Abstract
Although most human retrotransposons are inactive, both inactive and active retrotransposons drive genome evolution and may influence transcription through various mechanisms. In humans, three retrotransposon families are still active, but one of these, SVA, remains mysterious. Here we report the identification of a new subfamily of SVA, which apparently formed after an alternative splicing event where the first exon of the MAST2 gene spliced into an intronic SVA and subsequently retrotransposed. Additional examples of SVA retrotransposing upstream exons due to splicing into SVA were also identified in other primate genomes. After molecular and computational experiments, we found a number of functional 3′ splice sites within many different transcribed SVAs across the human and chimpanzee genomes. Using a minigene splicing construct containing an SVA, we observed splicing in cell culture, along with SVA exonization events that introduced premature termination codons (PTCs). These data imply that an SVA residing within an intron in the same orientation as the gene may alter normal gene transcription either by gene-trapping or by introducing PTCs through exonization, possibly creating differences within and across species.
Most eukaryotic genomes harbor retrotransposable elements (Malik et al. 1999). About 35% of the human genome is derived from retrotransposed sequences such as LINE-1, Alu, SVA, endogenous retroviruses, and processed pseudogenes (Lander et al. 2001). Although most human retroelement copies are no longer mobile, both active and inactive human elements have been shown to drive genome evolution and influence gene expression (Moran et al. 1999; Han et al. 2004; for reviews, see Belancio et al. 2008; Goodier and Kazazian 2008).
SVA RNAs are hominid-specific noncoding RNAs, which vary in size from 700–4000 bp, and are likely mobilized by the human LINE-1 in trans (Ono et al. 1987; Shen et al. 1994; Ostertag et al. 2003; Wang et al. 2005), similar to the human Alu (Dewannieux et al. 2003). There are roughly 2700 SVA copies in the human genome (Wang et al. 2005). A canonical full-length SVA (Fig. 1A) contains a number of sequence features proceeding from its 5′ end: (1) a CCCTCT hexameric repeat, ranging in repeat number from a few to as many as 71; (2) a sequence that shares homology with two antisense Alu fragments; (3) a variable number of tandem repeat sequence (VNTR); and (4) a sequence derived from the ENV gene and right LTR of an extinct HERV-K, hereafter referred to as SINE-R. SVAs typically terminate at their own polyA signal, with genomic insertions usually containing a number of adenines at the 3′ end. A target site duplication common to other LINE-1-driven retroelements (6–20 bp) flanks the inserted element (Ostertag et al. 2003).
Figure 1.
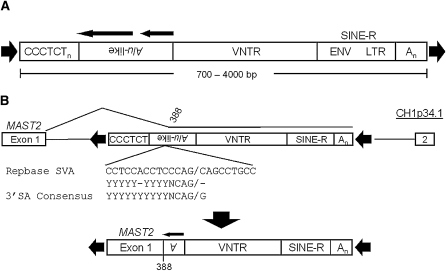
SVA and alternative splicing at the MAST2 locus. (A) The canonical SVA is displayed, consisting of some number of CCCTCT hexameric repeats followed by the Alu-like region, a variable number of tandem repeats, and then the SINE-R region, followed by a polyA signal with the entire SVA flanked by target-site duplications (black arrowheads). Two black arrows over the Alu-like region indicate sequence homology in the SVA to two ancestral Alu elements. (B) Shown is an ancestral CH1 MAST2 locus, sometime after the human–chimp split, containing a full-length SVA within intron 1 (top). An alternative splicing event into the Alu-like region (black line) likely occurred resulting in the original SVAF1 founder insertion (bottom) flanked by target-site duplications (black arrowheads) in the human genome. Note the loss of the entire CCCTCT hexamer and most of the Alu-like region after the alternative splicing event. The SVA splice site relative to SVARep is aligned to the known splice site sequence (middle).
Little is known about the biology of SVA apart from its structure. SVAs are currently active in the human genome, as indicated by the identification of de novo SVA insertions associated with disease (Hassoun et al. 1994; Rohrer et al. 1999; Makino et al. 2007). SVA disease insertions are associated with exon-skipping (Hassoun et al. 1994; Rohrer et al. 1999), deletion of genomic DNA (Takasu et al. 2007), and reduced or absent mRNA expression (Kobayashi et al. 1998; Wilund et al. 2002; Makino et al. 2007). In a manner similar to L1, SVAs have been shown to transduce 3′ flanking sequences to new genomic locations (Moran et al. 1999; Ostertag et al. 2003; Xing et al. 2006). SVAs are thought to be highly active due to the ratio of disease insertions to genomic copies. Furthermore, the high levels of insertion polymorphism of the human-specific subfamilies, E and F (Bennett et al. 2004; Wang et al. 2005), support the notion that SVAs are evolutionarily young and relatively active in the human population.
The mechanism of SVA transcription and the location of its promoter are unknown and are critical to our understanding of SVA retrotransposition. To date, experiments to characterize the SVA promoter have led to ambiguous results. Recently, we reexamined an SVA insertion on CH6 associated with a genomic deletion including the entire HLA-A gene that resulted in leukemia in three Japanese individuals (Takasu et al. 2007). The HLA-A insertion led to several interesting findings, including the identification of a new SVA subfamily formed by alternative splicing from the first exon of the MAST2 gene on CH1 into an SVA, and the identification of a SVA master element on CH10. We have also identified numerous functional 3′ splice sites (SSs) within SVA while analyzing human and chimp SVA 5′ transcriptional start sites (TSSs), and three more examples where splicing into the SVA followed by retrotransposition led to exon shuffling. Using a minigene construct containing an intronic SVA, we are able to show that splicing into the SVA is not rare and that this splicing results in both exonization and SVA gene-trapping. These data suggest that splicing into SVA elements enables their expression and can allow for adaptive evolution at the cost of altering the transcriptome in both humans and great apes.
