Abstract
SVA elements represent the youngest family of hominid non-LTR retrotransposons, which alter the human genome continuously. They stand out due to their organization as composite repetitive elements. To draw conclusions on the assembly process that led to the current organization of SVA elements and on their transcriptional regulation, we initiated our study by assessing differences in structures of the 116 SVA elements located on human chromosome 19. We classified SVA elements into seven structural variants, including novel variants like 3′-truncated elements and elements with 5′-flanking sequence transductions. We established a genome-wide inventory of 5′-transduced SVA elements encompassing ∼8% of all human SVA elements. The diversity of 5′ transduction events found indicates transcriptional control of their SVA source elements by a multitude of external cellular promoters in germ cells in the course of their evolution and suggests that SVA elements might be capable of acquiring 5′ promoter sequences. Our data indicate that SVA-mediated 5′ transduction events involve alternative RNA splicing at cryptic splice sites. We analyzed one remarkably successful human-specific SVA 5′ transduction group in detail because it includes at least 32% of all SVA subfamily F members. An ancient retrotransposition event brought an SVA insertion under transcriptional control of the MAST2 gene promoter, giving rise to the primal source element of this group. Members of this group are currently transcribed. Here we show that SVA-mediated 5′ transduction events lead to structural diversity of SVA elements and represent a novel source of genomic rearrangements contributing to genomic diversity.
SVA elements are nonautonomous non–long terminal repeat (non-LTR) retrotransposons that originated <25 million years ago (Mya) and represent the youngest retrotransposon family in primates. So far their copy number has increased to roughly 3000 in the human genome (Ostertag et al. 2003; Wang et al. 2005). Their ongoing activity in humans sporadically causes disease by randomly inserting into genes (for review, see Belancio et al. 2008). Considering the number of disease-causing insertions relative to their overall copy number, SVA elements are thought to represent a highly active retrotransposon family in humans (Ostertag et al. 2003; Wang et al. 2005). SVA elements stand out from the group of human non-LTR retrotransposons due to their composite structure, including modules derived from other primate repetitive elements. Starting at the 5′-end, a full-length SVA element is composed of a (CCCTCT)n hexamer repeat region; an Alu-like region consisting of three antisense Alu fragments adjacent to an additional sequence of unknown origin; a variable number of tandem repeats (VNTR) region, which is made up of copies of a 36- to 42-bp sequence or of a 49- to 51-bp sequence (Ostertag et al. 2003); and a short interspersed element of retroviral origin (SINE-R) region. The latter is derived from the 3′-end of the env gene and the 3′-LTR of the endogenous retrovirus HERVK-10 (Ono et al. 1987). A poly(A) tail is positioned downstream of the predicted conserved polyadenylation signal AATAAA (Ostertag et al. 2003).
The origin of SVA elements can be traced back to the beginnings of hominid primate evolution, only about 18–25 Mya. Their very young evolutionary age represents a unique opportunity to study the entire evolutionary history of a human retrotransposon. In addition, SVA elements may be valuable as markers for primate or human phylogenetic and population genetic studies, as has been the case for Alu elements (Bamshad et al. 2003; Watkins et al. 2003; Xing et al. 2007).
In order to draw conclusions on both the assembly process that led to the formation of SVA elements and the mechanism of their transcriptional regulation, we started out by assessing the different structural variants of SVA elements on a single chromosome. Thorough investigation of a single chromosome allows detailed structural inspection of individual elements. We chose chromosome 19 for the following reasons: (1) it is one of the best characterized single human chromosomes with regard to DNA sequence and biology (Grimwood et al. 2004), and (2) with an average density of one element per 0.62 Mb, it was reported to have the highest density of SVA insertions among human chromosomes (Wang et al. 2005). (3) Nearly 55% of this chromosome consists of repetitive elements (Grimwood et al. 2004), whereas the genome average is only 44.8% (Lander et al. 2001). This difference is due mainly to an unusually high content of short interspersed elements (SINEs) on this chromosome. Therefore, we reasoned that representatives of all human SVA variants should also be present on chromosome 19.
