Abstract
Hundreds of different factors adorn the eukaryotic genome, binding to it in large number. These DNA binding factors (DBFs) include nucleosomes, transcription factors (TFs), and other proteins and protein complexes, such as the origin recognition complex (ORC). DBFs compete with one another for binding along the genome, yet many current models of genome binding do not consider different types of DBFs together simultaneously. Additionally, binding is a stochastic process that results in a continuum of binding probabilities at any position along the genome, but many current models tend to consider positions as being either binding sites or not. Here, we present a model that allows a multitude of DBFs, each at different concentrations, to compete with one another for binding sites along the genome. The result is an “occupancy profile,” a probabilistic description of the DNA occupancy of each factor at each position. We implement our model efficiently as the software package COMPETE. We demonstrate genome-wide and at specific loci how modeling nucleosome binding alters TF binding, and vice versa, and illustrate how factor concentration influences binding occupancy. Binding cooperativity between nearby TFs arises implicitly via mutual competition with nucleosomes. Our method applies not only to TFs, but also recapitulates known occupancy profiles of a well-studied replication origin with and without ORC binding. Importantly, the sequence preferences our model takes as input are derived from in vitro experiments. This ensures that the calculated occupancy profiles are the result of the forces of competition represented explicitly in our model and the inherent sequence affinities of the constituent DBFs.
Hundreds of different factors adorn the eukaryotic genome, binding to it in large number. These DNA binding factors (DBFs) are quite diverse. For example, nucleosomes occupy 75%–90% of the genome (Van Holde 1989), and DNA and RNA polymerases traverse large swaths of it at a time. In contrast, transcription factors and other specialized proteins and protein complexes—like the origin recognition complex (ORC)—bind to very specific regions of the genome, often only at specific times or under specific conditions. Key cellular processes involving the genome—including replication, transcription, and chromatin packaging—are regulated by the spatio-temporal interactions of these various DBFs with DNA and with each other. Consequently, it is critical that we understand how these factors interact as they bind to the genome.
In particular, DBFs bind to the genome in competition with one another, jockeying for position along the DNA. As one example, to fit into the nucleus, DNA is highly compacted into chromatin in a hierarchy of levels. The lowest level is the formation of nucleosomes, where ∼150 base pairs of DNA are coiled around a histone octomer. Since transcription factors (TFs) bind primarily exclusively of nucleosomes, TF binding sites depend not only on TF sequence specificity, but also on competition with nucleosomes (and other DBFs, including other TFs). Yet, few current models of TF binding take competition with nucleosomes and other TFs into account, and few current models of nucleosome binding take competition with TFs into account. Current models that do consider nucleosomes and TFs together suffer from various drawbacks, such as being restricted to small genomic regions or coarse resolution (Teif 2007), or a lack of genome-wide improvement of positioning as a result of this incorporation (AV Morozov, K Fortney, DA Gaykalova, VM Studitsky, J Widom, ED Siggia, http://arxiv.org/abs/0805.4017). Although a few models of TF binding consider a small number of TFs at once at single-nucleotide resolution (Sinha 2006; Segal et al. 2008), these do not consider nucleosomes. We need approaches that can model the interactions along the entire genome of large numbers of DBFs of varying kinds, including both nucleosomes and arbitrary numbers of TFs.
A separate problem of current models is that most consider DBF binding to be a discrete, or binary, phenomenon. This leads to a view in which certain genomic positions are annotated as DBF binding sites, while the rest are assumed to be unbound. But the energetics of binding clearly indicates that DBF binding is a continuous phenomenon. This leads to a contrasting view in which genomic positions are not annotated as binding sites using the set {0, 1} but rather the interval [0, 1]. Under such a view, a DBF may bind anywhere in the genome, but more energetically favorable locations will be bound more frequently. Furthermore, the frequency of binding at any particular location will also depend on DBF concentration, a point ignored by nearly every model (some notable exceptions being Djordjevic et al. 2003; Bintu et al. 2005; Segal et al. 2008).
A continuum view of DBF binding is consistent with a wealth of recent experimental data. Large-scale transcript sequencing studies reveal that many genes do not have a single well-defined transcription start site (TSS), but rather a distribution over TSSs (Miura et al. 2006); that many genes are not spliced using well-defined acceptor/donor sites, but rather using a distribution over acceptor/donor sites (Chern et al. 2006); and that transcription is initiated not just at the TSSs of genes, but in many places in the genome (Suzuki et al. 2001). In addition, large-scale chromatin immunoprecipitation (ChIP) studies reveal that RNA polymerases are located not only upstream of genes, but in many other places in the genome (Steinmetz et al. 2006), and that TFs are bound not only to some promoters and not others, but rather in a continuum of detectable binding (Harbison et al. 2004). Computational studies have demonstrated that a continuum view of TF binding is useful in determining the probability of a promoter being bound somewhere by a single TF (Granek and Clarke 2005), in building models of TF sequence specificity (Berger et al. 2006; Foat et al. 2006), and in understanding the contributions of weak binding to transcriptional regulation (Tanay 2006; Segal et al. 2008).
In summary, we need models of continuous genome binding that take into account competition by many DBFs of various kinds and at varying concentrations and are capable of doing so at genome-wide scales and single-nucleotide resolutions. Here, we present a first such model. Our model takes as input the sequence specificities and concentrations of multiple kinds of DBFs, along with a genome sequence; it then computes as output the probability that each position in the genome will be occupied by each kind of DBF when these factors are in competition with one another for binding locations. This “occupancy profile” for each DBF across the genome is computed as the thermodynamic ensemble average over all valid binding configurations, each occurring with a different frequency in accordance with a Boltzmann distribution. We have implemented our model in a software package called COMPETE (Competitive Occupancy: Multi-factor Profile Evaluation of a Thermodynamic Ensemble). COMPETE is computationally efficient: Entire yeast chromosomes with hundreds of different kinds of DBFs can be processed in minutes. It is flexible and extensible, allowing incorporation of any DBF with sequence-specific binding preferences. We demonstrate this by considering simultaneously the binding of TFs, nucleosomes, and ORCs across the actual sequence of the yeast genome. Our model represents a significant early step toward the goal of understanding, at a mechanistic level, the interactions between factors regulating different genomic processes like replication, transcription, and chromatin packaging.
