glucosinolate analysis
Glucosinolate extraction and purification on parental and F2 leaf and seed material was carried out according to (Kroymann et al, 2001). High-pressure liquid chromatography (HPLC) with a diode-array detector was performed as described (Kliebenstein et al, 2001a; Reichelt et al, 2002) using an Agilent 1100 HPLC, with 96-cell extraction protocols and a 96-cell autoloader. Identification of desulfo-glucosinolates was carried out by comparison of retention times and UV absorption spectra at A229 nm with those of known standards and of peaks that were confirmed by electrospray mass spectrometry (LC/MS) or LC/MS/MS as reported in (Windsor et al, 2005).
Glucosinolate concentrations were inferred from HPLC peak areas and were normalized by dividing by the dry weight of the sample. The glucosinolate concentrations were then log-transformed before analysis. Ratios among compound classes were calculated to reflect enzyme function at key steps in glucosinolate biosynthesis (as described by Clauss et al, 2006).
QTL Analysis
QTL mapping of the log transformed F2 glucosinolate profiles and the least square model-adjusted means for herbivory based on the six replicated F3 lines was done using QTL Cartographer for Windows (Wang et al, 2001-2006) employing multiple interval mapping (MIM) (Kao et al, 1999; Zeng et al, 1999) and composite interval mapping (CIM) (Jiang and Zeng, 1995; Zeng, 1993; Zeng, 1994). CIM tests were performed at 1-cM steps, with a 10-cM window size. Genome-wide threshold values were determined at the 5% level by running 1000 permutations (Churchill and Doerge, 1994). The QTL detected in CIM analysis was used as an initial model for MIM mapping. Tests in MIM were performed in 1-cM steps, with Bayesian Information Criterion model selection. One- and two-LOD support limits for each QTL were calculated around the QTL positions. The results of QTL analysis, including the 1- and 2-LOD confidence intervals, were drawn using MapChart 2.2 (Voorrips, 2002).
RESULTS
Glucosinolate variation and QTL analysis
The leaves of the parental genotypes contained predominately aliphatic glucosinolates derived from either the amino acid methionine [4-methylsulfinylbutyl (4MSOB); 6-methylsulfinylhexyl (6MSOH); 7-methylsulfinylheptyl (7MSOH); 6-methylthiohexyl (6MTH); 7-methylthioheptyl (7MTH)] or from the branched-chain amino acids [1-methylethyl (1ME) from valine; 1-methylpropyl (1MP) from isoleucine; 2-methylpropyl (2MP) from leucine]. Both progenitor genotypes also displayed trace quantities of the tryptophan-derived indolic glucosinolate 4-methoxy-indol-3-yl-methyl (4MO-I3M). While the total quantities of glucosinolates were similar among genotypes, they differed in their relative quantities of particular compounds. The most prominent glucosinolate for the SAD12 parent was the methionine-derived 6MSOH (also known as glucohesperin), whereas the LTM parent had higher concentrations of BCAA-derived glucosinolates 1ME (also known as glucoputranjivin) and 1MP (also known as glucocochlearin). We have also observed a similar pattern of glucosinolate variation both within and between populations of B. stricta (Supplemental Data).
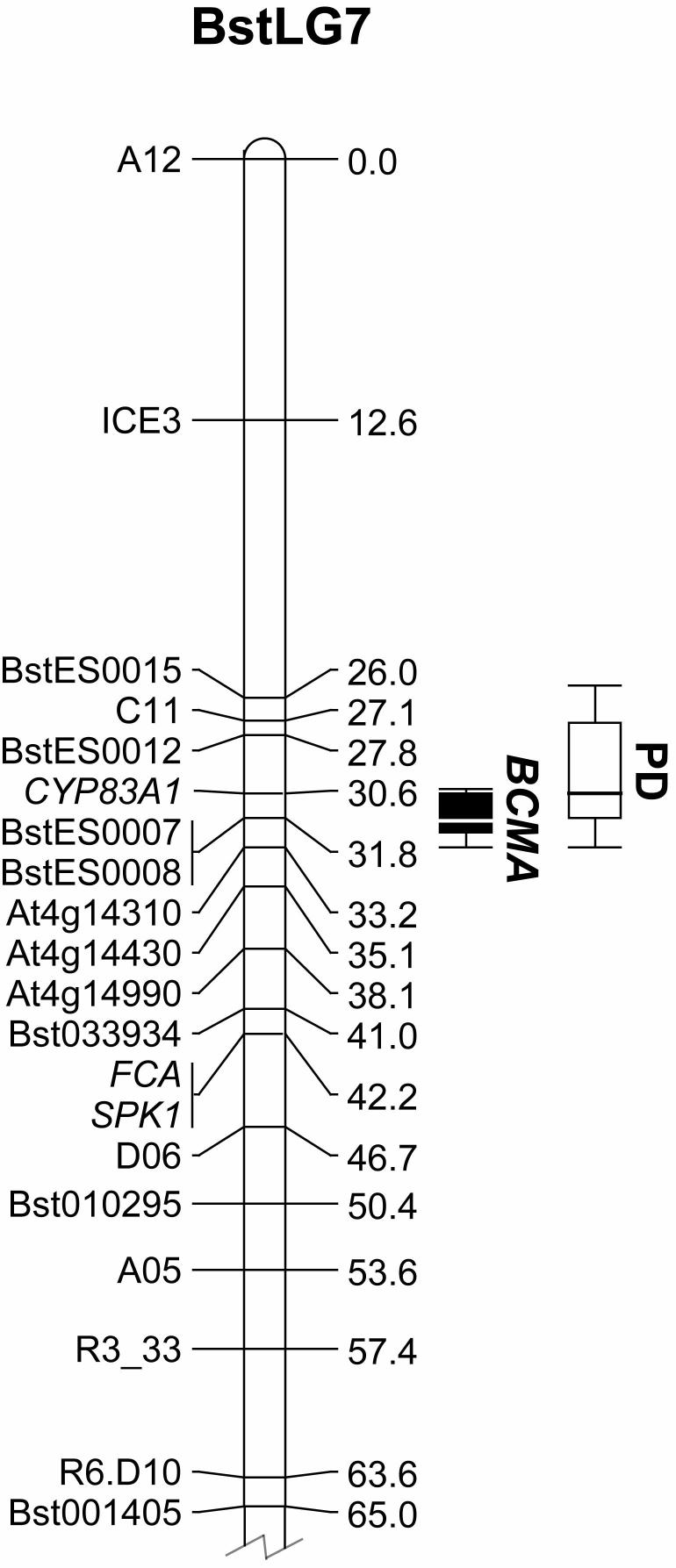