The comprehensive analysis of chromosome 19 uncovered 116 SVA elements that could be assorted into seven types of structural variants. Novel variants identified were 3′-truncated elements and a significant number of SVA elements with 5′-flanking sequence transductions. Our genome-wide analysis uncovered 220 5′-transducing SVA elements that originated from 93 diverse genomic source loci. The 93 resultant 5′ transduction groups are composed of one to 84 members. The organization of these elements demonstrates that SVA elements can recruit external heterologous promoters for their own transcriptional regulation, which in turn might affect mobilization frequencies. In a number of cases, 5′-transduced sequences are constituted by spliced cellular mRNAs. Therefore, SVA-mediated 5′ transduction represents a further mechanism of retrotransposon-mediated exon shuffling. SVA-driven transduction of 5′-flanking DNA is likely to be one mechanism of genome evolution via increasing genome plasticity and facilitating new combinations of coding and regulatory sequences. Acquisition of new 5′ sequences may also be a common theme in SVA evolution, with the objective of identifying new genomic components whose incorporation increases the efficiency of SVA mobilization.
Results
Chromosome 19 harbors seven structural variants of SVA elements
A comprehensive analysis of chromosome 19 of the human genome draft sequence (hg 17; May 2004) recovered 116 insertions that are members of the SVA family of non-LTR retrotransposons accounting for roughly 179 kb of genomic sequence. Except for one SVA element, all remaining copies were characterized by hallmarks of LINE-1–mediated retrotransposition: flanking target site duplications (TSDs) of 4–18 bp (one exception, 42 bp), a variable-length poly(A) tail of 2–72 nucleotides following putative polyadenylation signals, and a consensus target sequence AA↑TTTT. Poly(A) tails were composed of patterned repeats (Szak et al. 2002) in 60 cases, while homogenous poly(A) tails were found in 56 elements. Detailed features of each SVA insertion on chromosome 19 are listed in Supplemental Table 1.
Based on their SINE-R sequences (Wang et al. 2005), we were able to allocate 105 SVA elements to subfamilies A to F. Eleven elements could not be assigned unambiguously to any SVA subfamily because 3′ truncations of the SINE-R regions were too extensive or the SINE-R sequence was missing entirely. SVA_D represents the largest subfamily, accounting for about 44.8% of SVA elements on chromosome 19. The second and third largest subfamilies B and C comprise roughly 14.7 and roughly 12%, respectively. Human-specific subfamilies E and F are represented by six (5.1%) and 13 (11.2%) members, respectively.
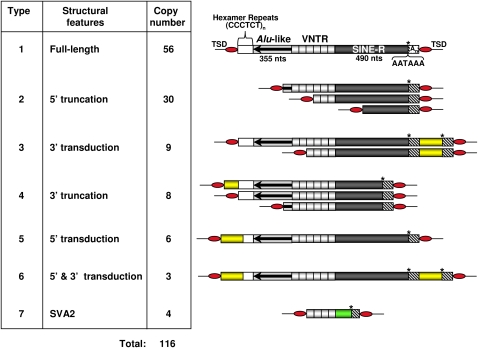
Due to differences in their structural organization, the 116 SVA elements can be grouped into seven types of structural variants (Fig. 1). The majority of elements are full-length (type 1, 48%) and 5′-truncated (type 2, 26%). Most of the 5′ truncation sites (19/30) are localized within VNTR sequences. Only in five and seven cases did 5′ truncation occur within the Alu-like and SINE-R regions, respectively. We also identified nine SVA insertions that display classical 3′ transduction events, and categorized them as type 3 elements. While these three SVA variants have been described earlier (Wang et al. 2005; Xing et al. 2006), we also found as yet unreported 3′-truncated elements (type 4) that feature the hallmarks of L1-mediated retrotransposition (Supplemental Table 1). Two out of the eight 3′-truncated SVA elements identified are also truncated at their 5′-ends. A genome-wide analysis of 3′-truncated SVAs is the subject of a separate study (A. Damert and G.G. Schumann, in prep.). Chromosome 19 also harbors four SVA2 elements (Fig. 1, type 7) (Jurka et al. 2005; Han et al. 2007), which are composed of VNTR units fused to a DNA-sequence that has no sequence homology with any known component of SVA elements (Supplemental Fig. 1). The SVA2 elements identified display hallmarks of L1-mediated retrotransposition: They are flanked by 5- to 16-bp TSDs, and their 3′-ends are terminated by poly(A) tails of 6–14 bp.
