Abstract
Transcription factors play an important role in orchestrating the activation of specific networks of genes through targeting their proximal promoter and distal enhancer regions. However, it is unclear how the specificity of downstream responses is maintained by individual members of transcription-factor families and, in most cases, what their target repertoire is. We have used ChIP-chip analysis to identify the target genes of the ETS-domain transcription factor ELK1. Two distinct modes of ELK1 target gene selection are identified; the first involves redundant promoter binding with other ETS-domain family members; the second occurs through combinatorial binding with a second transcription factor SRF, which specifies a unique group of target genes. One of the most prominent groups of genes forming the ELK1 target network includes classes involved in core gene expression control, namely, components of the basal transcriptional machinery, the spliceosome and the ribosome. Amongst the set of genes encoding the basal transcription machinery components, are a functionally linked subset of GTFs and TAFs. Our study, therefore, reveals an unsuspected level of coordinate regulation of components of the core gene expression control machinery and also identifies two different modes of promoter targeting through binding with a second transcription factor or redundant binding with other ETS-domain family members.
Eukaryotic transcriptional activator and repressor proteins control gene expression through altering the local chromatin structure around promoters and/or the recruitment of the basal transcription machinery and the subsequent engagement of RNA polymerases. Many transcriptional regulators respond to extracellular signals by acting as targets of signal-transduction cascades, which often leads to the modification of their activity through phosphorylation (for review, see Yang et al. 2003; Rosenfeld et al. 2006). The basal transcription machinery is thought to be a passive player with regard to responding to signaling pathways. However, recent studies suggest that the basal machinery is itself regulated, most notably demonstrated by the switching of TFIID for an alternative promoter recognition complex containing TBPL2 (also known as TRF3) and TAF3 during muscle differentiation (Deato and Tjian 2007). Other recent studies indicate that the expression of TBP, the core component of the TFIID promoter binding complex, is regulated in response to signaling via the MAP kinase pathways (Zhong et al. 2007). A key transcription factor controlling the expression of TBP under these conditions was identified as ELK1.
ELK1, along with ELK4 (also known as SAP1) and ELK3 (also known as SAP2/Net), is a member of the ternary complex factor (TCF) subfamily of ETS-domain transcription factors (for review, see Sharrocks 2002; Shaw and Saxton 2003). There are 27 different ETS-domain transcription factors expressed in mammalian cells, and their relative expression differs according to cell type (Hollenhorst et al. 2004). These transcription factors all bind to variants of the GGAA/T motif embedded in a larger 10-bp consensus sequence in vitro, suggesting that they might exhibit a high degree of redundancy in vivo. Indeed, chromatin immunoprecipitation with microarray hybridization (ChIP-chip) analysis suggested that this was the case for ETS1, ELF1, and GABPA (also known as GABPα) (Hollenhorst et al. 2007). This study noted, however, that many of the ETS-domain transcription-factor targets identified were apparently not bound by ELK1, suggesting that ELK1 might target a distinct subset of genes. This family of transcription factors is characterized by the presence of an ETS DNA-binding domain, but the TCF subfamily also contains a short protein interaction motif known as the B-box, which enables them to interact cooperatively with a second transcription factor SRF (Dalton and Treisman 1992; Shore and Sharrocks 1994; Hassler and Richmond 2001). The unique ability of the TCF proteins to bind to SRF is thought to confer distinctive promoter recognition specificity to these proteins. Several target genes have been identified for ELK1 (see Supplemental Table S1), which reflect its combinatorial interaction with SRF, although other genes are apparently regulated in an SRF-independent manner. However, more global analysis of ELK1-binding events is lacking, and our current models are extrapolated from a limited number of examples.
In this study we identified over 1000 new gene promoters that are bound by ELK1. Two distinct modes of binding were identified where ELK1 co-occupies promoters in combination with SRF and also binds a large proportion independently from SRF. Our data also uncover a previously unsuspected route for the coordinate regulation of gene expression, through ELK1 potentially controlling groups of genes involved in transcription, splicing, and translation.
Results
Identification of the ELK1 target gene network
ELK1 was originally identified as a key regulator of immediate-early genes, such as FOS, which are rapidly and transiently induced following exposure to extracellular ligands that activate the MAP kinase pathways (for review, see Sharrocks 2002; Shaw and Saxton 2003). To broaden our knowledge of the regulatory potential of ELK1, we performed ChIP-chip analysis to identify a more comprehensive ELK1 target gene network. ChIP analysis was performed for endogenous ELK1 in serum-starved HeLa cells (Supplemental Fig. S1). Bound material was detected using Affymetrix human promoter arrays that contain around 10 kb of DNA sequence surrounding each of 23,000 promoters. The raw ChIP-chip fluorescence intensity data were processed using Model-based Analysis of Tiling Arrays (MAT) (Johnson et al. 2006). The resulting MATscore is a statistical method for defining regions enriched on tiling arrays. A total of 1823 ELK1-binding regions corresponding to 1712 nonredundant promoters were identified, which corresponded to a MATscore of 2.94, a P-value of 0.00001 and an FDR value of 25.29 and were ranked according to the P-value (Supplemental Table S2A,B). To establish the validity of these targets, we randomly selected 32 binding regions from throughout the list and verified these by qPCR–ChIP; 20/32 (63%) of the tested genes were verified as true positive. The data set can be further partitioned based on computational FDR values, and 18/23 (78%) were validated within the binding regions with FDR values < 10 (FDR < 10 data set) (Fig. 1; Supplemental Fig S2). We therefore focused on the FDR < 10 data set as a high-confidence group of ELK1 targets (1053 ELK1-binding regions representing 1112 potential promoters), although there are clearly a large number of additional potential targets amongst the other binding regions identified by ChIP-chip analysis. Subsequent analysis of further genes in this data set confirmed the high true-positive rate of this subset of targets (see Fig. 3C, below). Further partitioning of the data set into lower FDR scores had little impact on the downstream analysis of the binding site features (data not shown); therefore, we selected regions scoring FDR < 10 for subsequent analysis to retain a large number of targets and refer to this group as the ELK1 FDR < 10 data set.
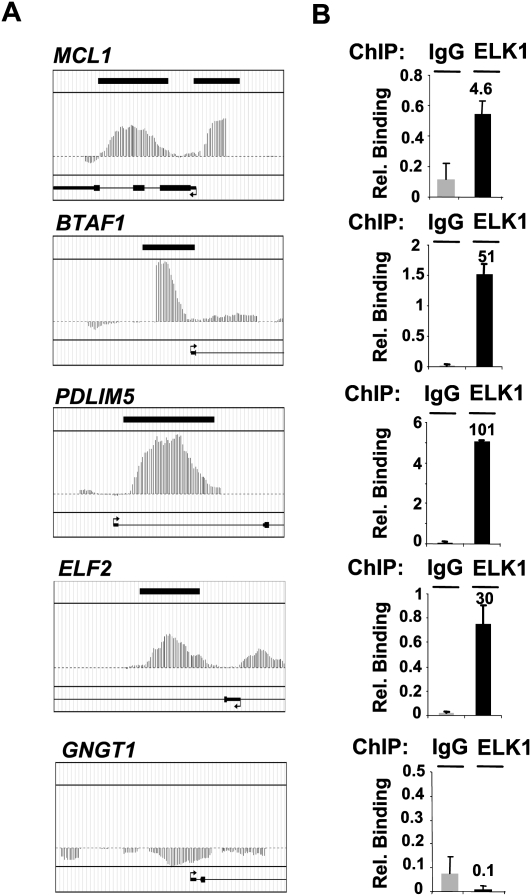
Figure 1.