Results
An extended “musical chairs” contest view of genome binding
Our model of the binding of DBFs along the genome can be likened to an extended “musical chairs” contest. Imagine the eponymous children's game in which children (DBFs) circle a finite set of chairs (genomic locations) and occupy the chairs in some configuration at the end of each round (sample a valid binding configuration for the genome). Further imagine that the children are grouped into teams (multiple copies of the same DBF) of different sizes (concentration levels); as a consequence, a team with more children (higher concentration) will occupy more chairs than a team with fewer children (lower concentration), all other things being equal. Finally, imagine that no chair is removed after each round, allowing an extended contest to proceed indefinitely.
In this extended contest, we record how often each chair is occupied by each team over time, resulting in an “occupancy distribution” for each chair. By considering these occupancy distributions across all chairs, we obtain an aggregate summary for each team, which we call an “occupancy profile.” Our model is almost perfectly analogous, but is more complicated in that we assume each DBF occupies some number of consecutive nucleotides along the genome, and exhibits varying preferences for different locations in the genome according to its sequence specificity. Because of this last point, the various valid binding configurations will be sampled with different frequency, with more energetically favorable configurations being sampled more frequently.
Efficient software for exact computation of ensemble-averaged occupancy profiles
We can represent the probability over valid binding configurations in the thermodynamic ensemble using a graphical model called a Boltzmann chain. A Boltzmann chain is a generalization of a hidden Markov model (HMM) in which values associated with state transitions and sequence emissions need not be normalized probabilities, but can be arbitrary non-negative numbers. This generalization allows for easier incorporation of information related to binding energy and concentration, but retains the convenient property of HMMs that exact posterior inference of genome occupancy can be performed efficiently using dynamic programming. Consequently, running time scales linearly with the length of the input sequence.
We have implemented this exact posterior inference of genome occupancy in our COMPETE software package. COMPETE can process entire yeast chromosomes being bound by hundreds of DBFs in a matter of minutes. To illustrate its usefulness and generality, in what follows, we present a series of examples illustrating distinctive features of our model of genome binding, along with demonstrations of how our model of DBF competition improves both TF positioning and nucleosome positioning genome-wide. These are all computed using COMPETE applied to the genome sequence of the yeast Saccharomyces cerevisiae, with binding specificities of DBFs determined by recent in vitro methods (Kaplan et al. 2009; Zhu et al. 2009).
This last point is quite important: By using binding specificities of TFs (Zhu et al. 2009) and nucleosomes (Kaplan et al. 2009) bound to naked DNA at low concentrations and in isolation of other factors, our model is able to distinguish between the inherent binding specificities of DBFs (which COMPETE takes as input) and the consequent locations that DBFs take up along the genome when in competition with one another (which COMPETE produces as output). In particular, methods that use TF or nucleosome specificities based on in vivo binding locations (such as those of Harbison et al. 2004 or Segal et al. 2006) will be unable to resolve the relative contributions of inherent sequence specificity versus the consequences of competitive binding.
Competition with transcription factors affects nucleosome occupancy
Direct competition between DBFs is an important guiding force in determining genome occupancy. The result of this competition depends on several factors, including concentration and sequence preferences of the DBFs involved, along with steric hindrance between nearby DBFs, all of which are accounted for in our approach. As one example, although nucleosomes and TFs may in some cases simultaneously bind the same stretch of DNA, their binding is primarily mutually exclusive. A consequence of this mutual exclusion is that nucleosome positions along on the genome are a result not only of inherent nucleosomal sequence preferences, but also competition with TFs and other DBFs.
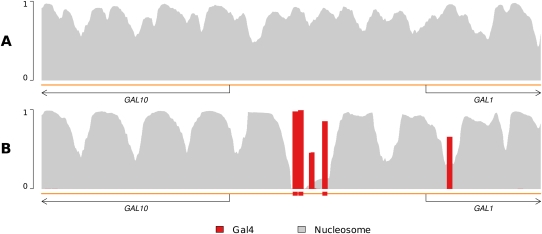
In Figure 1, we present an example showing how the binding of TFs can position nucleosomes more stably along the genome (by stable, we mean that a nucleosome appears in a well-defined location, as opposed to in one of a few different translated locations). Depicted is the region surrounding the GAL1-GAL10 promoter on chromosome II. In the top panel is the occupancy profile in which nucleosomes compete for positions along the genome without any other DBF competitors (as in the models of Segal et al. 2006; Kaplan et al. 2009). As is evident, nucleosome positions are largely ambiguous in this region, owing to a significant degree of uncertainty regarding how the nucleosomes are translated along the genome. In contrast, the bottom panel reveals how nucleosomes are positioned far more stably once the binding of Gal4 is taken into account. The strong binding of Gal4 to its cognate sites in the GAL1-GAL10 promoter (and even to a site within the coding region of GAL1) helps to establish nucleosome “boundaries.” These boundaries, in turn, reduce the probability of the various translational modes of the nucleosomes and thus increase their stability at specific locations.
Figure 1.
Transcription factor binding can stabilize nucleosomes. Shown are DBF occupancy profiles. The x-axis is the genome position, and the y-axis is the probability of occupancy. Plots with each legend color indicate the probability of the respective DBF occupying each position. The orange line underneath each plot represents the genome; oriented arrows demarcate coding regions of genes according to SGD (Cherry et al. 1998); colored boxes denote strong TF binding site matches according to MacIsaac et al. (2006). (A) Including only nucleosomes and unbound genomic sequence yields an occupancy profile similar to existing nucleosome-only positioning models like that of Segal et al. (2006). In this genomic region, the degenerate nature of nucleosome sequence preferences leads to significant uncertainty of nucleosome positioning. (B) Addition of Gal4 yields a strikingly different nucleosome occupancy profile, revealing stable nucleosome positioning. As Gal4 strongly binds its annotated binding sites, nucleosomes are outcompeted at those locations, and “boundaries” are established. These boundaries reduce the feasibility of nucleosome translation, so nucleosome positions coalesce into more stable locations in response.