Figure 1.
Structural variants of the SVA elements on human chromosome 19. Full-length SVA elements are composed of (CCCTCT)n hexamer simple repeats, an Alu-like region, a VNTR region, and a SINE-R sequence (type 1). Structures of 5′-truncated (type 2), 3′-transducing (type 3), 3′-truncated (type 4), 5′-transducing (type 5), 5′ as well as 3′-transducing (type 6), and SVA2 elements (type 7) are depicted. 5′- and 3′-transduced sequences are indicated as yellow boxes. Heterologous non-SVA sequences in SVA2 elements are illustrated as green box. Asterisks indicate polyadenylation signals. An, polyadenylate.
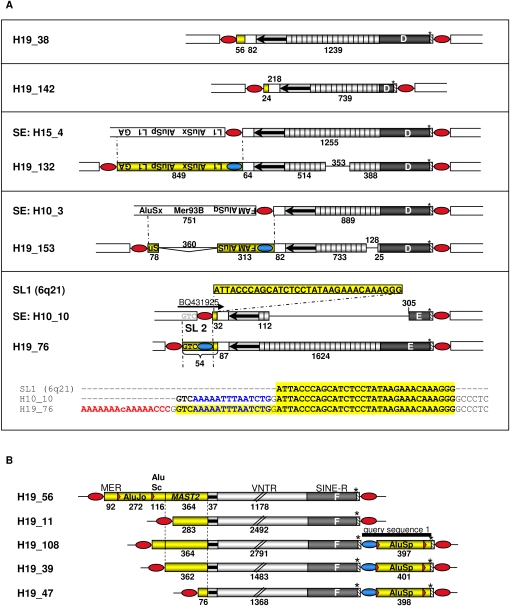
We also located SVA elements whose 5′ TSDs adjoined to additional genomic sequences bordering on the 5′-end of the original SVA sequence (Fig. 1; type 5 and 6 variants). The structures of these variants are consistent with a previous hypothesis suggesting that at least a fraction of SVA elements may be transcribed from external promoters (Wang et al. 2005): If a cellular promoter that is localized upstream of a full-length or 5′-truncated non-LTR retrotransposon reads into the element, the consequence would be a new 5′-end for the mRNA encoding the element. Retrotransposition of such an element will result in a new genomic copy that could contain additional genomic sequences at its 5′-end that originally adjoined the 5′-end of the source element. Chromosome 19 harbors 10 SVA insertions with such 5′-transduced sequences whose lengths ranged from 24 to 849 bp (Fig. 2).
Figure 2.
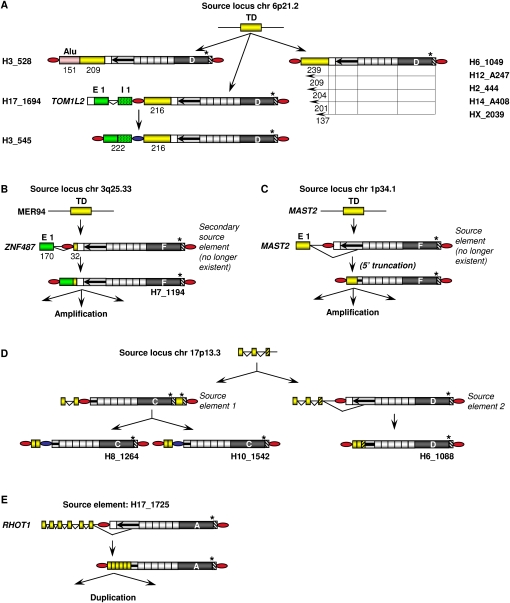
5′-Transducing SVA elements on chromosome 19 and their source elements. (A) Full-length elements and their 5′-transduced sequences. Source elements (SEs) of SVAs H19_132, H19_153, and H19_76 could be identified and were aligned with their respective descendant SVA copy. Cryptic splice sites located within oppositely oriented Alu sequences (Kreahling and Graveley 2004) of the 5′-transducing pre-mRNA of source element H10_3 led to alternative splicing, which resulted in a 360-bp deletion within the 5′ transduction of H19_153. Consecutive and nested SVA-mediated 5′ transduction events generated SVA H19_76. The primary 32-bp 5′ transduction in source element H10_10 originated from the SVA-free source locus (SL) on chromosome 6q21. Transcriptional control of this source element by an external promoter is indicated by EST BQ431925 covering the junction between H10_10 SVA element and the 5′-flanking genomic sequence. Subsequent transduction of a GTC trinucleotide and of the H10_10 5′ TSD during retrotransposition resulted in the formation of the 5′-transducing H19_76 element. 5′- and 3′-transduced sequences are shown as yellow boxes. Numbers indicate lengths of VNTR regions and 5′ transductions in nucleotides. Blue ovals, TSDs of source elements; red ovals, TSDs flanking chromosome 19 SVAs; GA, low complexity GA-rich sequence; TA, (TTAAA)n repeats; MER93B, endogenous retrovirus repetitive sequence; FAM and FLAM_C, Alu monomer from primates (Jurka et al. 2005). (B) Members of the MAST2 5′ transduction group. Identical junctions between the 5′-ends of the truncated Alu-like region and the shared 5′-transduced MAST2 sequences indicate that these SVAs originated from a common source element. MER, DNA transposon MER115; black bar, query sequence 1.