Results
Identification of MAST2-SVA
In 2007, Takasu et al. described a 14-kb deletion that included the entire HLA-A locus in three unrelated families, leading to leukemia in one individual from each of the families. Analysis of the deletion site identified an SVA insertion, hereafter referred to as SVAHLA-A. Using the SVAHLA-A DNA sequence, we located an SVA insertion on chromosome 3p21.31 as the likely progenitor of the SVAHLA-A insertion (Takasu et al. 2007). Further analysis of SVAHLA-A and its progenitor revealed several interesting details.
First, when using BLAST (Altschul et al. 1990) to align the SVAHLA-A sequence to the reference genome, many hits were obtained, most being SVAs. However, a few hits had a unique sequence upstream of the SVA. The unique sequence juxtaposed to the SVAHLA-A query sequence mapped with 99% identity (210/211 nucleotides [nt]) to the 5′ UTR and the first exon of the MAST2 (M2) gene on chromosome 1. Further analysis showed that 262 nt of the SVAHLA-A sequence mapped to MAST2; however, both the SVAHLA-A and CH3 insertions had a 40-bp deletion of nucleotides 210–249 relative to the MAST2 5′ UTR. Bioinformatics analysis identified 73 SVAs (Supplemental Table 1) containing some fraction of the MAST2 5′ UTR and first exon in the human genome. Subsequent analyses concluded that 3′ SVA sequences containing MAST2-derived 5′ ends cluster together phylogenetically in a clade, consistent with this subgroup being derived from a founder event. Moreover, sequence analysis grouped the MAST2-SVAs with the youngest human-specific SVA subfamily (F), a result consistent with the absence of SVAs containing MAST2 5′ transductions in the chimpanzee reference sequence. Hereafter, the MAST2-SVA subfamily will be referred to as SVAF1.
The number of nucleotides derived from MAST2 directly upstream of SVA varied from 35–382 (Supplemental Table 1). Given that the MAST2 5′ UTR and first exon combined is 460 nt, no SVAF1s present in the human reference genome contains the entire 5′ UTR-first exon.
The MAST2 sequence abutting the genomic copies of SVA in SVAF1 elements terminated directly at the 5′ SS of the MAST2 first exon. However, there is no SVA present in the reference MAST2 intron sequence. It is likely that an SVA with an allele frequency <1 resided in intron 1 of MAST2 (Fig. 1B) in the individual in which the first M2-SVA splicing and subsequent retrotransposition event occurred. We generated a consensus sequence from the SVAF1 in the human genome and aligned it to the SVA present in Repbase (Jurka 2000; Jurka et al. 2005), henceforth called SVARep, to determine whether the site where MAST2 and SVA intersect would have provided a suitable 3′ SS in a consensus SVAF1. The 3′ SS consensus sequence is YYYYYYYYYYNCAG/G, where Y represents a pyrimidine and N is any nucleotide (Wang and Cooper 2007). The SVA portion of SVAF1 aligns to SVARep beginning at position 388 of SVARep which is located in the 3′ region of the Alu-like fragment, 35 bp upstream of the VNTR. The sequence upstream of position 388 in SVARep is CCTCCACCTCCCAG (YYYYY-YYYYNCAG), a close match to the 3′ SS consensus sequence.
An SVA master element on CH10
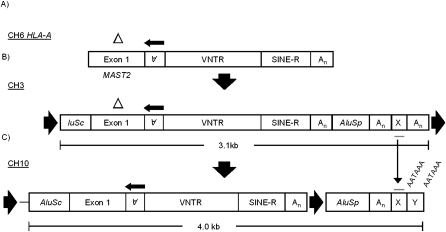
The SVA on CH3, the progenitor to SVAHLA-A (Fig. 2A), lacks a target-site duplication (TSD) directly flanking the SVAF1 (Fig. 2B). Given that retrotransposons are able to transduce sequences 5′ and 3′ of their location in the genome (Moran et al. 1999; Xing et al. 2006; Goodier and Kazazian 2008), we searched for a TSD further upstream and downstream from the CH3 SVAF1. We identified a 15-nt TSD, with the 5′ duplication directly in front of a truncated AluSc (153 nt) and the 3′ duplication following a polyA tail downstream from a non-RepeatMasker annotated sequence. The entire insertion between the AluSc and the terminal 3′ polyA tail nearest the 3′ TSD consisted of (1) the AluSc, followed by (2) a SVAF1, (3) an AluSp, and then (4) a 3′ transduction of 82 nt 3′ to the AluSp. When using BLAT (Kent 2002) to identify the source locus for the CH3 3′ transduction, 13 hits in addition to the CH3 query sequence were obtained (Supplemental Table 2). The source locus was identified on CH10 (Fig. 2C) due to the absence of a polyA tail 3′ of the transduced sequence present on CH3. Interestingly, the SVA on CH10 was flanked by a 5′ AluSc (320 nt) and a 3′ AluSp (299 nt). Overall, 13 SVAF1 insertions contained the 3′ transduction from chromosome 10 (Supplemental Table 2), and all 13 had the AluSp and were variably truncated with three containing the AluSc, four containing some portion of the MAST2 sequence and no AluSc, five truncated in the VNTR, and one truncated in the SVA polyA tail. Furthermore, one of the SVAs, which CH10 was the source locus for, had a 160-nt 3′ transduction. This element represents a transcript from the CH10 locus that bypassed the polyA signal at which the other 12 elements terminated. The sequence directly after the AluSp, the original source for these transductions, contains two canonical polyA signals, AATAAA (Colgan and Manley 1997), which are 15–20 nt upstream of the polyA tails of the SVAs derived from the CH10 locus (Fig. 2C).
Figure 2.