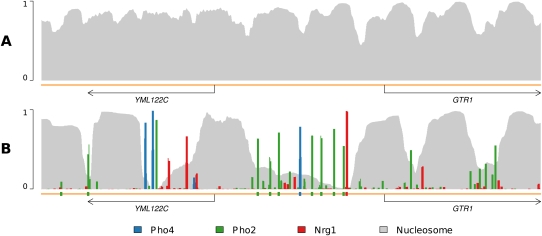
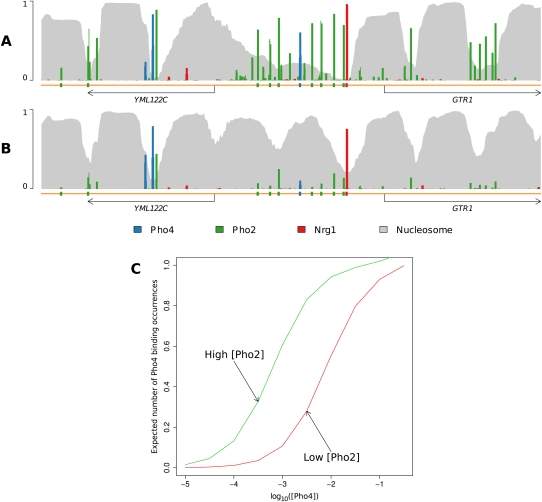
In Figure 2, we present another example showing how the binding of TFs can displace nucleosomes from the genome. Depicted is the region surrounding the GTR1 promoter on chromosome XIII. In the top panel again is the occupancy profile in which nucleosomes compete for positions along the genome without any other DBF competitors. Nucleosome positions at the left are somewhat ambiguous, but become more stably positioned to the right. Five to six reasonably stably positioned nucleosomes can be seen, starting near the start codon of YML122C. In contrast, the bottom panel reveals how nucleosomes are positioned once the binding of Pho4, Pho2, and Nrg1 is taken into account. The binding of Pho4, Pho2, and Nrg1 to sites in the GTR1 promoter displaces the two nucleosomes that would otherwise reside there. It also helps to stably position a nucleosome near the 3′-end of the coding region of YML122C and hems in a single nucleosome near the 5′-end. The strong binding of the TFs forms boundaries that prevent nucleosomes from translating upstream or downstream; instead, overall nucleosome occupancy in the region drops dramatically, illustrating the importance of modeling direct competition between nucleosomes and TFs.
Figure 2.
Transcription factor binding can displace nucleosomes. The promoter region of GTR1 is known to contain high-affinity binding sites of Pho4 and Nrg1, as well as several of Pho2. (A) An occupancy profile including only nucleosomes depicts five to six reasonably well-positioned nucleosomes beginning atop the start codon of YML122C. (B) Addition of Pho4, Pho2, and Nrg1 to the model results in the displacement of several of the previously bound nucleosomes, and the reduction of binding frequency of those remaining, to various extents. Additionally, a well-positioned nucleosome is established near the 3′-end of YML122C. The strong TF binding in this region precludes nucleosome repositioning upstream or downstream along the sequence; the strongly bound TFs displace some nucleosomes and hem in the remaining others, resulting in their well-defined positions.
Competition with nucleosomes affects transcription factor occupancy
Just as existing nucleosome positioning models do not generally take into account the binding of other DBFs, existing TF positioning models generally do not take other kinds of DBFs into account. In particular, existing TF positioning models do not generally consider competition with nucleosomes as ours does.
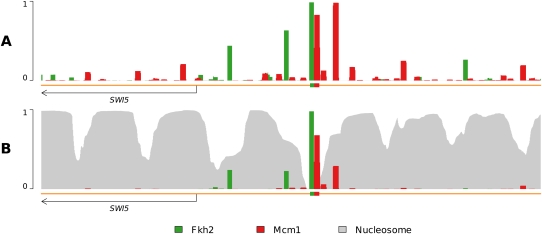
In Figure 3, we present an example demonstrating the role that nucleosomes play in reducing the nonspecific binding of TFs across the genome. Depicted is the region surrounding the SWI5 promoter on chromosome IV. In the top panel is the occupancy profile when only Fkh2 and Mcm1 are included in the model. Although the factors bind strongly to their annotated binding sites within the promoter, a great deal of weaker binding can also be observed. Of particular note is the fact that Mcm1 has a stronger match to a binding site just upstream (to the right) of its annotated site than it does to the annotated site itself. Because Fkh2 and Mcm1 form a ternary complex with the DNA, we know that Mcm1 should bind immediately adjacent to Fkh2 more frequently. How does the cell enforce this? Consider what happens when nucleosomes are included in the model, as shown in the bottom panel. Now, the vast majority of nonspecific binding of both Fkh2 and Mcm1 has been eliminated. In particular, because of direct competition between Mcm1 and nucleosomes, the upstream Mcm1 binding site is now occupied much less frequently than the site immediately adjacent to Fkh2, as expected.
Figure 3.
Nucleosome binding attenuates nonfunctional transcription factor binding. (A) A profile using a model of only TFs and DNA yields many (presumably nonfunctional) binding predictions throughout the genome. The binding of Fkh2 and Mcm1 is significant even away from their annotated binding sites; Mcm1 even displays an upstream (to the right) binding occurrence stronger than at its annotated site. However, Fkh2 and Mcm1 form a ternary complex with DNA and are known to bind adjacently to one another, making the observation of this additional, stronger, binding occurrence somewhat curious. (B) Addition of nucleosomes yields a dramatically altered TF occupancy profile. Fkh2 and Mcm1 binding is notably diminished at sites other than annotated binding sites, though not entirely eliminated. Additionally, the strong upstream Mcm1 binding event occurs at a much-reduced level, now approximately half as frequently as that of the annotated Mcm1 binding site.
Varying transcription factor concentration affects occupancy
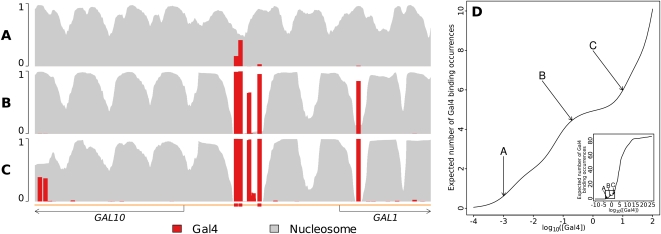
The binding frequency of a DBF to a particular segment of DNA will depend on its concentration. In Figure 4, we show an example in which Gal4 and nucleosomes compete for binding sites in the region surrounding the GAL1-GAL10 promoter, at varying concentrations of Gal4. Gal4 is known to bind strongly to several binding sites in this intergenic region (Giniger et al. 1985; Harbison et al. 2004), and our occupancy profiles recapitulate that behavior. At very low concentrations (Fig. 4A), Gal4 only binds at low levels, and essentially only at annotated binding sites, those being by definition the strongest in terms of sequence preference. An increased value of the concentration parameter for Gal4 results in an increased level of occupancy at the annotated binding sites (Fig. 4B), along with occupancy at two additional sites that can now be bound because of nucleosomal rearrangements introduced by strong binding to the three annotated sites. At even higher concentrations, additional sites start to become occupied (Fig. 4C). Throughout the process, as Gal4 binds more sites with increasing concentration, the nucleosome occupancy profile is dramatically reorganized. We can plot the total level of Gal4 occupancy in the promoter as a function of the log concentration parameter (Fig. 4D). The range of plausible concentrations is quite small, including B but not likely extending beyond the highlighted region of the overall occupancy versus concentration curve (Fig. 4D, inset). Interestingly, several well-positioned nucleosomes continue to occupy their positions with little change even as the increased concentration of Gal4 (C and beyond) yields strong binding away from its annotated sites, reiterating the importance of including nucleosomes in the model.