QTL analyses were performed using the weight adjusted and log transformed glucosinolate amounts, as well as by analyzing the log ratio of total BCAA-derived to total Met-derived glucosinolates. Previous quantitative work on glucosinolates has shown that ratios are an effective method for normalizing data (Clauss et al, 2006; Heidel et al, 2006). The predominant glucosinolate compounds, and the log ratio of BCAA-Met glucosinolates, had only one significant QTL located on the top of BstLG7 (Figure 1). The log ratio of BCAA-Met glucosinolates had the greatest level of support using MIM analysis, with a LOD score of 65.1 centered at 31.8 cM and had a very narrow 1-LOD confidence interval from 30.6-32.5 cM, encompassing the interval from markers Cyp83A to BstES007 and BstES0008 (Figure 1). CIM mapping gave very similar results with a LOD of 66.6. We have named this locus the Branched-Chain Methionine Allocation (BCMA) locus. The BCMA QTL has an R2=0.804, with the additive component accounting for 69.1% of the variance and the dominance component accounting for 11.3% of the variance.
Figure 1.
Positions of single QTLs detected for glucosinolate profile (BCMA) and levels of herbivory (PD), both located on linkage group BstLG7 of Boechera stricta. Marker names and cM positions for the top 65.0 cM of BstLG7 from the published F2 genetic map (Schranz et al. 2007b) are shown to the left. QTL analyses were performed using the log ratio of total BCAA-derived to total Met-derived glucosinolates for both leaves and seeds of F2 individuals, giving one significant QTL named the Branched Chain-Methionine Allocation or BCMA locus. BCMA is shown in black to the right of the linkage group, with the 2-LOD confidence interval represented by bars, the 1-LOD confidence interval by a box and the maximal LOD score (of 119.4) by the horizontal line. Plant Damage (PD) was calculated from 48-hour feeding trials conducted with second instar larvae of the generalist lepidopteran Trichoplusia ni. Analysis gave a single significant PD QTL, shown in white to the right of the linkage group, with the 2-LOD confidence interval represented by bars, the 1-LOD confidence interval by a box and the maximal LOD score (of 7.2) by the horizontal line.
The seeds of the F2 population had a similar spectrum of glucosinolates to that of the leaves. However, there were some major differences. In particular, there was a higher quantity of the thiol glucosinolates 6MTH and 7MTH than in the leaves (with more of the sulfinyl glucosinolates). MIM analysis showed a LOD score of 119.4 centered at 31.8 cM, with R2=0.934 for BCMA QTL. The additive component accounted for 73.7% of the variance and the dominance component for 19.7% of the variance. CIM mapping gave similar results, with a LOD score of 103.5 centered at 31.0 cM, and R2=0.923.
A summary of the variation in glucosinolate composition and how it varies with the genotype of BCMA (S alleles derived from SAD12 parent and L alleles from LTM parent) in the F2 population is presented in Figure 2. The figure shows how total leaf glucosinolate content remains nearly constant, but that genotypes carrying SAD12-derived genotypes have greater amounts of Met-derived glucosinolates whereas LTM genotypes have greater amounts of BCAA-derived glucosinolates (Figure 2a). In seeds, there is a more uniform level of Met-derived glucosinolates, with variable BCAA-derived glucosinolates (Figure 2b). The segregation of the log ratio of the BCAA/Met glucosinolates in leaves and seeds used for QTL analysis by genotype is shown in Figure 2c.