Validation of ELK1 targets identified by ChIP-chip. (A) MAT profiles of ELK1 binding to the novel BTAF1, ELF2, and PDLIM5 promoters, the known target MCL1, and the nontargeted promoter GNGT1. (B) qPCR-ChIP validation of ELK1 binding to the same set of promoters. Fold enrichment over IgG control precipitations is shown above each graph. Data are the average of duplicate samples and representative of three independent experiments.
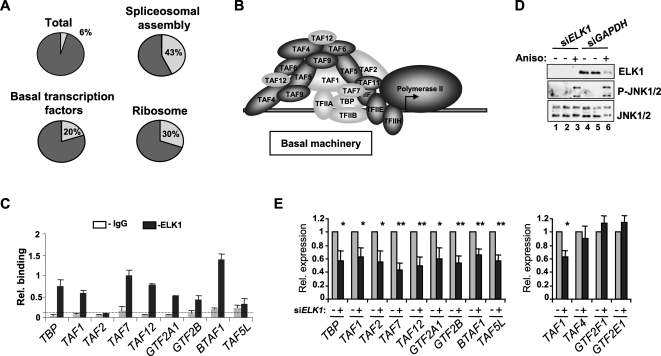
Figure 3.
Coordinate regulation of a subset of general transcription factors by ELK1. (A) Pie charts illustrating the frequency of occurrence of ELK1-binding events (light-shaded segment) in the total number of genes (represented by annotated gene symbols) on the promoter array (total), or in pathways identified by KEGG (“basal transcription factors” and “ribosome”) or BioCarta (“spliceosomal assembly”) pathway analysis. (B) Subunits of the basal transcription machinery are shaded, with shading representing subunits whose promoters are bound (light) or not bound (dark) by ELK1. (C) qPCR-ChIP verification of ELK1 binding to promoters of the indicated genes. The dotted line represents average binding across all IgG control precipitations. Data are representative of two independent experiments and the average of triplicate samples. (E) RT-PCR analysis of expression of the indicated genes following treatment with anisomycin for 60 min in HeLa cells in the presence and absence of control (GAPDH) (gray bars) or ELK1-specific (black bars) siRNA duplexes. A Western blot showing the expression of ELK1 and the levels of active JNK (P-JNK1/2) in each sample is shown in D. Error bars represent standard deviations calculated from three biologically independent replicates and the average of two samples. Asterisks denote differences with statistical significances (*P < 0.05; **P < 0.01) relative to the control sample for each gene (siGAPDH), as determined by the Students t-test.
Features of the ELK1 target regions
To gain possible insights into the mechanisms through which ELK1 might function on its target promoters, we searched for over-represented DNA-binding motifs in the FDR < 10 data set sequences. The most common motifs resembled those recognized by ETS-domain transcription factors (Supplemental Fig. S3). The top scoring 10-bp motif shows very high similarity to the in vitro selected optimal ELK1-binding site (Fig. 2A; Treisman et al. 1992; Shore and Sharrocks 1995). Indeed, ELK1-binding regions are significantly enriched for this motif compared with a random set of promoter sequences, and 3% contain exact matches to this 10-bp sequence (P < 0.0001) (Fig. 2B; Supplemental Table S4). Further searching with hexameric variants of this motif revealed even greater enrichment, with at least one motif present in over 89% of ELK1 target regions (P < 0.0001) (Supplemental Table S4). A total of 31% of ELK1-binding regions contain a motif that is an exact match to the composite octamer (CCGGAAGT). A previous study indicated that ELK1 can select for GGAT as well as GGAA in the central motif, especially when in combination with SRF (Treisman et al. 1992). However, although searching for GGAA and GGAT motifs gave little discriminatory power, there was a clear over-representation of GGAA motifs over GGAT in the ELK1-derived data set compared with a background promoter data set (Supplemental Fig. S4; Supplemental Table S4). Collectively, this analysis further emphasizes the quality of our data set and demonstrates that the in vitro and in vivo binding specificities of ELK1 are very similar.
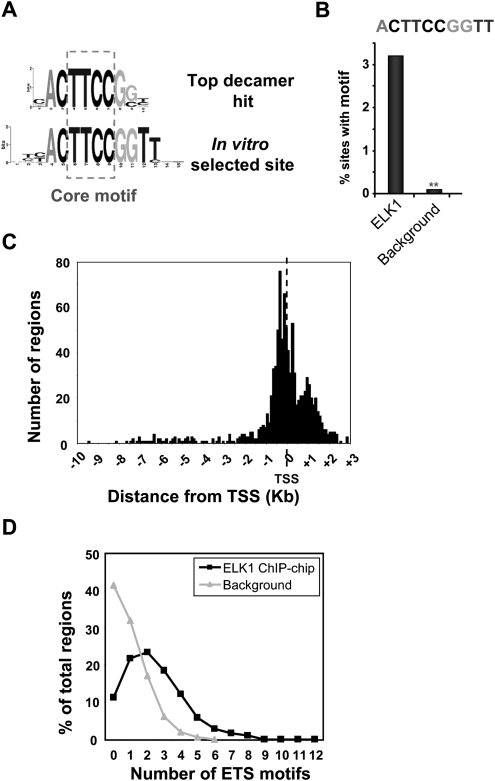
Figure 2.
Features of the ELK1-binding regions. (A) Sequence logo representation of the top 10-bp sequence identified by de novo searching the ELK1 FDR < 10 data set for over-represented motifs. The sequence of the optimum in vitro-selected ELK1-binding site (Shore and Sharrocks 1995) is shown and the inverted core GGAA “ETS binding motif” is boxed. (B) Over-representation of the optimal 10-bp ELK1-binding motif in the ELK1 FDR < 10 data set in comparison to a background data set. **P ≤ 1 × 10−4. (C) Location of the most significant probe of the ELK1-binding regions with respect to the closest transcriptional start site (TSS). The distances corresponding to each of the regions were grouped into bin sizes of 100 bp for quantification. (D) The number of nonredundant occurrences of the hexameric motifs CCGGAA, CGGAAG, and GGAAGT in each binding region in the ELK1 FDR < 10 (black lines) and background datasets (gray lines) was determined, and the distribution of the frequency of occurrence of numbers of motifs in each data set plotted.