Figure 4.
Binding occupancy is highly dependent on factor concentration. (A) At low concentration, Gal4 binding is virtually nonexistent apart from low levels of binding at its annotated binding sites. (B) As concentration increases, Gal4 binds strongly to its annotated sites, displacing nucleosomes there and leading to the stabilization of flanking nucleosome positions, as also illustrated in Figure 1. This stabilization and reorganization allows Gal4 to bind at two additional sites. (C) Increasing Gal4 concentration further eventually leads to strong binding even away from its annotated sites. However, even at this level, Gal4 cannot outcompete many of the strongly positioned nucleosomes, reiterating the importance of including many different DBFs concurrently in the model. (D) Gal4 binding occupancy as a function of Gal4 concentration. Note how occupancy changes more slowly around the seemingly plausible concentration used in B than in any other part of the overall occupancy versus concentration curve (inset), suggesting that while occupancy can be fairly sensitive to concentration on the whole, the cell may sometimes be operating in ranges that are relatively less sensitive.
Binding cooperativity emerges implicitly by considering competition explicitly
In addition to competing with one another, TFs are also known to bind cooperatively in some instances. A few models have shown the importance of TF cooperativity in establishing sharp expression patterns and have included explicit energetic “bonuses” to model this cooperativity (Segal et al. 2008). These explicit bonus terms may be added in our model, but binding cooperativity can also arise in our model implicitly, even without the inclusion of such terms. This can occur when multiple TFs are competing against the same nucleosome for nearby binding sites. In such a situation, the more frequently one of the factors is able to bind to a site on the genome, the less often the competing nucleosome will be present to occlude the binding of the other factor. This behavior has been termed “collaborative competition” and observed experimentally (Miller and Widom 2003) and characterized mathematically (Mirny 2009).
In Figure 5, we revisit the region surrounding the GTR1 promoter, this time focusing on the binding of Pho4 at different concentrations of Pho2. The top occupancy profile is computed at a low Pho2 concentration; the bottom profile is computed at a higher Pho2 concentration. Clearly, the higher Pho2 concentration will increase the overall occupancy of Pho2, but what is especially interesting is that it also increases the overall occupancy of Pho4, even though the concentration of Pho4 is unchanged. This is because as Pho2 increasingly binds sites in the promoter, it displaces nucleosomes from the promoter with greater frequency, thereby providing greater access for Pho4 to bind, even at the same level of Pho4. Indeed, we can plot Pho4 occupancy as a function of Pho4 concentration, at each of the two Pho2 concentrations. The occupancy is consistently higher across the entire range of Pho4 concentrations when Pho2 is present at the higher concentration. This upward shift of the Pho4 curve with increasing concentration of Pho2 is a form of binding cooperativity that has arisen entirely implicitly.
Figure 5.
Cooperative binding emerges implicitly from explicit competition between DBFs. Revisiting the GTR1 promoter, cooperativity among Pho4 and Pho2 can be observed via mutual competition with nucleosomes, particularly the nucleosome positioned atop the annotated Pho4 site and Pho2 sites adjacent to the strong Nrg1 binding site. (A) A profile of this region with a high concentration of Pho4 results in high occupancy of both Pho4 and Pho2. (B) A similar profile but with decreased concentration of Pho4 results in decreased occupancy of both Pho4 and Pho2, and a resulting increase in the occupancy of nucleosomes. (C) A plot of Pho4 binding occupancy as a function of Pho4 concentration, at both high and low concentrations of Pho2. The plot reveals that a higher concentration of Pho2 leads to higher occupancy of Pho4 at all concentrations of Pho4. Importantly, this cooperative effect is purely a consequence of inclusion of nucleosomes into the model. That is, Pho4 and Pho2 have an implicitly cooperative effect due to joint competition with the same nucleosome, as opposed to some explicit energetic cooperativity term.
Competitive occupancy of ARS sequences near origins of replication
Our examples thus far have focused on TFs and nucleosomes binding the genome, but DNA is occupied by a host of other proteins and protein complexes as well. Among these is the ORC, which is instrumental in the initiation of DNA replication. Like TFs, ORC binds exclusively of other DBFs and has a DNA sequence preference that guides its binding in yeast. However, ORC's sequence preferences are rather degenerate; scanning the ORC motif across the entire genome without consideration of other competing DBFs yields tens of thousands of matches (Breier et al. 2004), although only a few hundred sites seem to be bound by ORC or function as origins of replication in vivo (Raghuraman et al. 2001; Wyrick et al. 2001). This suggests a role for competition with other DBFs, including TFs and nucleosomes, in guiding ORC to functional replication origins. In particular, chromatin is known to play a role in prereplicative complex formation at origins of replication (Lipford and Bell 2001). Therefore, the ORC and nucleosomes must be considered together, along with other DBFs, to provide a full view of binding behavior at origins of replication.
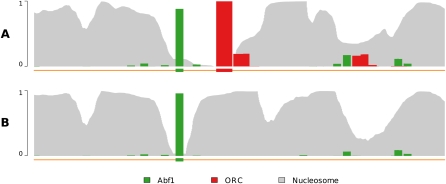
Figure 6 provides occupancy profiles for the region of the genome surrounding the well-studied ARS1 origin of replication. ARS1 has experimentally verified binding sites for ORC and Abf1 (Marahrens and Stillman 1992). Chromatin arrangement in ARS1 is fairly well characterized and changes depending on the binding of ORC (Lipford and Bell 2001). Our model is able to recapitulate this experimentally observed occupancy of ARS1 in both the presence and absence of ORC (Fig. 6A,B). In the presence of ORC, the strong experimentally observed nucleosome occupancy adjacent to the Abf1 and ORC binding sites is recapitulated, as is the nucleosome-free region between them. In the absence of ORC, however, a nucleosome has shifted to cover the ORC binding site, as was also observed experimentally.
Figure 6.