Figure 2.
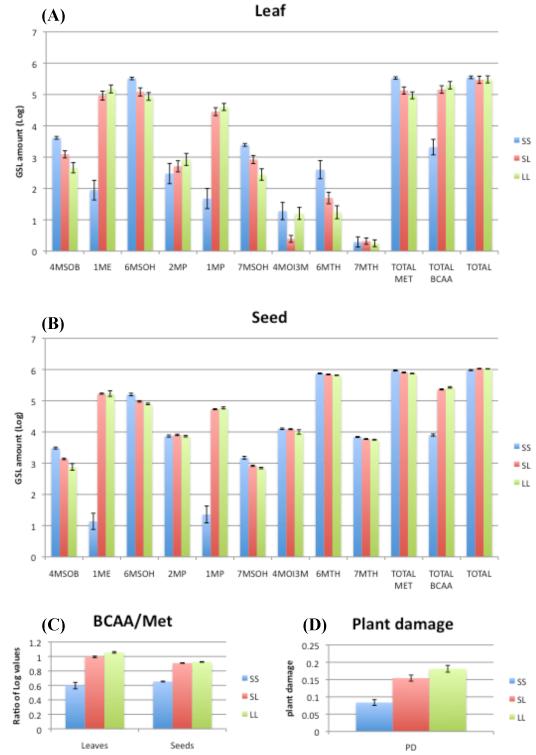
Summary of glucosinolate profiles and levels of herbivory given by genotype at BCMA locus of F2 individuals (SS=two alleles from maternal parent SAD12, LL=two alleles from paternal parent LTM, SL=heterozygote). Per gram dry weight adjusted Log-values of glucosinolates from Leaves (A) and Seeds (B), including methionine-deived [4-methylsulfinylbutyl (4MSOB); 6-methylsulfinylhexyl (6MSOH); 7-methylsulfinylheptyl (7MSOH); 6-methylthiohexyl (6MTH); 7-methylthioheptyl (7MTH)], BCAA-derived [1-methylethyl (1ME) from valine; 1-methylpropyl (1MP) from isoleucine; 2-methylpropyl (2MP) from leucine] and tryptophan-derived [4-methoxy-indol-3-yl-methyl (4MO-I3M)]. Also, shown are the sums for Met-derived, BCAA-derived and Total glucosinolates. (C) The Log-ratio of total BCAA-derived to Met-derived glucosinolates by genotype used for QTL analysis. (D) Levels of Plant Damage (PD) by genotype used for QTL analysis.
Insect feeding variation and QTL analysis
The analysis of Plant Damage (PD) by T. ni larvae showed significant differences among the 192 F3 families (GLMM result: F191, 928 = 24.83, P<0.0001), suggesting extensive variation in insect resistance among the lines for insect feeding on B. stricta leaves. PD was also significantly affected by variation in plant size (F = 4.19, P= 0.041; estimate ± 1SE: -0.0028 ± 0.0013) and Flat (Z = 0.047 P = 0.014). Least square mean values of PD for each genotype were used to QTL map insect feeding. We similarly detected a correlation between generalist insect feeding damage and glucosinolate profiles in an analysis of four B. stricta populations (Supplemental Data).
QTL analysis using PD had only one significant QTL by both MIM and CIM, located on the top of BstLG7 with a maximal LOD score of 7.2. The PD QTL is in the same position as BCMA, and is centered at 30.6 cM, and has a 1-LOD confidence interval from 27.2 - 31.8 cM (Figure 1). The PD QTL has R2=0.179, with the additive component accounting for 15.9% of the variance and the dominance component accounting for 2.1% of the variance. A summary of the variation in PD and how it varies with BCMA genotype is presented in Figure 2d.
Comparative genomics of BCMA region
The genetic map used in this study was previously utilized to establish a comparative genomics framework between B. stricta, Arabidopsis thaliana and n=8 chromosome relatives such as A. lyrata and Capsella (Schranz et al, 2007b). The BCMA genomic region is located within genomic block T (genomic blocks defined in (Schranz et al, 2006b)). The centromere of BstLG7 likely resides between blocks S and T based on comparison with A. lyrata (Schranz et al, 2006b). However, the 2-LOD confidence interval for BCMA lies within Block T (Figure 1), and there are normal levels of recombination between markers and nearly identical levels of recombination compared to the same interval of A. thaliana (which does not have a centromere near this genomic region). This supports the conclusion that the QTL does not reside within a low or non-recombining pericentromeric genomic region.