Studies on other transcription factors have indicated that they can be found in a variety of locations, often many kilobases away from the transcription start site (TSS), as exemplified by the estrogen receptor (Carroll et al. 2005). However, the peaks of the ELK1-binding regions were centered around the TSS with 71% being within 1 kb of the TSS (Fig. 2C), and this is consistent with the locations of the predicted ELK1-binding motifs that tightly cluster around the TSS (Supplemental Fig. S5B). While other transcription factors such as SMAD2/3 also show clustered binding close to the TSS, the distribution of binding region locations is broader than observed for ELK1 (Koinuma et al. 2009; Supplemental Fig. S5A). Moreover, when either a data set of random promoters (Supplemental Fig. S5C) or a series of random motifs with the same base composition as the ELK1-binding motifs (Supplemental Fig. S5D) was analyzed, a broader distribution of motif locations relative to the TSS was observed. Thus, promoter-proximal binding seems to be an important facet of ELK1 function and is in agreement with a recent in silico study that identified motifs resembling the ELK1-binding sites as common features located close to the TSS (Fitzgerald et al. 2004).
Previous studies indicated that a different ETS-domain transcription factor, GABPA, displays multiple ETS motifs in each of its binding regions (Valouev et al. 2008). Similarly, we find multiple potential binding motifs in the ELK1-binding regions with up to 12 hexameric potential ELK1-binding motifs, and the majority containing at least two motifs (Fig. 2D). This contrasted sharply with the lower frequency occurrence of these motifs in random datasets (Fig. 2D) or with a series of inverted motifs in the ELK1 FDR < 10 data set (Supplemental Fig. S6). For example, only 33% of ELK1-binding regions had one or no ELK1-binding motifs compared with over 70% of promoter regions from the random data set. Thus, an important characteristic of binding regions identified for ELK1 and other ETS-domain transcription factors is the presence of multiple binding motifs.
Coordinate control of a set of basal transcription factors
To classify the ELK1 target genes and identify potential pathways that might be controlled by ELK1, we carried out gene function analysis of the ELK1 target gene list using DAVID (Dennis et al. 2003). A number of pathways were identified as being significantly over-represented amongst the ELK1 target genes. These include several associated with controlling gene expression, such as the ribosome, basal transcription factors, and the spliceosomal pathway. Between 20% and 43% of each category was represented amongst the ELK1 target gene list compared with only 6% of total possible genes found on the array (Fig. 3A; Supplemental Fig. S7; Supplemental Table S5). Furthermore, several Gene Ontology (GO) terms associated with transcriptional and translational control were identified amongst the molecular function (level 3) GO categories, and the terms transcription and translation also ranked highly amongst the biological processes (level 5) GO categories (Supplemental Table S5). The specificity of ELK1 linkage to pathways associated with gene expression is further emphasized by analysis of a series of ChIP-chip experiments with different transcription factors that failed to identify the ribosome, basal transcription factors, and the spliceosomal pathway as enriched GO categories (Supplemental Table S6). Moreover, although genes from these pathways were identified as direct targets of different transcription factors, the overlap with ELK1-binding events was low. For example, although NFY binds to 10 genes in the basal transcription factor category, only two of these are in common with those bound by ELK1 (Supplemental Table S6). Further inspection of the basal transcription machinery class targeted by ELK1 revealed that these represented a subset of basal transcription factors involved in initial promoter recognition (TBP, GTF2A1/2, and GTF2B) and a subset of TAFs that mainly form peripheral parts of the TAF complex distinct from the core (Fig. 3B; Supplemental Fig. S7) (Wright et al. 2006). Many of these TAFs also have a role in promoter recognition (Albright and Tjian, 2000). Thus, ELK1-binding regions are preferentially found in the promoters of a distinct subclass of basal transcription factors.
To establish a functional link between ELK1 and the regulation of target gene expression, we focused on the genes in the basal transcription factor class. Verification of ELK1 binding by qPCR confirmed that seven out of nine genes were bound by ELK1 in vivo (Fig. 3C). TBP had previously been identified as an ELK1 target and its expression was shown to be enhanced in response to JNK pathway signaling (Zhong et al. 2007). The majority of genes within the basal transcription machinery class were up-regulated upon treatment with the JNK pathway activator anisomycin (Supplemental Fig. S8). Importantly, ELK1 was required for anisomycin-mediated up-regulation of this set of genes, as all of these genes showed reductions in expression upon knockdown of ELK1 with siRNA (Fig. 3D,E). In contrast, the expression of several other genes encoding basal transcription factor components that were not identified as direct ELK1 targets by ChIP analysis (TAF4, GTF2E1, and GTF2F1) were unaffected by ELK1 depletion (Fig. 3E).
In addition to the basal transcription factors, many other genes exhibit changes in gene expression upon anisomycin treatment. We therefore compared the ELK1 FDR < 10 data set with gene clusters containing genes whose expression is up-regulated following anisomycin treatment (Hori et al. 2008). A significant overlap was observed between these two data sets; 8% of all genes in these clusters (37/422; Z-score 3.12; P-value 0.0018) showed direct ELK1 binding by ChIP-chip analysis. Thus, our data indicate that ELK1 likely participates more widely in the response to cellular stresses such as anisomycin treatment. To probe associations with other datasets, we focused on melanomas, as these are associated with hyperactivated Ras/ERK pathway signaling, which is known to result in activation of ELK1 transactivation capacity (for review, see Sharrocks 2002; Shaw and Saxton 2003). Recent metadata analysis identified a signature of overexpressed genes that are characteristic of melanoma cell lines (Hoek 2007). There was significant overlap (13%, 148/1112) between genes in this data set and genes identified as being directly bound by ELK1 (FDR < 10 data set). This was substantially more than was observed in comparison to a set of a 1000 randomized gene lists of the same size (8%, 89/1112; Z-score = 8.01). Subsets of the promoters we identified as bound by ELK1 may therefore represent targets of deregulation in disease states such as melanomas.
Specificity and redundancy of ELK1 binding
Due to the overlapping DNA-binding specificities of different ETS-domain proteins, it is possible that some redundancy of function might be observed. Indeed, such a scenario was demonstrated in a recent ChIP-chip study that suggested a large overlap in promoter binding by the family members ETS1, ELF1, and GABPA (37% with respect to ETS1; Hollenhorst et al. 2007). However, qPCR-ChIP studies on candidate ETS target genes were unable to detect substantial ELK1 occupancy at any of the sites tested, suggesting that ELK1 either bound to a different set of target genes or binding was excluded in Jurkat cells (Hollenhorst et al. 2007). ELK1 is expressed in Jurkat cells, suggesting that the lack of binding was not due to its absence in this cell type (Hollenhorst et al. 2004).
ELK1 is a member of the distinct TCF subfamily of ETS-domain proteins, suggesting that redundancy of promoter binding might be operational amongst members of this subfamily. To investigate this possibility, we tested the binding of the closely related protein, ELK4, to a subset of ELK1 target regions. Binding of ELK4 could be detected on all the tested regions (Fig. 4A). However, the relative ratio of binding changed from favoring ELK1 on the highest ranking ELK1-binding regions (e.g., RPS27A) to favoring ELK4 on lower ranking regions (e.g., BCL10). Testing of additional sites occupied by ELK1 revealed that they were invariably scored positive for ELK4 binding in ChIP assays (J Boros, A O'Donnell, ZA Odrowaz, and AD Sharrocks, unpubl.). Thus, among the TCF subfamily, redundancy of binding can be observed, although the degree of selectivity differs amongst ELK1 target genes.