SVA-mediated 5′ transduction events are widespread and their source elements are still present in the human genome
Three out of five 5′-transducing full-length SVA elements on chromosome 19 could be traced back to their source elements (Fig. 2A). However, analysis of the genomic environment of these source elements did not uncover any putative external promoter sequences that might be responsible for the 5′ transductions. SVA H19_76 is the result of two consecutive 5′-transducing retrotransposition events with H10_10 (Fig. 2A) being the source element of H19_76. Identification of an expressed sequence tag (EST) covering the 5′ junction of H10_10 suggests that this SVA element is part of an actively transcribed region and might therefore serve currently as a source element for retrotransposition events.
Five 5′-transducing SVAs on chromosome 19 (Fig. 2B) must have originated from the same source element because (1) junctions between the 5′-ends of the truncated Alu-like region and the 5′ transductions are identical, and (2) 5′-transduced sequences are overlapping and can be traced back to the same source locus. 5′ Transductions comprise 76–364 bp of exon 1 of the MAST2 (microtubule-associated serine/threonine kinase 2; Entrez GeneID 23139) gene. SVA elements that originated from this ancestral, MAST2 sequence-transducing source element are defined as members of the MAST2 5′ transduction group. 5′-Ends of the MAST2 fragments of H19_56 and H19_108 are fused to identical AluSc sequences, indicating that both SVA insertions are derived from the same source element. Three members of the MAST2 5′ transduction group on chromosome 19 are characterized by identical 3′-transduced AluSp sequences in addition to their MAST2 5′ transductions (Fig. 2B).
Genome-wide analysis for SVA 5′ transductions
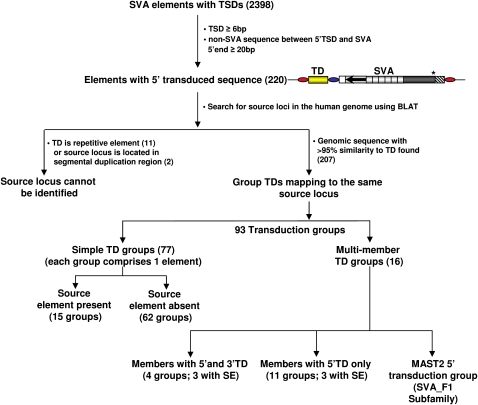
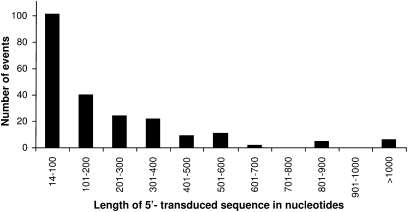
The identification of 10 5′-transducing SVA elements on chromosome 19 alone suggested that transcription from external promoters might be a more widespread phenomenon. As the mechanism of transcriptional regulation of SVA elements is unclear to date and transcription is an essential prerequisite for retrotransposition, a genome-wide analysis of SVA 5′ transductions would provide important insights into the regulation of SVA expansion. To establish a complete inventory of SVA 5′ transduction events, we screened the human genome reference sequence (hg17, May 2004) using the RepeatMasker pre-masked genome annotations. We identified 2398 SVA elements with TSDs comprising ≥6 bp, and 220 of them were found to carry additional sequences at their 5′-ends (Fig. 3), covering 49.1 kb of genomic sequences. The lengths of 5′ transductions range from 14–2161 bp, with an average of 223 bp. Almost 50% of all 5′-transduced sequences cover up to 100 bp (Fig. 4). In 31 cases, the final length of the 5′ transduction could be traced back to two consecutive transduction events. Detailed information on each SVA 5′ transduction identified is provided in Supplemental Table 2.