ORC and nucleosome binding at ARS1. The origin recognition complex (ORC) and chromatin play strong roles in the initiation of DNA replication. ARS1, an origin of replication in yeast, has a well-characterized DNA occupancy structure including binding by ORC, nucleosomes, and the transcription factor Abf1. This structure is observed to change in the absence of ORC binding, as can be seen in Figure 2A of Lipford and Bell (2001). The Abf1 binding site annotation is taken from Marahrens and Stillman (1992). The ORC binding site and sequence preferences are derived from Xu et al. (2006). (A) The wild-type occupancy profile recapitulates the strong binding of nucleosomes adjacent to the Abf1 and ORC binding sites, as well as the nucleosome-free region between them. (B) Without ORC present, the occupancy profile depicts a shifted nucleosome over the top of the ORC binding site, as is the case in the experimental observation of Lipford and Bell (2001).
DBF competition improves transcription factor binding predictions genome-wide
To this point, we have shown examples of various phenomena that are captured by our modeling framework, and we have done so by focusing on specific loci. However, we expect that predictions of genome-wide DBF positioning should also be improved by the inclusion of competition between nucleosomes and large numbers of TFs in our framework. To demonstrate this, we first examine the effect of DBF competition on the prediction of TF binding genome-wide; in the next section, we consider the effect on the prediction of nucleosome binding genome-wide.
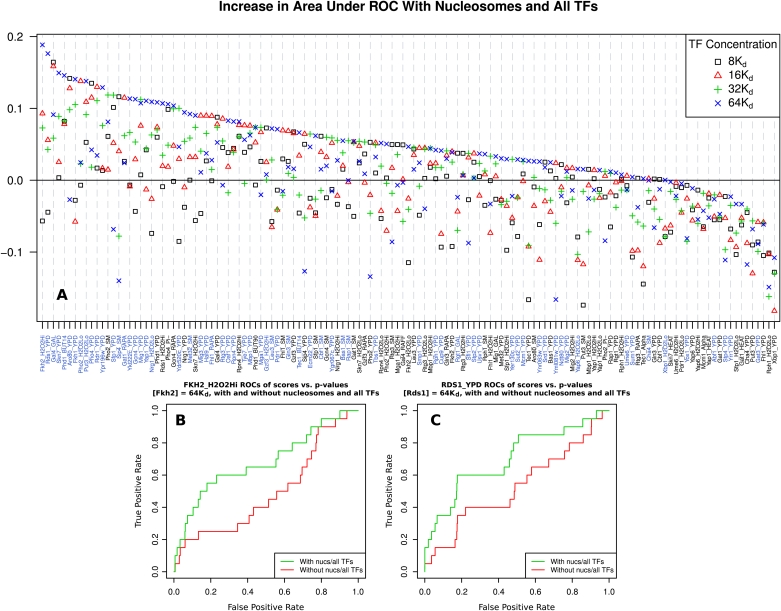
We analyzed the agreement between the binding of TFs to yeast promoters genome-wide as determined experimentally in vivo by ChIP with microarray hybridization (ChIP-chip) data (Harbison et al. 2004), and as predicted computationally by COMPETE under two settings: Each TF is modeled to bind by itself versus doing so in competition with nucleosomes and 88 other TFs. The improvement in agreement with experimental data under these two settings across 135 ChIP-chip experiments is shown in Figure 7A, with examples of how this improvement is computed for two particular factors shown in Figure 7, B and C.
Figure 7.
DBF competition improves transcription factor positioning predictions genome-wide. COMPETE is used to predict mean probabilities of TF binding in regions corresponding to probes from whole-genome ChIP-chip TF binding data (Harbison et al. 2004). The top and bottom 10% of regions as scored by COMPETE are respectively labeled as positively and negatively-bound promoters. ROCs are then constructed for probe P-values using these labels. The change in the area under the curve (AUC) of the ROCs is calculated between a model including only one TF and a model including that TF and nucleosomes and all other TFs, at various TF concentrations. The concentrations of all TFs are set to the same multiple of their respective Kd; the value of Kd for each TF is different and calculated from its PSSM, as in Granek and Clarke (2005). (A) Change in ROC AUC between TF-only and all TF and nucleosome models, across concentrations. Each point represents a pair of whole-genome analyses, with and without nucleosomes and all other TFs, totaling 272 decodings of the entire genome and 1080 individual analyses. Positive values signify improvement. Experimental conditions for each ChIP-chip experiment are given on the x-axis, with the best performing condition for each TF highlighted in blue. Most TF binding predictions either improve or behave somewhat similarly when including nucleosomes. As it is likely that different concentration choices are appropriate for each TF, several concentrations are included. Values of all points are given in Supplemental Table S1. (B) ROCs of Fkh2 ChIP-chip P-values with labels determined by binding probability as predicted by COMPETE in the FKH2_H2O2Hi experiment, with a model including Fkh2, nucleosomes, and all other TFs (green), and a model including Fkh2 alone (red). (C) Same as B, but using the RDS1_YPD experiment with models including Rds1 in place of Fkh2.
A clear trend can be observed in this plot, in that the majority of TF binding predictions either improve or behave similarly when competition with nucleosomes and other TFs is introduced. Because the ChIP-chip data are known to be somewhat noisy and might not always be a faithful representation of in vivo binding, small changes are perhaps not that conclusive. Calling inconclusive those experiments where the absolute change for the best of the four global concentration scalings is <0.025, ∼69% of the 135 experiments showed a positive change, while only ∼6% showed a negative change. Changes are unlikely to always be positive because of numerous complications involved in modeling actual in vivo DBF binding, including active chromatin remodeling, explicit binding cooperativity, and importantly, the challenge of correctly specifying 90 different DBF concentrations. Noticeably, the change is maximized at different concentrations for each TF, highlighting the difficulty of solving the issue of robustly determining appropriate DBF concentrations.
DBF competition improves nucleosome binding predictions genome-wide
As we expect that nucleosomes and TFs will mutually improve one another's positioning predictions, we also analyzed the agreement between the binding of nucleosomes across the yeast genome as recently determined experimentally in vivo (Kaplan et al. 2009), and as predicted computationally by COMPETE under two settings: Nucleosomes are modeled to bind by themselves versus doing so in competition with 89 TFs. To analyze our predictions, we consider the improvement under these two settings of the Spearman correlation between a ranked list of genomic regions enriched and depleted for nucleosome occupancy with a list of those regions ranked by predicted occupancy according to COMPETE.