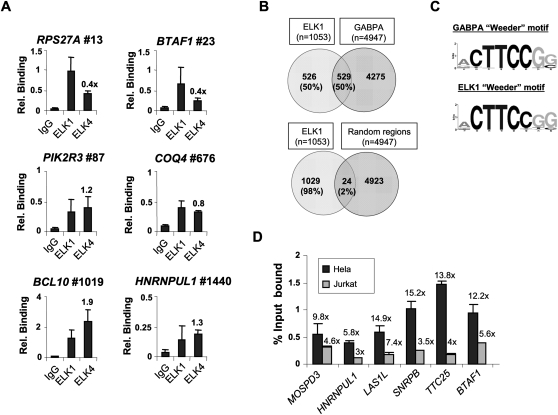
Figure 4.
Redundant promoter occupancy by ELK1 and other ETS-domain transcription factors. (A) qPCR-ChIP analysis of ELK1 and ELK4 binding to ELK1 target regions. The position of the binding region on the ranked target list is shown next to the gene name. Data are the average of duplicate samples and representative of three independent experiments. Relative binding ratios of ELK4 versus ELK1 are shown above the bars corresponding to ELK4. (B) Overlap in binding regions between ELK1 (FDR < 10) and GABPA ChIP-seq (Valouev et al. 2008) datasets. Comparisons were made with GABPA-binding regions that overlap with the tiled regions on the Affymetrix promoter arrays. The overlap between ELK1-binding regions and random equivalently sized promoter datasets is given for comparison. The total number of genes in each sector is shown, with percentages provided relative to the total number of ELK1 targets. (C) Sequence logo representation of the top 8-bp sequence identified by de novo searching the top 1000 binding regions identified as GABPA targets by ChIP-seq (±100 bp of midpoint) or the ELK1 FDR < 10 ChIP-chip datasets by Weeder. (D) qPCR-ChIP analysis of ELK1 binding to a set of promoters identified as binding to both ELK1 and GABPA. Data are presented as percent input bound in either HeLa (black bars) or Jurkat (gray bars) cells. Fold enrichment over IgG control precipitations is shown above each graph. Data are the average of duplicate samples from three independent experiments.
Recently, a chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) study identified the genomic binding regions for the ETS-domain transcription factor GABPA in Jurkat cells (Valouev et al. 2008). To establish whether we could detect more general overlaps in promoter occupancy between ELK1 and this more divergent ETS-domain protein, we compared the locations of the binding regions for ELK1 from our ChIP-chip data (FDR < 10) to the GABPA-binding regions in the ChIP-seq data set that overlap with the tiled regions on the promoter array we used. Unexpectedly, we found a large overlap, with over 50% of binding regions (with respect to ELK1) common to both factors (Fig. 4B; Supplemental Table S3). In keeping with this finding, de novo motif searching returned virtually identical top-scoring motifs, which closely resembled a generic ETS-domain transcription factor binding motif in both the ELK1 and GABPA datasets (Fig. 4C).
One explanation for the large overlap in promoter occupancy might be due to cell type specificity, with ELK1 binding predominating in HeLa cells and GABPA in Jurkat cells. Indeed, while GABPA levels are similar in both cell types, there are relatively much higher levels of ELK1 in HeLa cells (Supplemental Fig. S9A). We therefore compared ELK1 binding to a set of regions, identified as commonly bound by ELK1 and GABPA, in both HeLa and Jurkat cells. As expected, all of the regions were bound efficiently by ELK1 in HeLa cells (Fig. 4D, black bars). However, while significant promoter occupancy compared with background was observed, the overall magnitude of ELK1 binding was much lower in Jurkat cells (Fig. 4D, gray bars), consistent with the observation that GABPA occupies these promoters in this cell type. In contrast, a comparison of promoter occupancy by GABPA showed similar levels of binding in both HeLa and Jurkat cells, in keeping with its similar expression levels in these cell types (Supplemental Fig. S9B).
Together, these data demonstrate that a large proportion of promoters bound by ELK1 could also be occupied by alternative ETS transcription factors, suggesting a potential redundancy of function at these promoters. However, another set of promoters are bound by ELK1 independently from other ETS proteins such as GABPA, pointing to a different mode of regulation.
Combinatorial promoter targeting by ELK1 and SRF
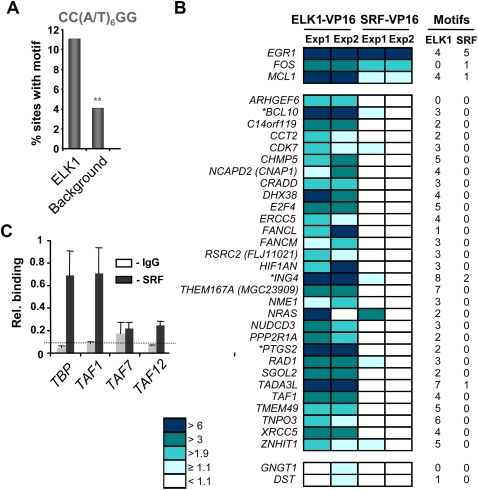
Many of the ELK1 target genes characterized to date contain composite binding sites for ELK1 (or other subfamily members) and for SRF (for review, see Sharrocks 2002; Shaw and Saxton 2003). However, a growing number of genes appear to be regulated by ELK1 independently from SRF. To probe for potential functional cooperativity with SRF, we examined our experimentally determined ELK1-binding regions for the presence of SRF CArG-box binding motifs. Using a stringent SRF consensus [CC(A/T6)GG] we detected statistically significant over-representation of potential binding sites in 11% of the ELK1-binding regions; this value increased to 34% with a less-stringent motif, which allows for a single non-A/T nucleotide anywhere in the core region (Fig. 5A; Supplemental Table S7). Thus, a co-occurrence of SRF-binding sites with ELK1-binding regions is potentially a common event, although a substantial number of regions do not appear to contain obvious consensus binding motifs for SRF.
Figure 5.
SRF-dependent and independent ELK1 target promoters. (A) Over-representation of the CC(W)6GG SRF-binding motif in the ELK1 FDR < 10 data set in comparison to a background data set. **P ≤ 1 × 10−4. (B) Reporter gene analysis in 293 cells of a panel of promoters containing ELK1-binding regions. Cells were cotransfected with the indicated reporter plasmids and plasmids encoding either ELK1–VP16 or SRF–VP16. Data are shown from two independent experiments (Exp1/2) and the levels of reporter activity are color coded according to fold activation by ELK1–VP16 or SRF–VP16. The numbers of ELK1 (nonredundant hexamers derived from CCGGAAGT motif) and SRF (CC(W)6GG) binding motifs in each promoter is shown on the right, and genes also found in the high confidence SRF ChIP-chip FDR < 1 data set are indicated by asterisks. (C) qPCR-ChIP analysis of SRF binding to the indicated promoters. The dotted line represents average binding across all IgG control precipitations. Data are the average of duplicate samples and representative of three independent experiments.