Figure 3.
Schematic diagram depicting the identification of SVA 5′ transductions and their source elements. Yellow box, 5′-transduced sequence; blue oval, TSD that originally flanked the source element and is now part of the 5′ transduction; red ovals, TSDs flanking the SVA element; TD, transduction; SE, source element.
Figure 4.
Length distribution of the 5′ transduction events identified. The number of SVA-mediated 5′ transduction events in the human genome in each 100-bp size interval is shown.
We were able to localize the source loci for 207 out of the 220 5′-transduced sequences. Source loci were either SVA-free or they were found to be localized upstream of the 5′ TSD of an SVA source element. Transduced sequences of 77 SVA elements could be uniquely assigned to their respective source locus (simple TD groups) (Fig. 3). Diverse SVA elements whose transductions could be mapped back to the same source locus were assigned to a single multimember transduction group.
Multimember transduction groups, sequential 5′ transduction events and the acquisition of spliced RNA sequences
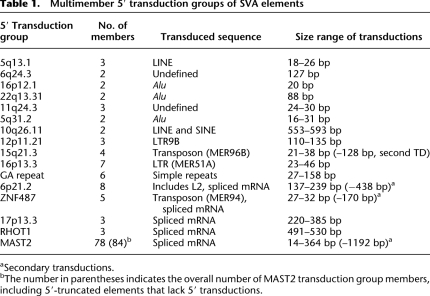
Four out of the 16 multimember transduction groups (Table 1) are characterized by both 5′ and 3′ transductions originating from the same source element (5q13.1, 6q24.3, 16p12.1, and 22q13.31). We found 11 multimember transduction groups genome-wide carrying exclusively 5′ transductions. They comprise two to eight individual SVA insertions each (Table 1). The MAST2 transduction group stands out because of its exceptionally high number of 78 elements. Transduced sequences of the majority of groups are repetitive and/or mobile DNA sequences like transposons and retroelements (Table 1).
Table 1.
Multimember 5′ transduction groups of SVA elements
aSecondary transductions.
bThe number in parentheses indicates the overall number of MAST2 transduction group members, including 5′-truncated elements that lack 5′ transductions.
Transduction groups 6p21.2 and ZNF 487 (Fig. 5A,B) are distinguished by a primary transduction event that was followed by a secondary transduction event in which spliced RNA sequences were acquired.
Figure 5.
Multimember transduction groups with sequential 5′ transductions involving splicing of 5′-transduced RNA sequences. Transduction groups with primary transduction events followed by secondary transductions of spliced host RNAs (A,B) and those with primary transductions of spliced host RNA sequences (C–E) are shown. Splice acceptor sites can be localized either within the SVA 5′ flanking intronic sequence, within primary transductions, or within the SVA Alu-like region. Exons and introns are not drawn to scale. Yellow boxes, primary transductions; green and pink boxes, secondary transductions; E1, exon 1; I1, intron 1; dotted green box, intronic sequence included in 5′ transduction; red ovals, TSDs; blue ovals, TSD sequence of the source element present in the 5′-transducing SVA; TD, transduction.