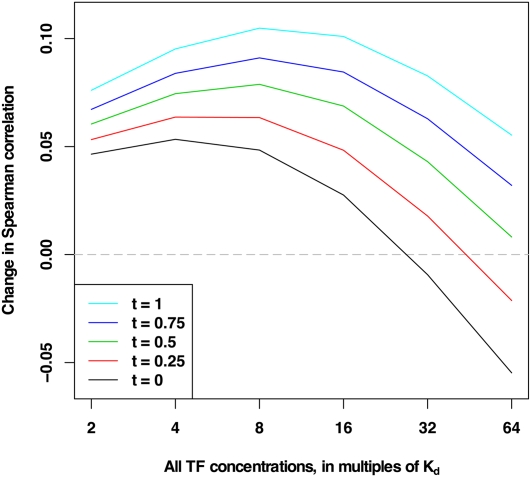
As shown in Figure 8, inclusion of competition with TFs is clearly beneficial in predicting nucleosome occupancy across a range of TF concentrations. This effect is demonstrated for various thresholds used for extraction of enriched and depleted regions and increases with the stringency of the threshold, as expected. In all cases, as TF concentrations are increased, the improvement reaches a plateau and then diminishes; for low-stringency thresholds, adding all 89 TFs at extremely high concentrations is even deleterious. This is not surprising because at these high concentrations, TFs are able to outcompete nucleosomes even at many enriched locations. Nevertheless, the vast majority of combinations of concentrations and stringency thresholds show improved prediction of nucleosome occupancy genome-wide when competition with TFs is incorporated.
Figure 8.
Change in Spearman correlation with and without TFs. DBF competition improves nucleosome positioning predictions genome-wide. Enriched and depleted regions across the genome are extracted from the in vivo experimental map of Kaplan et al. (2009) using different stringency thresholds t, with all positions in enriched regions having experimental nucleosome occupancy greater than t and all positions in depleted regions having experimental nucleosome occupancy less than −t. The Spearman correlation is computed between the experimentally measured nucleosome occupancy of the regions and the occupancy predicted by COMPETE using nucleosome models with and without all 89 TFs of Zhu et al. (2009). The TF concentrations are all set to the same multiples of their respective Kds. Each point corresponds to a pair of whole-genome analyses, with and without all 89 TFs, totaling 12 decodings of the genome and 60 individual analyses. The changes between Spearman correlations of experimental data and COMPETE predictions with and without nucleosomes are shown here, where positive values signify improvement. Inclusion of TFs is clearly beneficial to nucleosome positioning across TF concentrations. This effect ranges in extent and is demonstrated for various TF concentrations and various thresholds t. Raising TF concentrations to high levels imposes deleterious effects on nucleosome positioning, likely due to diminished nucleosome occupancy in competition with many different highly concentrated TFs. When TF concentrations are kept below these high levels, TF inclusion aids significantly in nucleosome binding predictions. Values of all points are given in Supplemental Table S2.
Discussion
The extended “musical chairs” contest view of DNA occupancy represents a fundamental shift in how DNA binding is typically modeled, while conforming to how it is currently understood. The simplifying assumptions often made in the past, such as discrete binding, have not harmonized well with our understanding of the thermodynamic nature of molecular interactions. By viewing binding as dynamic, probabilistic, and competitive, we move closer to our current understanding of the process, as revealed by numerous recent studies.
In particular, comprehension of binding behavior at an aggregate scale will facilitate mapping of binding to expression. Expression is a complex continuous signal resulting from many kinds of input and is difficult to explain on a large scale by a small number of strong binding targets alone (Tanay 2006). Capturing pervasive weak binding instances may aid in our understanding of this control, allowing discovery of contributions of transcriptional regulators for genes whose involvement has been hitherto unknown.
Some improvements may be made to our approach that might boost its accuracy and effectiveness without fundamentally altering its structure or applications. The largest immediate gain may be made by developing a more robust technique for determining appropriate values of DBF concentrations. Our concentration parameters are, in fact, a product of a true concentration and an unknown energetic constant unique to each DBF. Binding motifs (including those we use) are usually represented by position-specific scoring matrices (PSSMs), and each PSSM is normalized to a probability by a normalization constant. This normalization constant scales energies into probabilities. Experimentally recovering the normalization constants so as to convert the probabilities of PSSMs into energies is prohibitive.
Hence, we are developing a procedure to learn these constants automatically, although doing so is nontrivial in the absence of sufficient informative data. In the examples we presented, concentration parameters were chosen to recapitulate expected binding behaviors; namely, strong binding at literature annotated binding sites without overwhelmingly strong binding elsewhere. A suitable automated technique would attempt to likewise fit concentrations in such a way as to recapitulate existing nucleosome occupancy maps as well as annotated TF binding locations. As discussed elsewhere (Tanay 2006), ChIP-chip data contain information throughout the range of P-values and may be helpful to guide TF concentration parameter selection. Additionally, several experimentally determined or computationally predicted nucleosome positioning data sets exist at both genome-wide and locus-specific resolutions (Segal et al. 2006; Lee et al. 2007; Whitehouse et al. 2007; Shivaswamy et al. 2008; Kaplan et al. 2009). Training on nucleosome data is difficult given the noisy nature of existing nucleosome positioning data in conjunction with the mobility of the nucleosomes themselves. However, taking nucleosome and ChIP-chip data into account together may help to mitigate much of this uncertainty and yield the robust estimates of concentration that we desire.
Our modeling framework would allow wholly different methods of representing DBFs to be used. As the various DBFs are themselves modules of states, their specific implementation need not be Boltzmann chains (see Methods). Their replacement may be quite extreme, and our model could easily be converted into a generalized Boltzmann chain. For example, our current nucleosome model could be replaced with any of various existing models of nucleosome occupancy (Peckham et al. 2007; Field et al. 2008; Yuan and Liu 2008). When considering this type of modification, it should be noted again that models of in vitro sequence specificities are more appropriate than those obtained from in vivo data. Our approach focuses on understanding the resultant genomic binding occupancy given a set of DBFs and their inherent sequence specificities. In vitro models are likely to be more reflective of inherent specificity (without the influence of competition and other positioning pressures) than in vivo models. Indeed, our approach may be understood to be attempting to understand (and output) in vivo behavior in terms of in vitro inputs. For this reason, the sequence preferences used in our analyses are all in vitro where available.
Because our approach yields a full posterior distribution, we can sample from this distribution to answer increasingly complex questions. For example, it is possible to determine the frequency with which a given number of DBFs occupy sequence in a particular orientation, either with respect to one another or specified sequence elements, such as transcription start sites or TATA-boxes.