To experimentally test whether ELK1 and SRF coregulation of target genes is a common event, we investigated a series of ELK1 target promoters for activation by constitutively active versions of ELK1 or SRF (Fig. 5B; Supplemental Fig. S10; Supplemental Table 8). Activation was assessed in the context of promoter-reporter constructs, which contained ∼1 kb of sequence upstream from the TSS linked to a luciferase reporter gene. A total of 27/50 (54%) of the promoters were activated by Elk-VP16. In contrast, only 1/50 (2%) were consistently activated by SRF-VP16. This result was consistent with the lack of SRF-binding motifs found in these promoters (Fig. 5B). To further investigate the response to SRF-VP16, dose-response experiments were performed. While the ACTA2 (also known as SM-α actin) promoter was strongly activated, PTGS2 was only weakly activated and the E2F4 and CHMP5 promoters were barely affected (Supplemental Fig. S10C). These results provide additional validation of ELK1 target promoters identified by ChIP-chip analysis and suggest that only a small subset of these promoters are also targeted by SRF.
To provide more direct evidence for co-occurrence of SRF and ELK1-binding events, we tested a subset of four ELK1 target genes in the basal transcription factor cluster for SRF binding in vivo by ChIP analysis. While all of the targets tested were validated as ELK1 targets (see Fig. 3C), only two of these were also shown to be strong SRF targets (Fig. 5C), indicating that the ELK1 target promoters could be classified into two types: either bound by ELK1 alone or by ELK1 and SRF.
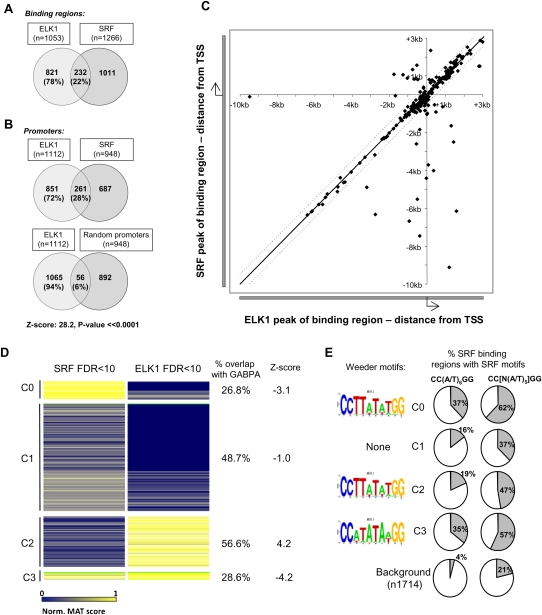
While this limited ChIP analysis demonstrated co-occurrence of SRF and ELK1-binding events on a subset of promoters, the wider significance of this finding was investigated by searching for in vivo SRF-binding events on a genome-wide scale. ChIP-chip analysis was performed for endogenous SRF in serum-starved HeLa cells to mirror the conditions we used for detecting ELK1-binding events. We focused on the most significant SRF-binding regions and used a cut-off FDR value of <10 to match the cutoff used to analyze the ELK1 data set. The SRF FDR < 10 data set contained a total of 1266 SRF-binding regions, corresponding to 948 nonredundant promoters (Supplemental Table 9). Comparison of the SRF FDR < 10 and ELK1 FDR < 10-binding regions demonstrated a substantial overlap with 22% of all ELK1-binding regions (232/1053) also bound by SRF (Fig. 6A). Moreover, comparison of nonredundant gene symbols associated with each set of binding data enabled us to assess co-occupancy by ELK1 and SRF at the promoter level. Again, a substantial amount of overlap was observed (28% of ELK1-bound promoters), which was considerably more than the 6% overlap of binding expected at random (Fig. 6B). Furthermore, when the locations of the ELK1 and SRF-binding events were plotted relative to their positions on the promoter, there was a very close colocalization, with 84% (196/234) of the regions being within 500 bp of each other, irrespective of their locations relative to the TSS (Fig. 6C). Thus, ELK1 and SRF represent a closely associating module found within promoters.
Figure 6.
Co-occupancy of promoters by SRF and ELK1 is a common event. (A,B) Overlap between binding events from the ELK1 FDR < 10 and SRF FDR < 10 datasets, when either binding regions (A) or co-occupancy of promoter regions (B) are considered. The overlap between ELK1-binding regions and random equivalently sized promoter datasets is given for comparison in B. The total number of genes in each sector is shown, with percentages also provided relative to the total number of ELK1 targets. (C) Scatter plot of the location of the peak binding positions of ELK1 FDR < 10 and SRF FDR < 10 binding regions relative to their closest and shared gene promoter TSS. The black and dotted lines indicate a distance of 0 and 500 nucleotides between the most significant probes in the ELK1- and SRF-binding region, respectively. (D) Cluster analysis of the ELK1 FDR < 10 and SRF FDR < 10 datasets (set to produce four clusters, C0–C3). The color variations between blue and yellow identify nonbound to strong binding events, respectively (as defined by normalized MAT scores). The percentage overlap of ELK1-binding regions in the four clusters that overlap with all GABPA ChIP-seq regions is shown on the right. The associated Z-score is based upon the chance of observing the same overlap using 1000 random sets of the ELK1 FDR < 10 binding regions compared with the observed overlap between the ELK1-binding regions in the cluster that overlap GABPA-binding regions. Clusters 0–3 contain 208, 1220, 577, and 86 binding regions, respectively. (E) SRF-like motifs identified in each of the four clusters (C0–C3) by Weeder analysis are shown on the left. The percentage occurrence of the CC(W)6GG and more degenerate CC[N(W)5]GG SRF binding motifs in each of the clusters (C0–C3) in comparison to a background data set is shown on the right.
To further partition our data, we used a novel clustering approach that uses MAT scores as surrogates for assessing the quality of the binding events where high scores correspond to high-probability binding events. Four clusters were identified (Fig. 6D): “SRF-specific” promoters (C0; high SRF binding and low ELK1 binding); “ELK1-specific” promoters (C2; high ELK1 binding and low SRF binding); promoters that are co-bound by ELK1 and SRF (C3; high SRF binding and high ELK1 binding); and an additional cluster, C1 that contained promoters with low-confidence binding events for both SRF and ELK1.
We searched for binding motifs in the SRF-binding regions contained in each of the four clusters; SRF motifs were readily identified using Weeder analysis in clusters C0, C2, and C3, but no obvious SRF-like motifs were identified in cluster C1 (Fig. 6E). These conclusions were further substantiated by searching for the occurrence of SRF-binding motifs in each of the clusters (Fig. 6E). Over-representation of SRF-like binding motifs was apparent in all clusters, but most noticeable in clusters C0 and C3. The latter observation is consistent with the designation of promoters in clusters C0 and C3 as high-confidence SRF target regions.
Cluster C3 appears to specify a high-confidence set of 86 promoters that are co-bound by ELK1 and SRF. This co-association could potentially provide a means for generating specificity of ELK1 binding compared with other ETS-domain transcription factors. To probe whether this is the case, we compared the overlap between each of the clusters with the binding events detected for GABPA by ChIP-seq analysis (Fig. 6D). Both clusters C3 and C0 (containing ELK1-binding regions associated with strong SRF-binding regions), showed significant under-representation of GABPA-binding events. The reciprocal was true for cluster C2, which contained weak SRF-binding events associated with strong ELK1-binding events, where overlap with GABPA-bound promoters was enriched. These comparisons therefore demonstrate that co-binding with SRF specifies a set of targets that are distinct from those that are bound by GABPA, whereas those regions bound by ELK1 in the absence of SRF tend to also be occupied by GABPA.