The 1-LOD confidence interval for BCMA spans only 1.9 cM. The genomic region shows high levels of synteny to the A. thaliana genomic region corresponding to the interval on chromosome 4 from approximately AT4G13410 - AT4G14250 (or nucleotide positions 7,790,000-8, 215,000). This interval in Arabidopsis only encompasses ∼100 annotated genes and ∼420 kb of sequence. Given that the Boechera genome is slightly larger (approximately 1.7x times the size of Arabidopsis (Schranz et al, 2006a)) this would still only equate to a genomic interval of around 700 kb. Additionally, the peak LOD scores for BCMA and PD suggests that the QTL may be restricted to the even smaller 1.1 cM window between markers Cyp83A (AT4G13770) and BstES0008 (AT4G14200), a region containing ∼50 genes and ∼200 kb in A. thaliana.
The genomic region on the top of BstLG7 near BCMA displays significant levels of gametic segregation distortion (Schranz et al, 2007b), with an under-representation of maternal SAD12 homozygous genotypes (17.9% of F2 progeny). So, although we did not find evidence of epistasis between BCMA and other genomic regions, segregation distortion within the genome could hamper our ability to detect such interacting loci. Ongoing back-crossing experiments of the BCMA region should help resolve this possibility.
DISCUSSION
The development of additional model species within the Brassicaceae which display novel physiological and ecological traits allows comparative genomic analysis of phenotypes that are not available in A. thaliana or Brassica crops (Schranz et al, 2007a). The diverse glucosinolate profiles that influence herbivory in Arabidopsis and other Brassicaceae arise largely from biosynthetic modifications in type and length of side chains and in the steps that determine the amino acid precursors of the glucosinolate skeleton. Species in the genera Arabidopsis and Brassica mostly produce chain-elongated glucosinolates derived from methionine in their leaves, and contain almost no glucosinolates derived from BCAA (Kliebenstein et al, 2001b; Windsor et al, 2005). In this report, we further the development of Boechera stricta as a system to study ecologically relevant phenotypic and genetic variation. In B. stricta we have found a major glucosinolate polymorphism that controls the allocation to BCAA- vs. methionine-derived glucosinolates. By quantitative trait locus analysis we have identified a single QTL with large effects on glucosinolate composition and small effects on insect resistance, which we have named the BCMA (Branched Chain Methionine Allocation) locus, influencing both the amino acid precursors of glucosinolate production and defense against a generalist herbivore. The future cloning of the locus will allow us to test evolutionary hypotheses, including the potential role of balancing selection and local adaptation.
Variation and QTL analysis of glucosinolates and insect feeding
Boechera shows a major glucosinolate polymorphism controlling the allocation to BCAA- vs. methionine-derived glucosinolates. The major methionine-derived glucosinolates are the six-carbon (6C) elongated 6MSOH and 6MTH in leaves and seeds, respectively. The distribution of carbon chain lengths of Met-derived glucosinolates is determined by allelic state of the GS-Elong locus. In A. thaliana this locus contains a tandem array of two or three genes (MAM1, MAM2 and/or MAM-L) and forms either 3C or 4C glucosinolates (Kroymann et al, 2003). Arabidopsis lyrata displays greater variation in longer-chain Met glucosinolates due to variation at the MAM locus (Heidel et al, 2006). It has been found that longer-chain glucosinolates tend to be more toxic than those with shorter chains (Borek et al, 1998). Hence, the predominantly 6C glucosinolates may provide a higher level of defense against herbaceous generalist insects. Boechera contains a tandem array of three MAM genes (MAMa, MAMb and MAMc) (Benderoth et al, 2006), which evidently control the chain-elongation process. In our cross we do not find variation or QTL for Met-derived chain-length differences. In other B. stricta accessions we have surveyed to date we also find little to no variation in methionine chain length variation (Mitchell-Olds, unpublished). Hence, unlike in Arabidopsis species (Clauss et al, 2006; Kroymann et al, 2003) variation at the MAM locus does not seem to be a major component of the natural variation of glucosinolates in B. stricta. However, we cannot rule out that some yet untested B. stricta populations will show significant differences.
In B. stricta the major BCAA-derived glucosinolates are 1ME derived from valine and 1MP derived from isoleucine. These glucosinolates are uncommon among members of the Brassicaceae (Windsor et al, 2005), but are found among other groups in the Brassicales. For example, within the genus Capparis (Capparaceae), C. flexuosa has high levels of the Ile-derived 1MP and C. spinosa (capers) has high levels of Val-derived 1ME (from which capers derive their characteristic flavor) (Mikkelsen and Halkier, 2003). In transgenic Arabidopsis plants (expressing the Cassava Cyp79D2 gene) that produced 1MP and 1ME glucosinolates, the authors found a strong correlation between total quantity of BCAA-glucosinolates and the ratio of 1MP:1ME ((Mikkelsen and Halkier, 2003). The authors hypothesized that this might be due to more rapid depletion of Val-pools. By contrast, in our B. stricta F2 lines carrying one or two copies of the LTM allele at BCMA we found a constant ratio of 1MP:1ME. This suggests that the flux through the pathway is either not rate limiting for Val or Ile precursors, or that regulation or enzyme kinetics are more finely controlled than in the Arabidopsis transgenic lines. The short-chained isothiocyanates 1MP and 1ME have a different bioactivity than the larger side chained and differently structured 6MOH as demonstrated by our feeding studies. A previous study had shown that shorter chain-length glucosinolates can be less toxic (Borek et al., 1998), however, the variable structure might also cause the differences in bioactivity. Potentially, the 1MP and 1ME glucosinolates have beneficial effects as biofumigants due to their high volatility, or alternatively, they may be more effective than Met-derived glucosinolates in combating bacterial or fungal pathogens (Brader et al, 2006).