The 6p21.2 transduction group comprises six members that carry exclusively 137- to 239-bp fragments of the 6p21.2 source locus (Fig. 5A). They could be direct descendants of the source element once existing at this site. Alternatively, any given transduction group member could have served as a source element of any 6p21.2 group member whose 5′ transduction is equal in length or a 5′-truncated version of its own. Two members of this group (H3_528, H3_545) acquired additional nonhomologous sequences by secondary 5′ transduction events. H3_528 transduced an Alu sequence from an SVA-free source locus on chromosome X. The secondary transduction of H3_545 could be traced back to SVA H17_1694 that had retrotransposed into intron 1 of the TOM1L2 (target of myb1-like2 [chicken]) gene. Transcription of H17_1694 from the TOM1L2 promoter, subsequent splicing of TOM1L2 exon 1 to a cryptic splice acceptor site upstream of the SVA integration site, and retrotransposition of the resulting processed RNA led to the formation of SVA insertion H3_545 (Fig. 5A).
The primary source locus of the ZNF487 transduction group is a fragment of the MER94 transposon on chromosome 3q25.33. The transduced MER94 sequence in an extinct secondary source element must then have provided the splice acceptor site for the fusion to exon 1 of the ZNF487 gene (Fig. 5B). H7_1194 is carrying the complete exon 1 sequence of the ZNF487 gene, while the remaining three group members include only 5′-truncated versions of the exon. Because the secondary source element was under transcriptional control of the testis-specific ZNF487 promoter, simultaneous presence of the L1-encoded protein machinery in germ cells was assured. This would have favored the amplification of this SVA transduction group in the germ line.
In multimember transduction group 17p13.3, splicing was involved in the primary transduction event. Transduced sequences of the three members of this group were mapped back to the same source locus on chromosome 17p13.3. However, examination of the junctions between the 5′ transduction and the 5′-end of the SVA elements and determination of their subfamily affiliation revealed that the three elements must have originated from two different source elements (Fig. 5D). 5′-Transducing SVA subfamily C members (H8_1264, H10_1542) resulted from transcription and retrotransposition of a 5′-truncated source element that is still present in the chimpanzee genome. After the human–chimpanzee divergence, an SVA_D element integrated further downstream into the same transcription unit. Cotranscription of the source locus with the SVA_D insertion, splicing to a cryptic splice acceptor site located within the SVA Alu-like region, and subsequent retrotransposition of the chimeric RNA led to the formation of SVA H6_1088 (Fig. 5D; Supplemental Fig. 2). MAST2 and RHOT1 transduction groups (Fig. 5C,E) are two additional examples for splicing of an exonic donor to an acceptor site localized within the Alu-like region of the SVA element. There are three genomic SVA elements that transduced spliced RHOT1 (ras homolog gene family, member T1) sequences whose source element is still present. It has been suggested that two of these were generated by duplication and do not represent individual retrotransposition events (Hancks et al. 2009).