Identification of the SVA master locus on CH10. (A) The SVA insertion on CH6. The 40-bp deletion in the MAST2 sequence (△) allowed the identification of the (B) CH6 progenitor element to be identified on CH3. The SVA insertion on CH3 contains both 5′ and 3′ tranductions. At the 5′ end, the SVA contains a truncated AluSc while at the 3′ end, an AluSp along with additional sequence, indicated by X, followed by a polyA tail, with the entire insertion flanked by target-site duplications. The 3′ transduction (X, red line) on CH3 allowed the identification of (C) the master element on CH10 along with 12 additional elements derived from the CH10 locus. The SVA on CH10 contains starting at it's 5′ end: 185 bp transduction derived from CH9, a full-length AluSc, the MAST2-SVA, an AluSp, polyA tail, and then a unique sequence, which was 3′ transduced, and which allowed the identification of CH10 as the source locus. A target-site duplication flanks the insertion, which inserted on CH10. Downstream polyA signals are displayed over the X and Y transduction sequences.
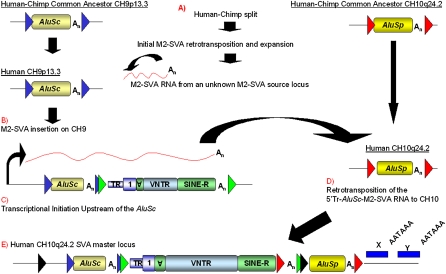
To distinguish whether the SVA on CH10 inserted alone, or with the AluSc and/or AluSp, we searched for TSDs of each element and examined the chimpanzee reference sequence (Chimpanzee Sequencing and Analysis Consortium 2005). Only the AluSp, hereafter referred to as 3′ Alu, was present in the chimp reference sequence, suggesting that it was the first insertion on CH10 (Fig. 3A) and that the SVA insertion occurred, with or without the AluSc, hereafter referred to as 5′ Alu, sometime since our last common ancestor with chimp. Furthermore, the 3′ Alu on CH10 is at least 25 million years (Myr) old because it is present in the Rhesus macaque genome sequence (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). The 5′ Alu on Ch10 could be traced back to a locus on CH9, due to 185 nt present directly upstream of the 5′ Alu on CH10 (Fig. 3E), which represents a 5′ transduction from the CH9 AluSc source locus (Fig. 3C). On CH10 there is a 13-nt TSD flanking the 5′ Alu containing the 5′ transduction and the SVA, suggesting that the 5′ Alu and SVA retrotransposed as one unit (Fig. 3C). However, at the CH9 locus there is no SVA downstream from the AluSc in the human reference sequence.
Figure 3.
Genesis of the SVA master locus on CH10. The AluSc on CH9 and the AluSp on CH10, hereafter 5′ Alu and 3′ Alu, respectively, are shown present in the human–chimp common ancestor. The original target-site duplications (TSDs) for the 5′ Alu and the 3′ Alu are indicated by blue and red arrowheads, respectively. (A) After the human–chimp split, the initial SVAF1 was formed due to an alternative splicing event. A SVA mRNA was transcribed from either the founder SVAF1 insertion or some other unknown SVAF1 locus in the human genome, derived from the original SVAF1 founder insertion. (B) The SVA mRNA is retrotransposed directly downstream from the 5′ Alu on CH9 as displayed by the SVA insertion flanked by TSDs (green arrowheads). (C) Transcriptional initiation occurred upstream of the 5′ Alu, generating a composite mRNA containing in order (1) 185 nt of 5′ transduced sequence from CH9, (2) the 5′ Alu, and (3) the SVAF1. (D) The composite mRNA from CH9 retrotransposed to CH10 directly upstream of the 3′ Alu. (E) The CH10 master element present in the human genome reference is shown. Directly downstream from the 3′ Alu TSD, unique sequence blocks (blue boxes), labeled X and Y, are present in the daughter insertions derived from this locus. The polyA signals utilized in the 3′ transductions are indicated by AATAAA relative to the unique sequence blocks.
Identification of multiple SVA TSSs
SVA is a nonautonomous retrotransposon and was previously thought to rely on an internal promoter to initiate its transcription, similar to LINE-1 (Swergold 1990) and Alu (Di Segni et al. 1981; Duncan et al. 1981; Fritsch et al. 1981). Previous attempts in our laboratory to locate the SVA promoter have led to ambiguous results (MC Seleme and HH Kazazian, unpubl.). We set out to identify the SVA TSS for insight into how SVA mRNA is transcribed.
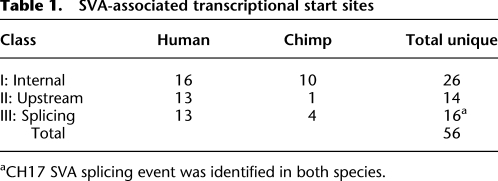
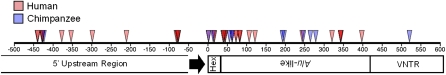
We used 5′ RACE to identify novel SVA 5′ ends from total RNA extracted from cell lines (see Methods) and chimpanzee testes. Currently, both the requirements for SVA transcription and the repertoire of expressed SVAs are unknown. We identified a total of 56 unique SVA-associated TSSs after sequencing and analysis of human and chimp SVA 5′ RACE products (Table 1). We grouped the TSSs into three classes: (1) internal SVA TSSs (Supplemental Table 3); (2) 5′ TSSs, defined as any position upstream of SVA annotated sequence (Supplemental Table 4); and (3) examples in which part of the sequence aligned within the SVA and part aligned upstream with a large gap in between, representing transcripts where 5′ sequences are spliced into the SVA part of the transcript (Table 2). The 26 class I TSS are scattered throughout the SVA but tend to cluster toward the 5′ end of the element (Fig. 4). The 14 class II TSSs start 76–440 bp upstream of SVAs in the human or chimp genome (Fig. 4; Supplemental Table 2).