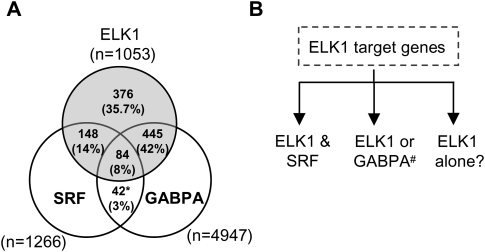
To extend these findings, we performed a three-way comparison of the overlaps between the regions identified by genome-wide ChIP studies as bound by ELK1 FDR < 10, GABPA, and the SRF FDR < 10 datasets (Fig. 7A; Supplemental Fig. S11). This demonstrated a clear partitioning of the data, where significant overlaps were seen between the ELK1- and SRF-binding regions, and between ELK1- and GABPA-binding regions, but little overlap was seen between the SRF- and GABPA-binding regions. This difference becomes even more apparent if a higher confidence SRF data set is used (FDR < 1), where 38% of SRF targets are co-bound by ELK1, while only 12% are co-bound by GABPA. Furthermore, if the ELK1 FDR < 10 data set is split into two subsets based on overlap with GABPA-binding events (Fig. 4B) and the sequence scanned using an SRF-binding matrix (represented by the consensus sequence CCATATAAGG), a clear over-representation of high-confidence matches emerges in the binding regions bound by ELK1 but not GABPA (P = 0.0016). To further underline the functional differences between the different classes of ELK1 target genes, we first tested the binding of GABPA to regions identified as bound by ELK1 and GABPA (but not SRF) or by ELK1 and SRF (but not GABPA). As predicted, GABPA occupancy was much higher on the first of these classes of target genes (Supplemental Fig. S12A). Conversely, we tested the effect of depleting SRF on the activities of genes in these two classes. ELK1 and SRF binding was first verified (Supplemental Fig. S12B). However, SRF depletion only affected the expression of genes identified as bound by ELK1 and SRF (but not GABPA) and did not affect those bound by ELK1 and GABPA (but not SRF) (Supplemental Fig. S12C–E). Finally, we compared our data to a study that identified SRF-dependent target genes through analyzing gene expression changes in srf knockout cells (Philippar et al. 2004), and found there was a statistically significant over-representation in the data set containing genes bound by ELK1 and SRF (but not GABPA) compared with the data set with genes bound by ELK1 and GABPA (but not SRF) (Z-score = 5.73). Together, these findings demonstrate that the SRF-ELK1 combination represents an important module that distinguishes a subset of ELK1 target genes from those that can also be occupied by a different ETS-domain protein, GABPA.
Figure 7.
Overlaps between promoter binding regions for ELK1, SRF, and GABPA. (A) Venn diagrams showing the overlap in target region binding between ELK1 FDR < 10, SRF FDR < 10, and GABPA ChIP-seq (Valouev et al. 2008) datasets. Comparisons were made with GABPA binding regions that overlap with the tiled regions on the Affymetrix promoter arrays. The total number of binding regions in each sector is shown, with percentages also provided relative to the total number of ELK1 targets. (B) Summary of the three classes of ELK1 target genes identified in this study, which are bound by (1) ELK1 and SRF, (2) ELK1 or GABPA, or (3) ELK1 alone. The # symbol indicates that redundancy with other ETS-domain proteins is likely.
Discussion
The ETS-domain transcription factor ELK1 plays a pivotal role in transducing extracellular signals into a transcriptional response through acting as a target of the MAP kinase signaling pathway (for review, see Sharrocks 2002; Shaw and Saxton 2003). However, as is the case with the majority of transcription factors, our knowledge of its regulatory potential is fragmentary due to the small number of target genes identified to date. Here, we have used ChIP-chip analysis to expand the number of confidently identified direct target genes to over 1000, although there are likely to be more bona fide targets amongst the lower confidence regions. Importantly, there are also a number of known targets that are not detected by our ChIP-chip analysis (Supplemental Table S1). The reasons for this are unclear and possibly reflect, in part, cell type-specific effects with several putative targets not bound by ELK1 in HeLa cells under the conditions analyzed. However, in other cases such as EGR1, qPCR-ChIP clearly identifies ELK1 targets in HeLa cells, but the same targets are completely negative in the ChIP-chip analysis (data not shown). False-negative rates can also be influenced by functional redundancy, where only partial promoter occupancy is observed due to competition with other family members. Thus, the high false-negative rate probably arises due to a combination of biological and technical reasons. Although we only interrogated a limited amount of the genome, the clustering of ELK1 target sites around the TSS suggests that the predominant mode of ELK1 function is within the context of the proximal promoter. However, we cannot rule out that there might be additional sites in distal enhancers, or within other noncoding regions in the genome. Furthermore, we cannot rule out that some of the signals at the proximal promoter are generated by looping from sites that are located long distances away, although the co-occurrence of ELK1-like binding motifs at the proximal promoter indicates that the majority of binding regions we have identified represent the actual location of ELK1 binding. Taken together, we therefore predict that there are likely to be significantly >1000 promoters directly targeted by ELK1.
The most prominent features of the ELK1 target gene network include components of the core gene regulation machinery: basal transcription complex components, spliceosome subunits, and ribosomal proteins. We confirmed a role for ELK1 in binding to and regulating the promoters of genes in the basal transcription-factor cluster. This is consistent with the recent finding that the TBP promoter is a target of ELK1 (Zhong et al. 2007), and demonstrates a more widespread role for ELK1 in coordinately regulating the basal transcription machinery. Previous studies have suggested that individual promoters can respond differently to basal transcription-factor components, such as TBP and GFT2B, in a concentration-dependent manner (George et al. 1995), thus providing a rationale for regulating a subset of these factors. Indeed, this appears to be the case for JUN regulation, where ELK1-mediated TBP up-regulation plays an important role (Zhong et al. 2007). It is also unclear why a subset of TAFs are coregulated, but as those targeted by ELK1 are generally not part of the stable core complex, it is possible that these might be subject to faster turnover in the cell (Wright et al. 2006). Furthermore, the basal transcription factors we have identified have roles that are focused on promoter recognition. This is most apparent for TFIID (which includes TBP), TFIIB, and TFIIA, which are the first three components that bind to TATA-box-containing promoters (the DAB complex) and initiate the subsequent recruitment of the rest of the basal machinery (Thomas and Chiang, 2006). As this is a key regulatory step, it makes sense to coordinately regulate all of the components from this complex. It is striking that none of the genes encoding proteins that are recruited subsequent to formation of the DAB complex, such as other basal transcription factors (TFIIE, TFIIF, and TFIIH) or RNA polymerase II subunits, are identified as ELK1 targets. In this study, we use anisomycin as an inducer that impacts on JNK pathway signaling, and we have shown this up-regulates a subset of basal transcription-factor components (Supplemental Fig. S8). Others have used oncogenic Ras, and have shown that this also acts through ELK1 to activate TBP (Zhong et al. 2007). Importantly, the JNK pathway is up-regulated by a range of cytokines and stress signals (Weston and Davis 2007), suggesting that our findings have more widespread biological implications for responses to various signaling events.