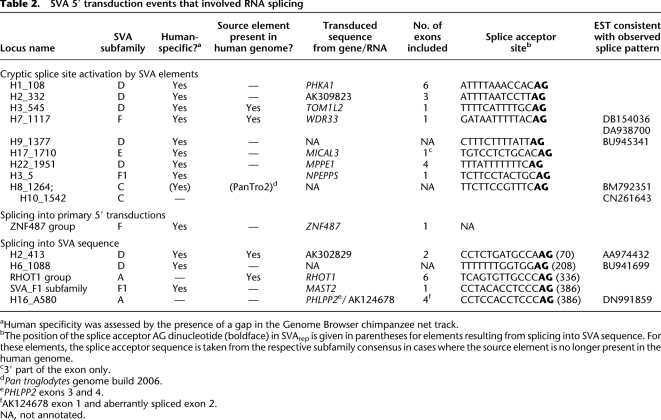
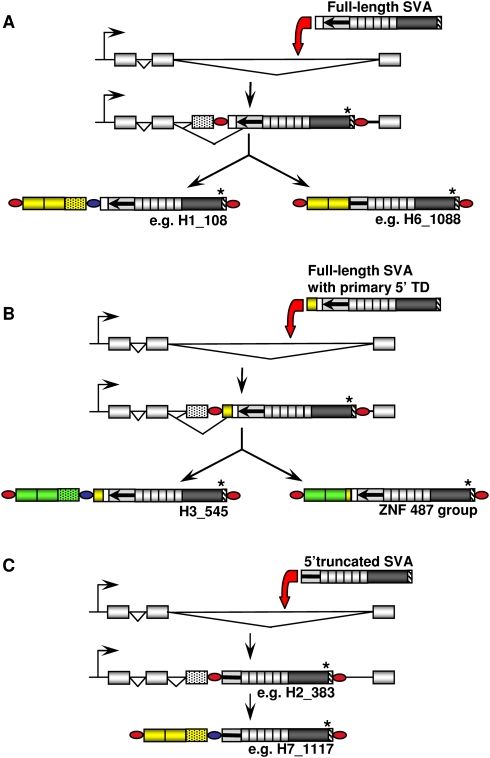
In total, we found 15 5′ transduction groups whose transduced sequences resulted from splicing of an exonic splice donor to a cryptic splice acceptor that is localized either upstream of an SVA 5′-end, within a primary 5′ transduction or within an SVA sequence (Table 2). Source elements of four of these 15 transduction groups are still present in the human genome. Different scenarios of SVA-mediated 5′ transduction events involving pre-mRNA splicing into SVA sequences and into sequences upstream of SVA sequences are depicted in Figure 6. In all three scenarios, the consequences of retrotransposition are 3′ truncation, 3′ modification, and translocation of host-derived RNA sequences that comprised coding sequences in several cases. Thus, SVA 5′ transduction represents an additional mechanism for exon shuffling.
Table 2.
SVA 5′ transduction events that involved RNA splicing
aHuman specificity was assessed by the presence of a gap in the Genome Browser chimpanzee net track.
bThe position of the splice acceptor AG dinucleotide (boldface) in SVArep is given in parentheses for elements resulting from splicing into SVA sequence. For these elements, the splice acceptor sequence is taken from the respective subfamily consensus in cases where the source element is no longer present in the human genome.
c3′ part of the exon only.
dPan troglodytes genome build 2006.
ePHLPP2 exons 3 and 4.
fAK124678 exon 1 and aberrantly spliced exon 2.
NA, not annotated.
Figure 6.
SVA 5′ transduction events involving pre-mRNA splicing. (A) After insertion of a full-length SVA element into an intron, cryptic splice acceptor sites upstream of the element can be activated. The retrotransposed copy includes both exon and intron sequences as 5′ transduction (e.g., H1_108). Alternatively, an upstream exon can be spliced to cryptic acceptor sites within the SVA sequence itself, resulting in a 5′-truncated SVA with exon sequences as 5′ transduction. (B) Retrotransposition of a full-length SVA with a 5′-transduced segment into intronic sequences can have two alternative outcomes: Activation of intronic splice acceptors leads to the inclusion of intron sequences (stippled box) in the secondary transduction (green boxes) (H3_545). Alternatively, the 5′-transduced sequence is fused to a splice acceptor localized in the primary transduction (e.g., ZNF487 group, cf. Fig. 5B). (C) After retrotransposition of 5′-truncated SVA elements into an intron (e.g., H2_383), only activation of intronic splice sites could be observed (e.g., H7_1117). A two-exon setting was chosen for the schematic representation. The actual number of exons transduced differs between elements. Yellow boxes, primary transductions; green boxes, secondary transductions; dotted green boxes, intronic sequence included in 5′ transductions; red ovals, TSDs; blue ovals, TSD sequence of the source element present in the 5′-transducing SVA; TD, transduction.