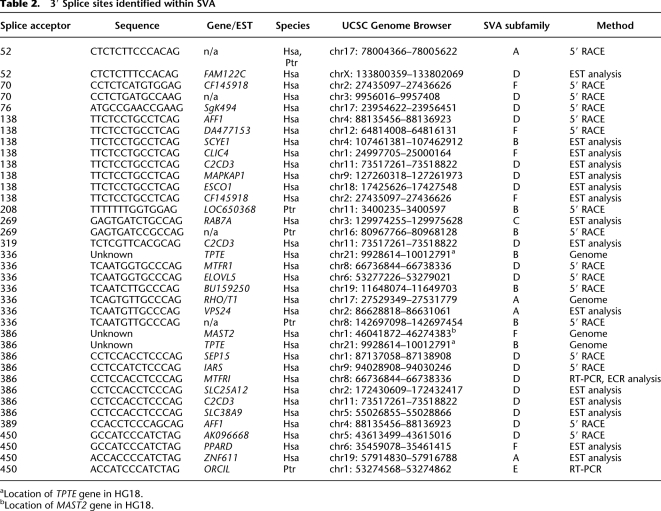
Table 1.
SVA-associated transcriptional start sites
aCH17 SVA splicing event was identified in both species.
Table 2.
3′ Splice sites identified within SVA
aLocation of TPTE gene in HG18.
bLocation of MAST2 gene in HG18.
Figure 4.
SVAs contain many transcriptional start sites (TSSs). 5′ RACE was performed on total RNA extracted from 293T, HeLa, and nTera cell lines, along with chimp testes. A nested PCR was performed followed by Sanger sequencing of Topo clones. The 5′ RACE adaptor was identified, and the first nucleotide after it was annotated as the TSS. Human (red triangles) and chimp TSS (blue triangles) within SVA are aligned relative to the position in SVARep. The CCCTCT hexamer (Hex), Alu-like region, and part of the VNTR are shown. TSSs aligning upstream of SVA are displayed relative to the first nucleotide of the SVA insertion in the genome. Both human (red triangles) and chimp (blue triangles) TSSs identified by 5′ RACE are shown.
Class III SVA-associated TSSs represent splicing into the SVA
We identified 17 class III TSSs, 16 of which are unique (13 human and four chimp) where the 5′ ends mapped upstream and represented splicing into SVA at 10 different 3′ SS (Table 2). SVA splicing events are listed in Table 2, with the 3′ SS position and 3′ SS sequence annotated relative to SVARep. Twelve of 17 class III TSSs involved exons from known genes/ESTs. One gene, AFF1 on CH4 had two SVA alternative splicing events identified by 5′ RACE, suggesting the same SVA may contain multiple functional 3′ SS.
Many SVA splicing events are present in human ESTs
To identify further examples of SVA splicing, we performed a computational survey to identify splicing events. We focused on splicing events involving the 228 SVA elements present in intronic regions oriented in the same direction as the surrounding gene. EST databases (Boguski et al. 1993) were mined for uniquely aligned sequences with evidence of intronic SVA expression. Spliced ESTs were selected where one or more blocks aligned within the intronic SVA sequence, and the exon–SVA junctions were examined. The SVA sequences upstream and downstream from the EST junctions were compared to the SS consensus sequence, and those containing the canonical “(T/C)AG” trinucleotide (Wang and Cooper 2007) at the junction preceded by a reasonable poly-pyrimidine tract were kept. We defined an event as a unique exon 5′ SS and a unique SVA 3′ SS. If multiple overlapping ESTs having the same 5′ SS and SVA 3′ SS existed, we called it one event. ESTs with the same 5′ SS and two different SVA 3′ SS were called two events.
In total, 16 events, involving 14 genes, at eight different SVA 3′ SS were detected, supporting the notion that splicing into SVAs occurs with some frequency across the genome (Table 2). We found one gene, C2CD3, which had ESTs aligning to three different 3′ SS locations at the C2CD3 locus, AG138, AG319, and AG386, further indicating that multiple functional 3′ SS exist within SVA.
Gene-trapping occurs in primates
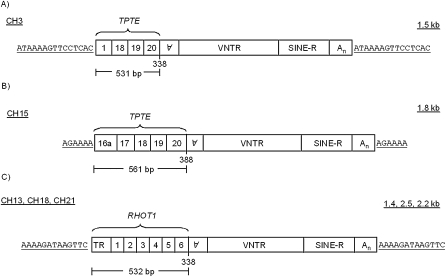
The upstream sequence of all class II TSSs were aligned to either the human or chimp reference sequence using BLAT (Kent 2002) to determine whether the sequence was present elsewhere in the genome, which would indicate potential SVA retrotransposed 5′ transductions. Of the 14 class II TSSs, only the lone example from chimp aligned elsewhere in the chimp reference sequence. The sequence consisted of 423 bp upstream of a truncated SVA on 3q11.2 (Fig. 5A) that aligned to multiple genomic locations with 91%–95% identity, one of which was a 342-bp hit on 15q14 (Fig. 5B). Further analysis revealed that the SVAs on CH3 and CH15 were insertions derived from two different SVA alternative splicing events, and that the transduced exons were derived from the transmembrane phosphatase with tensin homology (TPTE) gene on CH22 in chimp (CH21 in humans). TPTE is a testis-specific gene that shares significant homology with PTEN (Chen et al. 1999). However, similar to MAST2, no SVA is present in the TPTE gene reference sequence. These SVA insertions differ in that the 5′ transduction of the CH3 insertion is 531 bp and contains exons 1, 18, 19, and 20, while the 5′ transduction of the CH15 insertion is 561 bp and contains five exons, an unannotated exon in intron 16 referred to as 16a, and exons 17, 18, 19, and 20 (Fig. 5B). The CH3 insertion spliced into AG 336 of SVA, while the CH15 insertion spliced into AG 386. Both 3′ SS were identified previously in our data. Both the CH3 and CH15 SVA insertions are present in the human reference genome; however, both are absent from the orangutan reference genome. We used the ensemble browser to identify the presence of the CH15 SVATPTE insertion in the gorilla genome sequence, while the CH3 SVATPTE insertion was not located.