ELK1 is thought to work primarily through complexes with SRF. This paradigm is chiefly based on its regulation of FOS, where interactions with SRF are required for the recruitment of ELK1 to suboptimal DNA recognition sites (Gille et al. 1992; Treisman et al. 1992). Here, we have used ChIP-chip analysis to significantly extend our knowledge of this combinatorially acting transcription factor pair with over 22% of ELK1-binding regions being co-bound by SRF. Their close colocalization within promoters is consistent with there being a core regulatory module involving ELK1 and SRF (Fig. 6C). Despite this substantive overlap, there are over 77% of the ELK1-binding regions that do not appear to be occupied by SRF. Indeed, reporter gene analysis confirmed that a large proportion of the ELK1 target genes are not responsive to SRF. Thus, ELK1 occupancy of promoters occurs in combination with, or independently from, SRF. Interestingly, cluster analysis of SRF and ELK1-bound regions, and subsequent analysis of overlaps with the clusters with GABPA-binding regions, indicates that co-occupancy of ELK1 and SRF (Figs. 6D, 7A) defines a distinct category of target genes that are largely distinct from those that are bound by ELK1 or GABPA. Thus SRF appears to specify a unique set of ELK1 targets that are distinct from those that can be bound by both ELK1 and the different ETS-domain protein, GABPA.
The most closely related ETS-domain transcription-factor family member to ELK1 is the TCF subfamily member ELK4, which also works through complexes with SRF (for review, see Shaw and Saxton 2003). ELK4 binding is detectable at all of the targets we have tested for ELK1 binding (Fig. 4A; J Boros, A O'Donnell, and AD Sharrocks, unpubl.), suggesting functional redundancy within this subfamily, at least at the level of promoter occupancy. Indeed, reductions in levels of ELK4 lead to increased promoter binding by ELK1 (J Boros, A O'Donnell, and AD Sharrocks, unpubl.). Interestingly, the relative binding of ELK1 and ELK4 varies according to the bound promoter. This suggests that there are specificity mechanisms that differentiate between them, most likely at the level of DNA sequence recognition, which subtly differs in vitro (Shore and Sharrocks 1995). This differential promoter occupancy might also influence our ChIP results, as genes scoring highly for ELK4 binding are likely to then score less highly for ELK1 binding, and might therefore be excluded from our target gene list due to low enrichment values. Indeed, we see a reciprocal preferential binding of ELK1 at the high-scoring target sites identified by ChIP-chip (Fig. 4A). It is therefore likely that the number of target genes bound by the TCF subfamily is higher than that observed for ELK1 alone.
In addition to the apparent redundancy in binding between ELK1 and ELK4, there is also substantial overlap in promoter occupancy with another ETS-domain transcription factor GABPA (Valouev et al. 2008; Fig. 4B), suggesting a high degree of binding redundancy amongst ETS-domain family members. Indeed, our data are broadly consistent with the conclusions of a recent study that showed substantial overlap in promoter binding between the ETS-domain proteins ETS1, ELF1, and GABPA in Jurkat cells (Hollenhorst et al. 2007). However, the latter study was unable to show redundancy of binding with ELK1, most likely due to the lower levels of binding seen in Jurkat cells (see Fig. 4D). Interestingly, however, despite the differing cell types, HeLa and Jurkat, a large overlap in ELK1-binding regions is observed when comparing our data set to GABPA-bound regions identified by ChIP-seq (Valouev et al. 2008). Thus, depending on the relative abundance of ETS proteins (ELK1 is expressed at relatively high levels in HeLa, whereas GABPA levels are similar in both HeLa and Jurkat cells; Supplemental Fig. 9), a similar set of promoters can be targeted by different ETS-domain family members, thereby providing potentially different regulatory opportunities.
In summary, we have identified three modes of action of ELK1 (Fig. 7B): the first involves coordinate binding with a second transcription factor SRF; the second set of promoters can be bound by either ELK1 or a more divergent ETS-domain family member GABPA; lastly, there are a large number of promoters that are apparently uniquely bound by ELK1, although it is likely that additional combinatorial interactions might occur on this class of target promoters. These findings have important implications for thinking about how other transcription-factor families operate in redundant or combinatorial ways. Importantly, here, we have uncovered a previously unsuspected level of coordinate regulation of a subset of genes encoding basal transcriptional regulators and other core gene regulatory machines, including the splicing and translational machinery. As ELK1 and other ETS-domain proteins are targets of the Ras–ERK signaling pathway (Sharrocks 2001), these findings might have important implications for understanding tumourigenesis, as this would represent a convenient route for tumor cells to up-regulate their biosynthetic capacity.
Methods
Plasmid constructs
pRSV-Elk-1-VP16 (pAS348) encoding full-length ELK1 fused to residues 410–490 of VP16 C-terminal sequence (Price et al. 1995), pMLV-SRF-VP16 (kindly provided by Richard Treisman, Cancer Research UK, London), pGL3-SM α-actin-Luc/pAS2268 (containing the rat ACTA2 promoter sequences −713 to +51 bp; kindly provided by S. Phan; Hu et al. 2003) and pRL encoding Renilla luciferase (Promega), have been described previously. All reporter vectors were generated by SwitchGear Genomics, and contain ∼1 kb of promoter fragment cloned into pGL3 vector (Promega) (Supplemental Table S8).
Tissue culture, cell transfection, reporter gene assays, RT-PCR, and RNA interference
HeLa and 293 cells were grown in DMEM supplemented with 10% fetal bovine serum. Jurkat cells were grown in RPMI supplemented with 10% fetal bovine serum. Where indicated, cells were serum starved for 24 h and either analyzed immediately or stimulated with anisomycin (250 ng/mL) for an additional time period.
For luciferase assays, 293T cells were plated on 96-well plates (1 × 104 cells/well). Typically, 50 ng of reporter plasmid and 5 ng of pRL were cotransfected with 10 ng of expression plasmids. Transfections were carried out using PEI reagent (polyethylenimine, Polysciences, Inc.). The Dual-Glo Luciferase Reporter Assay System (Promega) was used according to the supplier protocol. The relative luciferase activity was measured as a ratio of firefly luciferase to control Renilla luciferase of ELK1-VP16 or SRF-VP16 activated promoters and presented relative to the values of each individual basal promoter activity (cells cotransfected with pcDNA3.1). The threshold for activated promoters was defined as three standard deviations above the mean ratio of the negative controls GNGT1 and DST (which were not identified in the ELK1 FDR > 10 data set) and the negative controls R1–R4 provided by SwitchGear genomics).