More generally, our model of competitive multi-factor binding will be able to guide biological experiments by providing testable hypotheses. Mutations may be introduced to both sequence and DBFs in our model to predict novel and experimentally verifiable binding behavior. Experiments such as these may help elucidate the true roles of TFs whose functionality is not entirely known, or disambiguate the mechanism by which these roles arise. As one such example, we can test whether a set of TFs that are known to have a mutually cooperative relationship achieve this cooperativity through explicit interactions with one another or implicit cooperativity achieved via nucleosome displacement.
Methods
Boltzmann chains and model structure
Our approach is implemented via a Boltzmann chain (Saul and Jordan 1995), a statistical framework that allows calculation of posterior probabilities of all valid binding configurations under a Boltzmann distribution. A Boltzmann chain is a generalization of a hidden Markov model (HMM) that relaxes constraints on transition and emission weights, allowing them to be any non-negative real number. This characteristic lends itself well to our system since binding energies and factor concentrations need not be constrained to probabilities. Additionally, a system temperature can be explicitly modeled with this framework, although in all the results presented here, we have not used such a temperature.
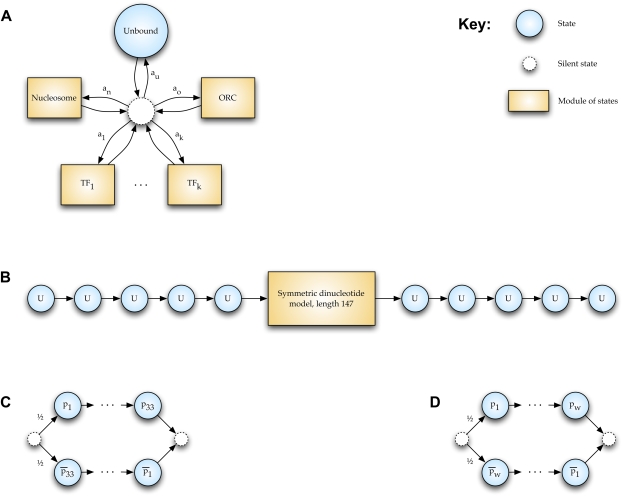
As illustrated in Figure 9, our model is conceptually simple and quite flexible and extensible. It consists of a central silent state, from which transitions are possible to states that emit DNA not bound by any factor or bound by one of any number of modeled transcription factors, ORC, or a nucleosome, each of which, in turn, returns to the central silent state. Unbound DNA is represented by a single state with emission probabilities reflecting genomic DNA content. TFs, ORC, and nucleosomes are “modules” of several states. TFs are represented as one silent state transitioning into two linear sets of states that proceed through the factor's motif in forward or reverse-complement orientations. Our TF motifs are taken from position-specific scoring matrices (PSSMs) learned from protein binding microarray data and trimmed according to an information content threshold of 0.3 (Zhu et al. 2009), except for the analyses of the GTR1 promoter, in which the PSSMs are taken from MacIsaac et al. (2006). The ORC module consists of a PSSM-based EACS+B1 motif (Xu et al. 2006) in either forward or reverse-complement orientation. Our nucleosome model is a 147-position dinucleotide HMM flanked by five positions of unbound sequence on either side to enforce a minimum length of linker regions, as described in Segal et al. (2006). However, we have constructed the parameters of the dinucleotide model from recent in vitro nucleosome binding data (Kaplan et al. 2009), using more than 4.7 million sequences instead of the 199 used originally in Segal et al. (2006). Following Kaplan et al. (2009), we replace the outermost 10 positions of either side of the 147-position model with background distribution to avoid potential micrococcal nuclease sequence-specificity biases. Thus, the final model has a 127-position dinucleotide core, flanked by 15 positions of background sequence on either side.
Figure 9.
State transition diagram for Boltzmann chain implementation in COMPETE. Blue circular nodes are states and correspond to single nucleotides. Dashed circular nodes are silent states because they do not correspond to any nucleotide. Orange rectangular nodes are modules of states and correspond to a sequence of nucleotides of some length. Edges represent probabilistic transitions between states, with transition probabilities labeled; to reduce clutter, if a node has only one outbound edge, it is unlabeled (the probability is exactly 1). (A) At the highest level, the model represents each position in the genome either as being in an unbound state or as being bound by one of any number of DNA binding proteins or protein complexes. For instance, we might choose to model the genome as being bound by nucleosomes, the origin recognition complex (ORC), and k different transcription factors (TFs). Each DBF is represented by a module of an appropriate length. Transition probabilities from the central silent state are proportional to concentrations of the respective DBFs. (B) The nucleosome module consists of a symmetric dinucleotide model of length 147 flanked by 5 unbound nucleotides on either end to enforce a spacing between nucleosomes of at least length 10. To reduce clutter, we do not represent the states within the central module in this figure. (C) The ORC module consists of a PSSM-based EACS+B1 motif of length 33 (Xu et al. 2006) in either a forward or reverse-complement orientation (the origin might be on either strand, and we assume that each occurs with probability 1/2). (D) A TF module consists of a PSSM-based motif of length w, a value that varies for each TF. Because a TF can bind to either strand, the w nucleotides arise either from the PSSM or from its reverse complement, each with probability 1/2.
Given the modular nature of our model, any combination of DBFs may be represented, so our approach generalizes a wide range of previous techniques. Inclusion of only one TF at a specific concentration can reproduce results from GOMER (Granek and Clarke 2005). Using only a few TFs results in a model similar to that of Sinha (2006). A model with only nucleosomes recapitulates the model described in Segal et al. (2006).
COMPETE: Implementing our model in efficient software
As with HMMs, posterior decodings of Boltzmann chains can be calculated by the forward–backward algorithm (Rabiner 1989). COMPETE (Competitive Occupancy: Multi-factor Profile Evaluation of a Thermodynamic Ensemble) implements this algorithm to compute its occupancy profiles in a highly efficient manner. It uses probability scaling with some slight mathematical extensions to allow states other than the begin or end state to be silent, and it breaks the traditional dependence between calculation of the forward and backward tables that arises during usage of probability scaling. Additionally, a prescan of transition weights allows the calculation of only the non-zero elements of each table, those corresponding to feasible state transitions. Our model connectivity is especially sparse, so this particular optimization provides an enormous performance boost. The software implementation, written in C for speed, includes other practical optimizations, such as multi-threading to allow simultaneous computation of the forward and backward tables on multi-core systems. In terms of performance, occupancy profiles of entire yeast chromosomes can be computed in tens of minutes even when simultaneously using nucleosomes and all TFs for which we have PSSMs (around 125). The memory required to store the forward and backward tables is the only practically limiting resource, and this can easily be alleviated by periodically writing the tables to disk as they are being computed.