Figure 5.
SVA gene-trapping has occurred in other hominids. (A,B) Two different SVA splicing events followed by retrotransposition derived from the TPTE locus on chromosome 21 flanked by a target-site duplication. (A) CH3 SVA containing four processed TPTE exons that utilized AG 336. (B) CH15 SVA containing five processed exons that utilized AG 386. (C) The structure of SVA insertions on CH13, CH18, and CH21 containing the six processed exons from the RHOT1 gene on CH17 with identical target-site duplications.
After finding SVAF1 and SVATPTE, we examined the human genome to see if we could identify additional examples of splicing followed by retrotransposition. We searched the human genome reference sequence (Lander et al. 2001) for SVAs that had a non-SVA sequence upstream present multiple times in the genome. This approach would identify examples where an SVA provided a 3′ SS and this mRNA retrotransposed and then likely jumped again. Computational analysis identified an SVA subgroup, SVARHOT1, where the first six exons, 532 bp, of the RHOT1 gene on chromosome 17, also known as mitochondrial Rho GTPase 1 (MIRO-1) (Fransson et al. 2003), were spliced into SVA at AG 336 and subsequently retrotransposed (Supplemental Fig. 1). However, unlike the MAST2 and TPTE examples, there is an SVA residing in the sixth intron of RHOT1 in the human reference genome. There are three SVAs in the human genome containing upstream RHOT1 processed exons, located on 13q11, 18p11.21, and 21q11.2. Two SVARHOT1 copies were identified in the chimp genome on CH13 and CH18 and one on CH13 in the gorilla genome. Both the human and chimp SVARHOT1 insertions are missing the first 36 nt of the RHOT1 5′ UTR and share identical 13-nt TSDs. Surprisingly, these different insertions represent duplications and not individual retrotransposition events. The human SVARHOT1 insertions are within large CNVs, present in both human and chimp several times, yet only the CH13, CH18, CH21 CNVs contain the SVARHOT1 insertion (Supplemental Fig. 1). We concluded that the CH13 SVARHOT1 insertion most likely represents the original SVARHOT1 insertion based upon its presence in the gorilla genome draft sequence; however, this is contingent upon the CNVs containing the other SVARHOT1 insertions not being polymorphic in these species. Both the SVA insertion in intron 6 of RHOT1 and all of the SVARHOT1 insertions are absent from the orangutan genome sequence, suggesting that the SVA at the RHOT1 locus and the original SVARHOT1 event occurred some time after the orangutan diverged from the human–gorilla last common ancestor, aging the insertion between 8 and 15 Myr.
SVA splicing is not rare
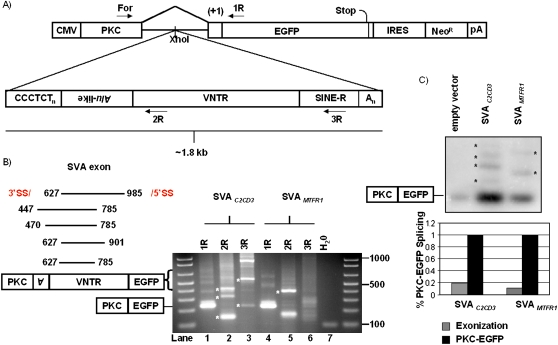
To study the potential mutagenic potential of SVA splicing, we cloned two SVAs from the human genome, SVAC2CD3 and SVAMTFR1, containing multiple functional 3′ SS. We cloned the SVAs into the intron of a splicing minigene construct named pPKC-EGFP (Fig. 6A), hereafter pPKC-SVA. 293T cells were transfected with pPKC-SVA, and total RNA was harvested after 1 d. To characterize SVA splicing and identify functional 3′ SSs, we performed RT-PCR with a forward primer in the first exon, PKC, and then used three reverse primers (6A) in independent reactions to answer three questions: (1) are SVAs exonized (PKC For + 1R primers); (2) what 3′ SS exist in SVA (PKC For + 2R); and (3) can we detect SVA gene-trapping (PKC For + 3R).
Figure 6.
SVA gene-trapping and exonization are not rare. (A) Two SVAs were cloned into PKC-EGFP (Newman et al. 2006) to test the mutagenic potential of SVA splicing. Primers used for RT-PCR are marked. (B) RT-PCR was performed on total RNA extracted from 293T cells transiently transfected with pPKC-EGFP containing one of two different SVAs cloned into the intron. SVA exonization events (left panel) are annotated with the first and last nucleotide of the SVA exon, all of which occur within the Alu-like and VNTR domains. A representative agarose gel displaying SVA alternative splicing events is shown (right panel) (see Table 3). (*) Indicates bands verified by DNA sequencing to be SVA splicing events. (C) Semi-quantitative PCR to determine the frequency of SVA exonization. Ten cycles of PCR on cDNA from individual pPKC-EGFP, pPKC-SVAC2CD3, and pPKC-SVAMTFR1 transfections were carried out using PKC For and 1R. PCR products were resolved on a 2% agarose gel, followed by overnight transfer to a membrane, and subsequent probing using a DNA probe targeting the PKC exon (top panel). (*) Indicates bands quantified by a phosphorimager. Total SVA exonization was normalized to PKC-EGFP splicing within each respective lane and graphed (bottom panel).