We observed higher concentrations and altered ratios of glucosinolates in seeds than in leaves. Most notably, there much higher levels in the amount of the reduced 6MTH glucosinolate and higher levels in the amount of indolic glucosinolate 4MO-I3M. The larger concentration and greater amount of reduced Met-derived and non-aliphatic glucosinolates is similar to the difference between leaves and seeds observed in the Columbia ecotype of Arabidopsis thaliana (Brown et al, 2003). The conversion of reduced thiol- to the oxygenated sulfinyl-glucosinolates in Arabidopsis leaves has recently been found to be controlled by a flavin-monooxygenase as the S-oxygenating enzyme FMOGS-OX1 (Hansen et al, 2007). Expression of this locus and/or other FMO genes may be restricted to leaf tissues and absent in developing seeds. Also, the ratio of 1MP:1ME was slightly higher than in leaves when the lines had at least one LTM allele at BCMA, potentially due to different substrate pools and/or gene(s) expression between the two tissues. The differences in glucosinolates in seeds versus leaves could be of ecological importance. There are a number of insect species within the host range of B. stricta that do feed upon its the silques and/or seeds (Mitchell-Olds, personal observation). The potential role of the specific seed glucosinolates in the defense of these tissues will be an important future research objective.
To test whether the difference in allocation between Met- and BCAA-derived glucosinolates had any potential ecological significance due to altered defense, we preformed a feeding assay with a generalist herbivorous insect. QTL analysis revealed a single major additive QTL that co-segregated with the BCMA glucosinolate locus. From this result, we conclude that the long-chained Met-derived glucosinolates provide a higher level of defense against a generalist herbivore than the shorter-chain BCAA glucosinolates. We also observed a similar pattern in glucosinolate profiles and effects on generalist insect feeding by analysis of individuals from four populations of B. stricta (Supplemental Data), suggesting that the single major QTL identified in our cross is segregating within and between populations.
Potential genetic control of BCMA
There are several potential mechanisms for the differences in glucosinolate profiles controlled by the BCMA locus: a polymorphism in glucosinolate biosynthetic genes; a change in precursor amino acid substrate availability; a transporter between cellular compartments; or a regulatory change influencing biosynthesis. The analysis of glucosinolate biosynthetic mutants in Arabidopsis suggests that components of the core glucosinolate biosynthetic pathway are partly modular (Kliebenstein et al, 2001b; Windsor et al, 2005). In other words, the components are non-redundant and catalyze equivalent reactions while displaying varying substrate specificities. In A. thaliana, the conversion of Met-derivative amino acids to aldoximes is catalyzed by CYP79F1 and CYP79F2 (Chen et al, 2003; Hansen et al, 2001; Reintanz et al, 2001) with the resultant Met-aldoximes being metabolized mostly by CYP83A1 (Hemm et al, 2003; Naur et al, 2003). Even the final reaction in glucosinolate biosynthesis, the conversion of desulfoglucosinolates to the core glucosinolate structure is catalyzed by a family of sulfotransferases that display a level of substrate specificity (Piotrowski et al, 2004). Thus, alterations involving CYP79, CYP83, or AtST5 homologs, or the occurrence of a gene of unknown function could be an explanation for a difference between Met- and BCAA-glucosinolate phenotypes.
Variation in the availability of glucosinolate precursor substrates could also underlie the BCMA QTL. A number of biochemical changes could lead to differences in available substrate pools such as the redox state of the cells or available levels of sulfur needed for both Met and glucosinolate production. An interesting trade-off between levels of Met and branched chain amino acids was recently established, when it was shown that the catabolism of Met leads to increased Ile production (Rebeille et al, 2006). Furthermore, there are several strong connections and similarities between the synthesis of branched chain amino acids and the chain elongation reactions of Met leading to glucosinolate production (recently reviewed by Binder et al, 2007). For example, the isopropylmalate synthase (IPMS) genes involved in the elongation of Val to Leu are evolutionarily and biochemically related to the MAM genes involved in Met-chain elongation (de Kraker et al, 2007; Field et al, 2004). Similarly, the branched-chain aminotransferase 3 (BCAT3) gene has been shown to be directly involved in both BCAA- and glucosinolate-formation (Knill et al, 2007). Hence, the alteration in the specificity or function of a number of genes effecting precursor molecules could lead to the trade-off in Met- vs. BCAA-derived glucosinolates controlled by the BCMA locus in Boechera.