Genome-wide analysis of transcription factor binding
Several steps are involved in carrying out our analysis of the genome-wide effect of DBF competition on TF binding. First, the mean probability of starting a binding site for a given TF is calculated for every region corresponding to the ChIP-chip probes of Harbison et al. (2004) across the genome. Under the premise that high probability regions should correspond to bound promoters and low probability regions should correspond to unbound promoters, the top and bottom 10% of regions by mean predicted binding probability are labeled as bound and unbound, respectively. These top and bottom 10% sets are selected by setting thresholds scaled against the maximum value in the genome, and contain either the points above (or below, for the bottom set) that threshold or the 20 highest (or lowest) points, whichever is larger. The receiver operating characteristic (ROC) curve is then constructed for the probe P-values using the aforementioned labels. The area under the curve (AUC) of the ROCs is used as a measure of binding prediction. The change in these AUCs is calculated (by subtraction) between a model including only one TF, and a model including that TF in competition with nucleosomes and 88 other TFs, at various TF concentrations. Intersecting the set of TFs from the Harbison et al. (2004) ChIP-chip data set with those for which PBM binding motifs exist in Zhu et al. (2009) yields 67 TFs and 135 binding experiments for which this change can be computed. Although only these 67 TFs are analyzed for this metric, all 89 TFs from the PBM data of Zhu et al. (2009) are included in the competition model.
The choice of TF concentrations is a difficult challenge, in that allowing 89 TFs to compete for binding sites along the genome at once implies the need for selection of 89 different concentration parameters. Selecting these parameters independently can result in excessive parameter tuning or over-fitting. However, cross-validation strategies for learning these parameters from data present their own complications because they would require searching in an 89-dimensional real concentration parameter space, even setting aside the many additional parameters relating to binding specificity that could be optimized. To address these challenges and simplify the TF concentration parameter space to a one-dimensional subspace, we first calculate the Kd of each TF's PSSM, as detailed in Granek and Clarke (2005). Each TF's concentration is then set to its Kd multiplied by a single global scaling factor, using the same factor for all TFs. To simplify the space of concentrations further, only six discrete values ({2, 4, 8, 16, 32, 64}) are used instead of the full range of real values for this global scaling factor. By setting concentrations for all TFs to be the same multiple of their respective Kds, the probability of each TF binding its consensus site is equal under a simple scan of the motifs across the genome without any competition.
Genome-wide analysis of nucleosome binding
The experimental map of in vivo nucleosome positions is the YPD map taken from Kaplan et al. (2009). Following Kaplan et al., we identify maximal regions along the genome of at least 50 consecutive nucleotides in which the nucleosome occupancy of each position in the region is either greater than some threshold t (for enriched regions) or less than −t (for depleted regions). These regions are then sorted by their mean nucleosome occupancy as experimentally determined, as well as by their mean nucleosome binding probability as computed by COMPETE, to produce a pair of ranked lists. The Spearman correlation between the experimental ranking and the ranking produced by COMPETE is then computed under two settings: one in which COMPETE models nucleosomes to bind the genome in isolation, and one in which it models them to bind in competition with 89 TFs whose in vitro specificities are known (Zhu et al. 2009). The TF concentrations in the competition setting are globally scaled by multiples of their respective Kd, as discussed above. The improvement in Spearman correlation under the two settings for various global scalings and various choices of threshold parameter t is computed in each case by subtraction.
Acknowledgments
The research presented here was supported by a CAREER award from the National Science Foundation (NSF 0347801), an Alfred P. Sloan Research Fellowship, and grants from NIH (P50-GM081883-01 and R01-ES015165-01) and DARPA (HR0011-08-1-0023 and HR0011-09-1-0040) to A.J.H.
Footnotes
[Supplemental material is available online at http://www.genome.org. The COMPETE software is available at http://www.cs.duke.edu/∼amink/software/compete/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093450.109.
References
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R. Transcriptional regulation by the numbers: Applications. Curr Opin Genet Dev. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Chatterji S, Cozzarelli NR. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 2004;5:R22. doi: 10.1186/gb-2004-5-4-r22. http://genomebiology.com/2004/5/4/R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern TM, van Nimwegen E, Kai C, Kawai J, Carninci P, Hayashizaki Y, Zavolan M. A simple physical model predicts small exon length variations. PLoS Genet. 2006;2:e45. doi: 10.1371/journal.pgen.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M, Sengupta AM, Shraiman BI. A biophysical approach to transcription factor binding site discovery. Genome Res. 2003;13:2381–2390. doi: 10.1101/gr.1271603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore I, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat B, Morozov A, Bussemaker H. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics. 2006;22:e141–e149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- Giniger E, Varnum S, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Granek J, Clarke N. Explicit equilibrium modeling of transcription-factor binding and gene regulation. Genome Biol. 2005;6:R87. doi: 10.1186/gb-2005-6-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C, Gordon D, Lee T, Rinaldi N, Macisaac K, Danford T, Hannett N, Tagne J, Reynolds D, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse R, Davis R, Hughes T, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- MacIsaac K, Wang T, Gordon D, Gifford D, Stormo G, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Miller J, Widom J. Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol. 2003;23:1623–1632. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Nat Preced. 2009. http://hdl.handle.net/10101/npre.2009.2796.1. [DOI] [PMC free article] [PubMed]
- Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham H, Thurman R, Fu Y, Stamatoyannopoulos J, Noble W, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989;77:257–286. [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Saul LK, Jordan MI. Boltzmann chains and hidden Markov models. In: Tesauro G, et al., editors. Advances in neural information processing systems. Vol. 7. The MIT Press; Cambridge, MA: 1995. pp. 435–442. [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore I, Wang J, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer V. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. On counting position weight matrix matches in a sequence, with application to discriminative motif finding. Bioinformatics. 2006;22:e454–e463. doi: 10.1093/bioinformatics/btl227. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Taira H, Tsunoda T, Mizushima-Sugano J, Sese J, Hata H, Ota T, Isogai T, Tanaka T, Morishita S, et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001;2:388–393. doi: 10.1093/embo-reports/kve085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif VB. General transfer matrix formalism to calculate DNA-protein-drug binding in gene regulation: Application to OR operator of phage lambda. Nucleic Acids Res. 2007;35:e80. doi: 10.1093/nar/gkm268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde K. Chromatin. Springer-Verlag; New York: 1989. [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavar S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Liu J. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]