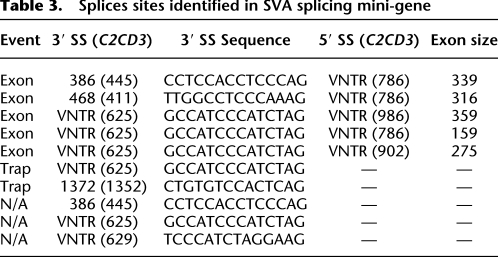
PCR products were analyzed on a 2% agarose gel (Fig. 6B); bands were cloned and sequenced. Bands from the lane labeled 1R for pPKC-SVAC2CD3 corresponded to the normal splicing, PKC exon to EGFP exon, and also to SVA exonization events. Five SVA exonization events utilizing three different 3′ SS and three different 5′ SS with SVA exons ranging from 159–359 nt (Fig. 6B; Table 3) were identified. Three out of the five SVA exonizations shift the reading frame, while all five SVA exonization events introduce premature stop codons (PTCs) located in the exonized SVA sequence. SVA splicing events using pPKC-SVA and verified by sequencing are listed in Table 3. It is noteworthy that a 3′ SS site was identified in the SINE-R domain of SVA (Fig. 6B, lane 3, lower band). These PCR results suggest that SVA splicing is not rare and that both SVA exonization and SVA gene-trapping can occur in the same SVA.
Table 3.
Splices sites identified in SVA splicing mini-gene
Semi-quantitative RT-PCR followed by Southern blotting using amplicons from a PKC for and 1R PCR (Fig. 6A,C) was carried out for pPKC-EGFP, pPKC-SVAC2CD3, and pPKC-SVAMTFR1 to estimate SVA exonization. The intensities for normal splicing varied across the samples, so the Southern blot was exposed overnight in order to ensure no bands were present in the vector-only lane (data not shown). The ratio of the higher molecular weight bands indicative of SVA exonization relative to PKC-EGFP splicing within that lane was determined using a phosphorimager (Fig. 6C, bottom panel). The ratio of total SVAC2CD3 exonization relative to PKC-EGFP splicing was 0.19:1, while total SVAMTFR1 exonization to PKC-EGFP was 0.12:1.
Discussion
These data are the first to provide insight into how SVA retrotransposons are expressed and how they might impact gene expression. Our data suggest that SVAs are expressed in a variety of ways in humans and chimps. Recently, a study identified many TSSs in LINE-1s and SINEs in human and mouse tissues and cell lines. (Faulkner et al. 2009). Whether or not internal TSSs identified here represent retrotransposition-competent SVA transcripts is unknown. Many SVAs have transduced sequence 5′ of their location in the genome to other locations (Damert et al. 2009); this is consistent with our observation of upstream TSSs. What is unclear is whether upstream TSSs represent solely upstream promoters driving SVA expression or whether something inherent to SVA directs transcriptional initiation upstream.
The SVAF1 subfamily, SVATPTE, and SVARHOT1 together indicate that if an SVA loses the CCCTCT hexamer and most of the Alu-like region due to alternative splicing into it, the remaining SVA sequence is able to retrotranspose. Furthermore, the lack of most of the Alu-like region suggests that the model suggested by Mills et al. (2007), adapted from Boeke's model (Boeke 1997), where the SVA Alu-like region hybridizes to Alu RNAs at the ribosome in order to compete for the LINE-1 ORF2 reverse transcriptase, may not be case. However, it is possible that SVA RNA may be located at the ribosome where competition for the LINE-1 ORF2 takes place, but that it is not hybridizing to Alu RNA.
The lack of SVAF1s with a complete MAST2 5′ UTR and first exon suggests that a full-length MAST2 5′ UTR first exon is not required for transcription or retrotransposition. Exactly, how the MAST2 sequence contributed to the expansion of SVAF1s in the absence of the CCCTCT hexamer and the Alu-like region still needs to be determined. One possibility is that the MAST2 sequence, in combination with certain SVA sequence variants or in a specific genomic context, enhances transcription or retrotransposition relative to a canonical SVA. It is worthwhile to note that TPTE is a testis-specific gene (Chen et al. 1999) and that we found the CH3 SVATPTE by 5′ RACE, and the TSS was in exon 1 of the transduced TPTE sequence.
We have identified 11 3′ SS throughout SVA, in addition to multiple 3′ SS in the VNTR, including examples from all subfamilies except E, and we have shown that exonization can occur. Whether or not older SVAs are still retrotransposing in humans is currently unknown; however, the older SVAs are still able to be transcribed and may influence transcription if residing within an intron in the same orientation as the gene.
SSs within retrotransposons are not uncommon. Alus are primate-specific retrotransposons that are known to exonized (Sorek et al. 2002; Lev-Maor et al. 2003). In addition to Alu, internal splicing has been observed in the human L1 (Belancio et al. 2006) and the zebrafish LINE (Tamura et al. 2007). Although, it appears that if an SVA undergoes a splicing event, it can still carry out subsequent rounds of retrotransposition, as indicated by SVAF1.
SVA splicing followed by retrotransposition may be rare based on only four examples identified in the human genome, three of which are present in the chimpanzee. Additional splicing followed by retrotransposition events may have occurred, but the results are undetectable due to truncation upon insertion or low allele frequency. On the contrary, splicing into the SVA is not rare, as indicated by our semi-quantitative PCR data, which suggest that SVA exonization events may occur 12%–19% of the time relative to normal splicing in our minigene.
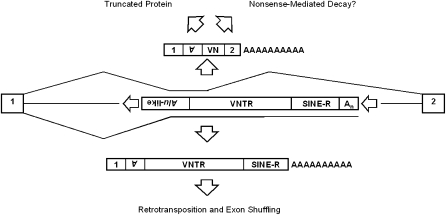
Currently, the ratio of SVA gene-trapping to SVA exonization has not been determined. Here we provide a low-end estimate for SVA splicing by assessing SVA exonization using a splicing minigene. Our SVA exonization estimate may be an underestimate because SVAs contain more than 10 nonsense codons in each reading frame on the sense strand and exonization of these sequences may induce nonsense-mediated decay if the exonized SVA sequence is more than 50–55 nt upstream of the 3′ most exon–exon junction (Nagy and Maquat 1998). Be that as it may, an SVA splicing event, exonization or trapping (Fig. 7), will likely lead to a dead-end to the protein-coding capacity of the mRNA because either event has the capability to produce truncated proteins.