The third possibility is that there are regulatory changes affecting glucosinolate and/or BCAA biosynthetic genes. Once again, recent studies have found that transcription factors effecting Met-glucosinolate formation often also regulate genes involved in BCAA-formation. For example, gene expression analyses of mutants and transgenic lines containing the Myb transcription factors (Myb28/HAG1, Myb29/HAG3 and Myb76/HAG2) have shown co-regulation of methionine chain elongation and glucosinolate biosynthetic genes (e.g. MAM, Cyp79F, Cyp83A1, BCAT4) and BCAA biosynthetic genes (IPMS, BCAT3 and several additional genes involved in Leu production such as AtLeuC1) (Gigolashvili et al, 2007a; Gigolashvili et al, 2007b; Hirai et al, 2007).
Comparative Genomics and potential candidate genes
In our F2 mapping population we delimited the BCMA QTL to a small genomic region with a maximal LOD score of 119.4, and with a 1.0 LOD confidence interval just 1.9 cM wide, which in A. thaliana contains ∼100 genes. We used eight tightly linked markers to dissect this interval. Analysis of chromosome rearrangements among four crucifer species finds no evidence for a centromere within the QTL interval (Lysak et al, 2006; Schranz et al, 2006b), providing further support for normal recombination in our target region. Markers that are separated by 1 cM in B. stricta contain on average <30 genes in the corresponding region of A. thaliana, suggesting a gene density and recombination rate similar to A. thaliana.
Based on syntenic relationships between Boechera and Arabidopsis (Schranz et al, 2007b), the only predicted glucosinolate biosynthetic gene in the BCMA region is Cyp83A1 (Bak and Feyereisen, 2001; Naur et al, 2003). The Boechera homolog of this gene was used as a molecular marker, confirming its position within the QTL confidence interval. It is plausible that allelic differences in the Cyp83A1 might lead to substrate specificity as Cyp83A1 shows high affinities for other non-Met derived aldoximes in Arabidopsis (Brader et al, 2006; Mikkelsen and Halkier, 2003). The knockout phenotype of Cyp83A1 in Arabidopsis is a pronounced reduction of Met-derived glucosinolates and an increased production of indolic glucosinolates derived from Tryptophan via the activity of the Cyp79Bx and Cyp83B1 genes (Grubb and Abel, 2006; Hemm et al, 2003; Naur et al, 2003). Since we observe an increased production of BCAA-derived glucosinolates, and not increased Trp-derived glucosinolates, it seems unlikely that the BCMA phenotype is due to a non-functional Cyp83A1 allele from the LTM parent. An alternative is that genes associated with substrate availability might influence our glucosinolate polymorphism. At least one BCAA biosynthetic gene (AtLeuC1; AT4G13430) is near the QTL peak. Alternatively, a gene such as a methionine-tRNA ligase (AT4G13780) could be involved in altering the available pools of Met, or Selenium binding proteins (AT4G14030) might regulate sulfur availability (Van Hoewyk et al, 2008). Ultimately, the identification of the biochemical mechanism of this insect resistance polymorphism is conditional upon positional cloning of BCMA.
Future Directions
Despite recent progress in identifying genes controlling complex trait variation (Alonso-Blanco et al, 2005; Koornneef et al, 2004), we have little understanding of the frequencies and effects of QTL alleles or the evolutionary processes that maintain this variation (Mitchell-Olds and Schmitt, 2006; Mitchell-Olds et al, 2007). Similar to the variation found within our mapping population, we have also found large differences in the ratio of Methionine and Branched-Chain amino acid derived glucosinolates within and between populations of Boechera (Supplemental Data). The future cloning of the locus will allow us to test evolutionary hypotheses, including the potential role of balancing selection and local adaptation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kasavajhala Prasad, Bao-Hua Song, Kathy Springer, Petra Eissmann, Kerstin Weniger, Michael Reichelt, and Jonathan Gershenzon for advice and assistance. A.J.M. was funded by Spanish Ministry of Education and Science Grant EX-2006-0652. Support was provided by Duke University, the University of Amsterdam and the National Institutes of Health Award R01 GM086496-01 to T.M.O.