Real time RT-PCR was carried out as described previously (O'Donnell et al. 2008). The primer-pairs used for RT-PCR experiments are listed in Supplemental Table S10.
siRNA against ELK1 and a matched GAPDH control were constructed by the Silencer siRNA construction kit (Ambion). For ELK1 silencing, a mix of two siRNA was used, using the templates ADS1926/11927 (5′-AATTCAAGCTGGTGGATGCAGCCTGTCTC-3′, 5′-AACTGCATCCACCAGCTTGAAcctgtctc-3′) and ADS1928/1929 (5′-AAGGCAATGGCCACATCATCTCCTGTCTC-3′, 5′-AAAGATGATGTGGCCATTGCCCCTGTCTC-3′). To carry out RNA interference (RNAi), cells were transfected with 50 nM siRNA using oligofectamine (Invitrogen) in Optimem (Invitrogen). Transfection was repeated 24 h later. Cells were left for 24 h and then treated with anisomycin (250 ng/mL) where required. For SRF knockdown experiments, HeLa cells were transfected with 50 nM siRNA (siSRF (h): SantaCruz; siGAPDH (h): Dharmacon) using Lipofectamine RNAiMax in a mixture of 90% DMEM only and 10% OptiMEM. Cells were cultured in serum-free DMEM for 48 h post-transfection, followed by a 30-min stimulation with 10% FBS, and harvesting.
Western blot analysis
Western blotting was carried out with the primary antibodies; ELK1 (Epitomics), phosho-JNK1/2 (Cell Signaling), JNK2 (BD Pharminogen), GABPA (Santa Cruz), and GAPDH (Abcam). The proteins were detected by chemiluminescence with SuperSignal West Dura Substrate (Pierce) and visualized with a Fluor-S MultiImager (Bio-Rad).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays using control IgG (Upstate) or antisera specific to ELK1 (Epitomics), ELK4 (Santa Cruz), GABPA (Santa Cruz), and SRF (Santa Cruz) were performed as described previously (O'Donnell et al. 2008) except that cells were serum starved for 24 h before harvesting. Bound promoters were detected by quantitative PCR (using primers listed in Supplemental Table S10), at least in duplicate, from at least two independent experiments using Quantitect SYBR green PCR reagent (Qiagen). Results were analyzed with Rotorgene 6.0 software (Corbett Research) relative to input using the standard curve method.
ChIP-chip assays
Immunoprecipitated DNA was amplified using the random amplification method as described previously (Bohlander et al. 1992 and modified according to DeRisi and Rando labs; Kim et al. 2004) with minor modifications. A total of 40 μL of ChIP-DNA (∼20 ng) was taken for each random priming reaction. Fragmented and labeled DNA targets (7.5 μg) were hybridized to Affymetrix Human Promoter 1.0R arrays according to the manufacturers' instructions. The hybridized arrays were then processed using fluidics protocol FS450_001 and scanned using a GeneChip Scanner 3000 7G.
Bioinformatics analysis.
Details of bioinformatic analysis of the ChIP-chip data, the derivation of background datasets, and subsequent motif searching can be found in the supplemental information.
Statistical analysis
Fisher's exact χ2 for 2 × 2 contingency tables (http://www.quantitativeskills.com/sisa/) was applied to determine whether ELK1-binding region sequences where enriched with ELK1, SRF, or ETS-factor core sequences compared with the other datasets. Statistical analysis for qRT-PCR studies and luciferase assays were performed using paired, 2-tailed Student's t-test. The error bars in all graphs represent standard deviation.
Acknowledgments
We thank Anne Clancy, Karren Palmer, and Leanne Wardleworth for excellent technical assistance; Andy Hayes in the core microarray facility; Casey Bergman and Stefan Roberts and members of our laboratory for comments on the manuscript and stimulating discussions; Richard Treisman for reagents; Andrew Smith (Cold Spring Harbor Laboratory) for help, advice, and access to DME2; and Takashi Kondo and Takeshi Hori for access to anisomycin microarray cluster analysis gene lists. This work was supported by grants from the Wellcome Trust, the AICR, Cancer Research UK and a Royal Society-Wolfson award to A.D.S.
Footnotes
[Supplemental material is available online at http://www.genome.org. The ChIP-chip data from this study have been submitted to ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae) under accession nos. E-MEXP-1527 and E-MEXP-2084.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093047.109.
References
- Albright SR, Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Bohlander SK, Espinosa R, III, Le Beau MM, Rowley JD, Díaz MO. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics. 1992;13:1322–1324. doi: 10.1016/0888-7543(92)90057-y. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes & Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Fitzgerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–1574. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CP, Lira-DeVito LM, Wampler SL, Kadonaga JT. A spectrum of mechanisms for the assembly of the RNA polymerase II transcription preinitiation complex. Mol Cell Biol. 1995;15:1049–1059. doi: 10.1128/mcb.15.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille H, Sharrocks AD, Shaw PE. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- Hassler M, Richmond TJ. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 2001;20:3018–3028. doi: 10.1093/emboj/20.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS. DNA microarray analyses of melanoma gene expression: A decade in the mines. Pigment Cell Res. 2007;20:466–484. doi: 10.1111/j.1600-0749.2007.00412.x. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes & Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Kondo T, Tabuchi Y, Takasaki I, Zhao QL, Kanamori M, Yasuda T, Kimura T. Molecular mechanism of apoptosis and gene expressions in human lymphoma U937 cells treated with anisomycin. Chem Biol Interact. 2008;172:125–140. doi: 10.1016/j.cbi.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Hu B, Wu Z, Phan SH. Smad3 mediates transforming growth factor-beta-induced alpha-smooth muscle actin expression. Am J Respir Cell Mol Biol. 2003;29:397–404. doi: 10.1165/rcmb.2003-0063OC. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol Cell Biol. 2009;29:172–186. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell A, Yang SH, Sharrocks AD. MAP kinase-mediated c-fos regulation relies on a histone acetylation relay switch. Mol Cell. 2008;29:780–785. doi: 10.1016/j.molcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U, Schratt G, Dieterich C, Müller JM, Galgóczy P, Engel FB, Keating MT, Gertler F, Schüle R, Vingron M, et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Price MA, Rogers AE, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. Complexities in ETS-domain transcription factor function and regulation: Lessons from the TCF (ternary complex factor) subfamily. The Colworth Medal Lecture. Biochem Soc Trans. 2002;30:1–9. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- Shaw PE, Saxton J. Ternary complex factors: Prime nuclear targets for mitogen-activated protein kinases. Int J Biochem Cell Biol. 2003;35:1210–1226. doi: 10.1016/s1357-2725(03)00031-1. [DOI] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol Cell Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The ETS-domain transcription factors Elk-1 and SAP-1 exhibit differential DNA binding specificities. Nucleic Acids Res. 1995;23:4698–4706. doi: 10.1093/nar/23.22.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19:142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Wright KJ, Marr MT, II, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD, Whitmarsh AJ. Transcriptional regulation by the MAP kinase signaling cascades. Gene. 2003;320:3–21. doi: 10.1016/s0378-1119(03)00816-3. [DOI] [PubMed] [Google Scholar]
- Zhong S, Fromm J, Johnson DL. TBP is differentially regulated by c-Jun N-terminal kinase 1 (JNK1) and JNK2 through Elk-1, controlling c-Jun expression and cell proliferation. Mol Cell Biol. 2007;27:54–64. doi: 10.1128/MCB.01365-06. [DOI] [PMC free article] [PubMed] [Google Scholar]