Figure 7.
SVA alternative splicing outcomes. An intronic truncated SVA is shown (middle). The SVA is truncated because these SVAs are still likely to be spliced. If SVAs are exonized, they will likely generate a truncated protein or subject the mRNA to nonsense-mediated decay due to the inclusion of SVA nonsense codons (top). If SVAs mimic an endogenous gene-trap, that is provide a 3′ SS followed by termination at the SVA or downstream polyA signal, this may result in truncated proteins, but more importantly the retrotransposition of exons.
Most SSs identified using the splicing minigene were not identified by 5′ RACE, such as the 3′ SSs in the VNTR and SINE-R. This is likely due to the nested PCR approach utilized in 5′ RACE. However, downstream SVA 3′ SS may be selected for in the splicing minigene due to the small size of the intron. Each SVA was cloned into pPKC-EGFP with less than 100 bp of flanking DNA to ensure splicing was inherent to the SVA and not due to intronic splicing enhancers. If SVA is cloned in as a larger fragment, one may see 3′ SS selection shift toward the 5′ end of SVA residing in the Alu-like domain.
If SVAs impact gene expression by being alternatively spliced, then one would expect to observe either a depletion of SVAs in genes or on the coding strand of genes. An underrepresentation was observed for SVAs on the coding-strand in the human genome; 1060/2772 SVAs are in RefSeq genes, with 228/1060 on the coding strand (introns or exons) and 832/1060 on the antisense strand. This underrepresentation of intronic SVA insertions on the coding strand is highly significant (P < 2.2 × 10−16) under a null hypothesis of random orientation. Likewise, a similar significant underrepresentation is observed in the chimp reference genome, with 228 (partial overlap with the human 228) of 1024 intronic SVAs oriented on the sense strand with respect to the surrounding gene. Nevertheless, this SVA strand bias may be due to a factor other than selection, such as SVA insertional preference.
Altogether, these data show that SVAs are alternatively spliced in cell culture, in tissue, and in vivo. We speculate that SVAs may influence local gene expression by providing alternative SSs and might even account for some of the variation in gene expression observed within and across hominids. As more primate genomes are sequenced along with more studies on SVA, the impact of this retrotransposon will become clear. Thus, although SVAs effect on genome evolution may be less than that of L1 and Alu because of their smaller numbers, SVA has had recent effects that are likely growing with their continued expansion as indicated by the SVAF1 subfamily and the CH10 subgroup. In another 50 Myr, the SVA effect on genome evolution may be much greater than that of L1 and Alu.
Methods
Sequence analysis
BLAT (Kent 2002) and BLAST (Altshul et al. 1990) were used in mapping sequences to the reference genomes. Censor (Kohany et al. 2006) and RepeatMasker (Smit et al. 1996) were used to identify relative positions in SVA and subfamily classification, respectively.
Cell culture
293T and HeLa cells were grown in a humidified, 5% CO2 incubator at 37°C in DMEM (GIBCO) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 100 U/mL penicillin, 0.1 mg/mL streptomycin. nTera cells were grown as described above except that the media was supplemented with nonessential amino acids.
5′ RACE, cDNA synthesis, and PCR
RNA extraction was performed using the RNeasy kit (Qiagen) according to the manufacturer's instructions. DNase treatment consisted of using twice the recommended amount of RQ1 RNase-Free DNase (Promega) followed by ethanol precipitation of the RNA. Chimp testis was used for RNA extraction (Department of Veterinary Medicine and Surgery, University of Texas M.D. Anderson Cancer Center, Houston, TX).
5′ RACE was performed using the GeneRacer Kit (Invitrogen) with 5 μg DNase-treated RNA as the starting material. First-strand cDNA synthesis was performed using the supplied SuperScript III RT kit with random hexamer primers or Array Script Reverse Transcriptase (Ambion). All steps were carried out according to the manufacturer's instructions. A two-round PCR scheme was utilized in order to enrich for SVA containing transcripts using GoTaq (Promega) or Expand Long (Roche) according to the manufacturer's instructions with 1 μL of cDNA containing the 5′ RACE adaptor. The first round of PCR consisted of reverse primers complementary to the SINE-R region of SVARep. Primers complementary to the Alu-like region were used for the second round of PCR. PCR cycling parameters consisted of variations on touchdown PCR with the initial annealing temperature at 60°C and cycled down to 50°C over 40 cycles. PCR reactions were analyzed on 1%–1.5% agarose gels. Bands of varying size were cut out, gel purified using QIAquick Gel Extraction kit (Qiagen), Topo cloned (Invitrogen), and sequenced.
RT-PCR
DNase-treated RNA was reverse transcribed using random primers with the SuperScript III First-Strand Synthesis SuperMix (Invitrogen) according to the manufacturer's instructions. One microliter of cDNA was used in PCR with GoTaq (Promega) or Expand Long (Roche).
EST analysis
EST locations corresponding to human genome assembly hg18 were obtained from the UCSC Genome Browser and stored in a local relational database along with SVA and RefSeq gene locations. ESTs with blocks aligning unambiguously within SVA locations present on the same strand as a RefSeq intron were further analyzed using Perl scripts locate EST splice junctions within the SVA. Junctions corresponding to splicing patterns consistent with SVA gene-trapping were compared to the SS consensus sequence to ensure presence of the relevant nucleotides.
SVA splicing minigene, transfection, and RT-PCR
pPKC-EGFP has been previously reported (Newman et al. 2006). SVAs were amplified from human genomic DNA and subcloned into Topo (Invitrogen). The SVA was then amplified as a XhoI fragment and cloned into the XhoI site within the intron.