REFERENCES
- Alonso-Blanco C, Mendez-Vigo B, Koornneef M. From phenotypic to molecular polymorphisms involved in naturally occurring variation of plant development. International Journal of Developmental Biology. 2005;49(5-6):717–732. doi: 10.1387/ijdb.051994ca. [DOI] [PubMed] [Google Scholar]
- Bak S, Feyereisen R. The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiology. 2001;127(1):108–118. doi: 10.1104/pp.127.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J, van Leeuwen W, van Dam NM, Bertossi M, Grandi V, Mizzi L, et al. The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE. 2008;3(4):e2068. doi: 10.1371/journal.pone.0002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J. Positive selection driving diversification in plant secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24):9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S, Knill T, Schuster J. Branched-chain amino acid metabolism in higher plants. Physiologia Plantarum. 2007;129(1):68–78. [Google Scholar]
- Borek V, Elberson LR, McCaffrey JP, Morra MJ. Toxicity of isothiocyanates produced by glucosinolates in Brassicaceae species to black vine weevil eggs. Journal of Agricultural and Food Chemistry. 1998;46(12):5318–5323. [Google Scholar]
- Brader G, Mikkelsen MD, Halkier BA, Palva ET. Altering glucosinolate profiles modulates disease resistance in plants. Plant Journal. 2006;46(5):758–767. doi: 10.1111/j.1365-313X.2006.02743.x. [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry. 2003;62(3):471–481. doi: 10.1016/s0031-9422(02)00549-6. [DOI] [PubMed] [Google Scholar]
- Burow M, Muller R, Gershenzon J, Wittstock U. Altered glucosinolate hydrolysis in genetically engineered Arabidopsis thaliana and its influence on the larval development of Spodoptera littoralis. Journal of Chemical Ecology. 2006;32(11):2333–2349. doi: 10.1007/s10886-006-9149-1. [DOI] [PubMed] [Google Scholar]
- Chen SX, Glawischnig E, Jorgensen K, Naur P, Jorgensen B, Olsen CE, et al. CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant Journal. 2003;33(5):923–937. doi: 10.1046/j.1365-313x.2003.01679.x. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss MJ, Dietel S, Schubert G, Mitchell-Olds T. Glucosinolate and trichome defenses in a natural Arabidopsis lyrata population. Journal of Chemical Ecology. 2006;32(11):2351–2373. doi: 10.1007/s10886-006-9150-8. [DOI] [PubMed] [Google Scholar]
- Clauss MJ, Koch MA. Poorly known relatives of Arabidopsis thaliana. Trends in Plant Science. 2006;11(9):449–459. doi: 10.1016/j.tplants.2006.07.005. [DOI] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J. Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiology. 2007;143(2):970–986. doi: 10.1104/pp.106.085555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobeš CH, Mitchell-Olds T, Koch MA. Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A. x divaricarpa, and A. holboellii (Brassicaceae) Molecular Ecology. 2004;13(2):349–370. doi: 10.1046/j.1365-294x.2003.02064.x. [DOI] [PubMed] [Google Scholar]
- Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nature Reviews Genetics. 2003;4(8):651–657. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- Field B, Cardon G, Traka M, Botterman J, Vancanneyt G, Mithen R. Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiology. 2004;135(2):828–839. doi: 10.1104/pp.104.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao MQ, Li GY, Potter D, McCombie WR, Quiros CF. Comparative analysis of methylthioalkylmalate synthase (MAM) gene family and flanking DNA sequences in Brassica oleracea and Arabidopsis thaliana. Plant Cell Reports. 2006;25(6):592–598. doi: 10.1007/s00299-005-0078-1. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, Fluegge UI. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant Journal. 2007a;50(5):886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant Journal. 2007b;51(2):247–261. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S. Glucosinolate metabolism and its control. Trends in Plant Science. 2006;11(2):89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annual Review of Plant Biology. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kliebenstein DJ, Halkier BA. Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant Journal. 2007;50(5):902–910. doi: 10.1111/j.1365-313X.2007.03101.x. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA. Cytochrome P450CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. Journal of Biological Chemistry. 2001;276(14):11078–11085. doi: 10.1074/jbc.M010123200. [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Clauss MJ, Kroymann J, Savolainen O, Mitchell-Olds T. Natural variation in MAM within and between populations of Arabidopsis lyrata determines glucosinolate phenotype. Genetics. 2006;173(3):1629–1636. doi: 10.1534/genetics.106.056986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell. 2003;15(1):179–194. doi: 10.1105/tpc.006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(15):6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CJ, Zeng ZB. Multiple-trait analysis of genetic-mapping for quantitative trait loci. Genetics. 1995;140(3):1111–1127. doi: 10.1093/genetics/140.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantama L, Sharbel TF, Schranz ME, Mitchell-Olds T, de Vries S, de Jong H. Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(35):14026–14031. doi: 10.1073/pnas.0706647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CH, Zeng ZB, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152(3):1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidapsis thaliana. Genetics. 2002;161(1):325–332. doi: 10.1093/genetics/161.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics. 2001a;159(1):359–370. doi: 10.1093/genetics/159.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, et al. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiology. 2001b;126(2):811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Mitchell-Olds T. The glucosinolate-myrosinase system in an ecological and evolutionary context. Current Opinion in Plant Biology. 2005;8(3):264–271. doi: 10.1016/j.pbi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Knill T, Schuster J, Reichelt M, Gershenzon J, Binder S. Arabidopsis thaliana branched-chain aminotransferase 3 (BCAT3) functions in bothamino acid and glucosinolate biosynthesis. Plant Physiol. 2007:107.111609. doi: 10.1104/pp.107.111609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, et al. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiology. 2001;127(3):1077–1088. [PMC free article] [PubMed] [Google Scholar]