293T cells were seeded into T-75 flasks in order to be 50%–80% confluent upon transfection. Twenty-four hours later, 8 μg of each splicing minigene was transfected using 24 μL of Fugene6 (Roche) according to the manufacturer's instructions. Total RNA was isolated 1 d after transfection as described above. Five micrograms of DNase-treated RNA was reverse-transcribed with Array Script (Ambion) using an olio dT primer. Touchdown PCR from 59°C to 51°C over 40 cycles was performed with elongation at 72°C for 2 min using GoTaq Master Mix (Promega) with 1 μL of cDNA as template and primers at a final concentration of 0.2 μM per reaction. Amplicons were analyzed on 2% agarose gels.
Semi-quantitative PCR
One microliter of oligo dT primed cDNA derived from total RNA from splicing minigene transfections was amplified by 10 cycles of PCR (20 sec at 94°C, 30 sec at 57°C, 1 min at 72°C) using GoTaq MasterMix (Promega) with PKC forward and 1R primers in a 25 μL reaction. The entire reaction was resolved on a 2% agarose gel. Overnight alkaline transfer to N+ hybond membrane (Amersham) was performed followed by overnight hybridization with a 182-bp DNA probe labeled with [α-32P]dCTP targeting the PKC exon at 65°C. Reaction products were imaged using a Storm 840 phosphorimager (GE Healthcare) and quantified with ImageQuant 5.2 (GE Healthcare). The intensity of each band was determined followed by the subtraction of background. SVA exonization band intensities were summed followed by normalization to PKC-EGFP splicing.
Acknowledgments
This work was supported by grants from the NIH. D.C.H. is funded by NIH training grant T32-M007229-27. We thank Dr. Yoshihide Ishikawa, Japanese Red Cross Central Blood Institute, for HLA-A deletion samples. We also thank Claude Warzecha and Dr. Russ Carstens for the PKC-EGFP construct.
Footnotes
[Supplemental material is available online at http://www.genome.org. The 5′ RACE sequence data from this study have been submitted to dbEST (http://www.ncbi.nlm.nih.gov/dbEST) under accession nos. GR564526–GR564716.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093153.109.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD. LINEs and Alus—the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TMJ, Tolstoshev CM. dbEST—database for “expressed.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Chen H, Rossier C, Morris MA, Scott HS, Gos A, Bairoch A, Antonarakis SE. A testis-specific gene, TPTE, encodes a putative transmembrane tyrosine phosphatase and maps to the pericentromeric region of human chromosomes 21 and 13, and to chromosomes 15, 22, and Y. Hum Genet. 1999;105:399–409. doi: 10.1007/s004390051122. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Damert A, Raiz J, Horn AV, Löwer J, Wang H, Xing J, Batzer MA, Löwer R, Schumann GG. 5′-Transduced retrotransposons groups spread efficiently throughout the human genome. Genome Res. 2009 doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Di Segni G, Carrara G, Tocchini-Valentini GR, Shoulders CC, Bralle FE. Selective in vitro transcription of one of the two Alu family repeats present in the 5′ flanking region of the human epsilon-globin gene. Nucleic Acids Res. 1981;9:6709–6722. doi: 10.1093/nar/9.24.6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CH, Jagadeeswaran P, Wang RR, Weissman SM. Structural analysis of templates and RNA polymerase III transcripts of Alu family sequences interspersed among the human beta-like globin genes. Gene. 1981;13:185–196. doi: 10.1016/0378-1119(81)90007-x. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;5:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Fransson A, Ruusala A, Aspenström P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J Biol Chem. 2003;278:6495–6502. doi: 10.1074/jbc.M208609200. [DOI] [PubMed] [Google Scholar]
- Fritsch EF, Shen CK, Lawn RM, Maniatis T. The organization of repetitive sequences in mammalian globin gene clusters. Cold Spring Harb Symp Quant Biol. 1981;45:761–765. doi: 10.1101/sqb.1981.045.01.095. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Hassoun H, Coetzer TL, Vassiliadis JN, Sahr KE, Maalouf GJ, Saad ST, Catanzariti L, Palek J. A novel mobile element inserted in the alpha spectrin gene: Spectrin dayton. A truncated a spectrin associated with hereditary elliptocytosis. J Clin Invest. 1994;94:643–648. doi: 10.1172/JCI117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- Makino S, Kaji R, Ando S, Tomizawa M, Yasuno K, Goto S, Matsumoto S, Tabuena D, Maranon E, Dantes M, et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80:393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Newman EA, Muh SJ, Hovhannisyan RH, Warzecha CC, Jones RB, McKeehan WL, Carstens RP. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Kawakami M, Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15:8725–8737. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. Evolutionary and biomedical insights from the Rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Minegishi Y, Richter D, Eguiguren J, Conley ME. Unusual mutations in Btk: An insertion, a duplication, an inversion, and four large deletions. Clin Immunol. 1999;90:28–37. doi: 10.1006/clim.1998.4629. [DOI] [PubMed] [Google Scholar]
- Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region: Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996. http://www.repeatmasker.org.
- Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu M, Hayashi R, Maruya E, Ota M, Imura K, Kougo K, Kobayashi C, Saji H, Ishikawa Y, Asai T, et al. Deletion of entire HLA-A gene accompanied by an insertion of a retrotransposon. Tissue Antigens. 2007;70:144–150. doi: 10.1111/j.1399-0039.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- Tamura M, Kajikawa M, Okada N. Functional splice sites in a zebrafish LINE and their influence on zebrafish gene expression. Gene. 2007;390:221–231. doi: 10.1016/j.gene.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: A hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Wilund KR, Ming Y, Campagna F, Arca M, Zuliani G, Fellin R, Ho Y, Garcia JV, Hobbs HH, Cohen JC. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum Mol Genet. 2002;11:3019–3030. doi: 10.1093/hmg/11.24.3019. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]