- Li GY, Quiros CF. Genetic analysis, expression and molecular characterization of BoGSL-ELONG, a major gene involved in the aliphatic glucosinolate pathway of Brassica species. Genetics. 2002;162(4):1937–1943. doi: 10.1093/genetics/162.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(13):5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio R, Rausher MD. Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution. 1997;51(5):1435–1444. doi: 10.1111/j.1558-5646.1997.tb01467.x. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Halkier BA. Metabolic engineering of valine- and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from cassava. Plant Physiology. 2003;131(2):773–779. doi: 10.1104/pp.013425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441(7096):947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nature Reviews Genetics. 2007;8(11):845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, et al. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiology. 2003;133(1):63–72. doi: 10.1104/pp.102.019240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouborg NJ, Vriezen WH. An ecologist’s guide to ecogenomics. Journal of Ecology. 2007;95(1):8–16. [Google Scholar]
- Padilla G, Cartea ME, Velasco P, de Haro A, Ordas A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry. 2007;68(4):536–545. doi: 10.1016/j.phytochem.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Petersen BL, Chen SX, Hansen CH, Olsen CE, Halkier BA. Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta. 2002;214(4):562–571. doi: 10.1007/s004250100659. [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Schemenewitz A, Lopukhina A, Muller A, Janowitz T, Weiler EW, et al. Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. Journal of Biological Chemistry. 2004;279(49):50717–50725. doi: 10.1074/jbc.M407681200. [DOI] [PubMed] [Google Scholar]
- Rebeille F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, et al. Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(42):15687–15692. doi: 10.1073/pnas.0606195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt M, Brown PD, Schneider B, Oldham NJ, Stauber E, Tokuhisa J, et al. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry. 2002;59(6):663–671. doi: 10.1016/s0031-9422(02)00014-6. [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, et al. Bus, a bushy arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13(2):351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman JE, Soltis PS, Soltis DE, Sytsma KJ, Karol KG. Parallel evolution of glucosinolate biosynthesis inferred from congruent nuclear and plastid gene phylogenies. American Journal of Botany. 1998;85(7):997–1006. [PubMed] [Google Scholar]
- Schranz ME, Dobeš C, Koch MA, Mitchell-Olds T. Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae) American Journal of Botany. 2005;92(11):1797–1810. doi: 10.3732/ajb.92.11.1797. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Kantama L, de Jong H, Mitchell-Olds T. Asexual reproduction in a close relative of Arabidopsis: a genetic investigation of apornixis in Boechera (Brassicaceae) New Phytologist. 2006a;171(2):425–438. doi: 10.1111/j.1469-8137.2006.01765.x. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Science. 2006b;11(11):535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Song BH, Windsor AJ, Mitchell-Olds T. Comparative genomics in the Brassicaceae: a family-wide perspective. Current Opinion in Plant Biology. 2007a;10(2):168–175. doi: 10.1016/j.pbi.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Windsor AJ, Song B-h, Lawton-Rauh A, Mitchell-Olds T. Comparative genetic mapping in Boechera stricta, a close relative of Arabidopsis. Plant Physiology. 2007b;144(1):286–298. doi: 10.1104/pp.107.096685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Johnson LC, Herman MA. Ecological genomics: understanding gene and genome function in the natural environment. Heredity. 2007 doi: 10.1038/sj.hdy.6800992. [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D, Takahashi H, Inoue E, Hess A, Tamaoki M, Pilon-Smits EAH. Transcriptome analyses give insights into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiologia Plantarum. 2008;132(2):236–253. doi: 10.1111/j.1399-3054.2007.01002.x. [DOI] [PubMed] [Google Scholar]
- van Straalen NM, Roelofs D. An Introduction to Ecological Genomics. Oxford University Press; Oxford, UK: 2006. [Google Scholar]
- Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. Journal of Heredity. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten C, Zeng ZB. Department of Statistics, North Carolina State University; Raleigh, NC: 2001-2006. [Google Scholar]
- Windsor AJ, Reichelt M, Figuth A, Svatos A, Kroymann J, Kliebenstein DJ, et al. Geographic and evolutionary diversification of glucosinolates among near relatives of Arabidopsis thaliana (Brassicaceae) Phytochemistry. 2005;66(11):1321–1333. doi: 10.1016/j.phytochem.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Windsor AJ, Schranz ME, Formanova N, Gebauer-Jung S, Bishop JG, Schnabelrauch D, et al. Partial shotgun sequencing of the Boechera stricta genome reveals extensive microsynteny and promoter conservation with Arabidopsis. Plant Physiology. 2006;140(4):1169–1182. doi: 10.1104/pp.105.073981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(23):10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Precision Mapping of Quantitative Trait Loci. Genetics. 1994;136(4):1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB, Kao CH, Basten CJ. Estimating the genetic architecture of quantitative traits. Genetical Research. 1999;74(3):279–289. doi: 10.1017/s0016672399004255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.