The genome-wide success of the human-specific MAST2 5′ transduction group
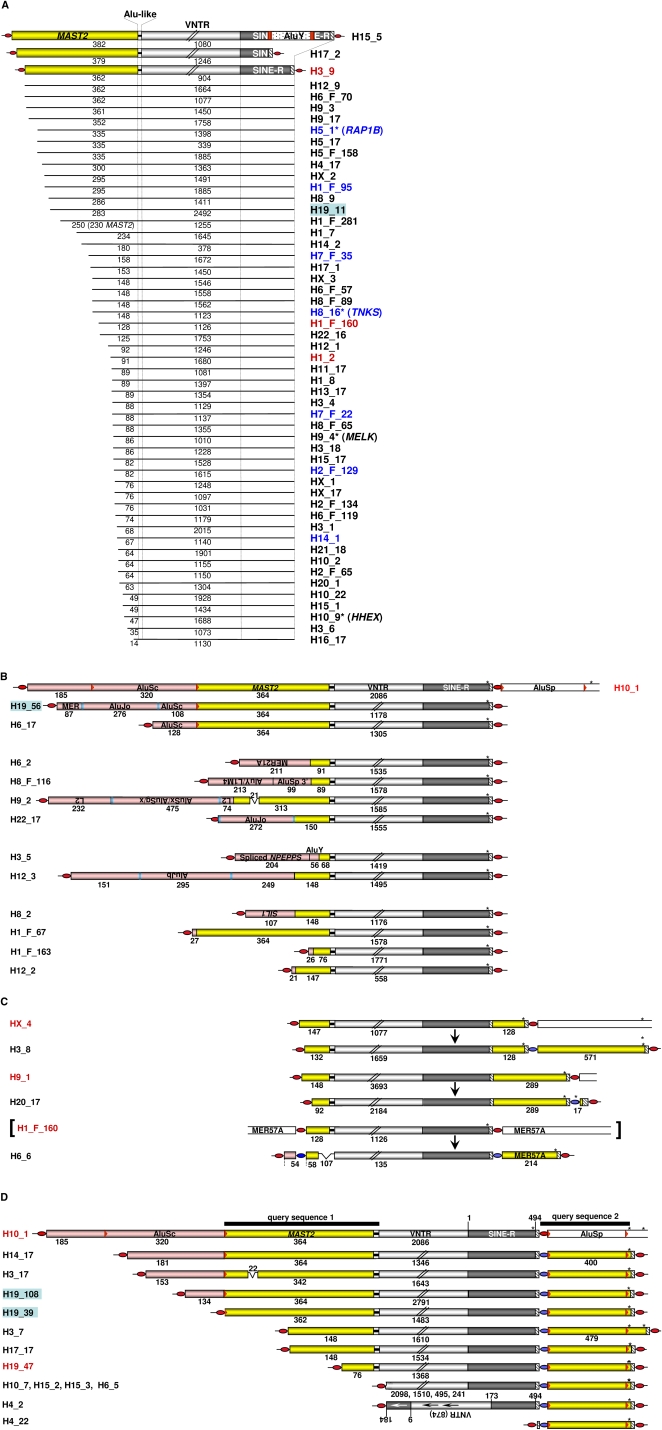
The MAST2 5′ transduction group stands out due to its unsurpassed number of members (Fig. 5C; Supplemental Table 3). In order to understand this exceptionally successful amplification process, we set out to investigate this group in more detail. Because a subset of the identified members of the MAST2 5′ transduction group shares a ∼400-bp 3′ transduction including an AluSp element (Fig. 2B, query sequence 1), we used this sequence as a query to find those members of the MAST2 - transduction subgroup that are devoid of any MAST2 sequences as a result of 5′ truncation events. In this way we retrieved six additional SVA copies that lack any MAST2 sequence but are clearly derived from the same 3′-transducing source element H10_1 (Fig. 7D).
Figure 7.
SVA subfamily F1 can be subdivided into four transduction groups. (A) Transduction group 1 members carry exclusively MAST2 sequences as 5′ transduction. They could be direct descendants of source elements 1A, 1B, 2, or 3 (Fig. 8) except for H17_2 and H15_5. The latter can only be derived from source elements 1A or B due to the extension of their transduced MAST2 sequence. In theory, within this group any given SVA_F1 member could have served as a source element of any other SVA_F1 member whose MAST2 sequence is equal in length or shorter than its own. Source elements and polymorphic SVA_F1 members are marked by red and blue lettering, respectively. Yellow boxes; MAST2 sequence; red boxes, TSDs flanking AluY insertion; *, element was originally described by Bennett et al. 2004 and named as denoted in brackets. (B) Transduction group 2. In addition to MAST2 sequences, members of this group include heterologous sequences (pink boxes) as part of their 5′ transductions. Heterologous sequences indicating consecutive transduction events are derived from source locus 2 (Fig. 8B) on chromosome 9p13.3 (H10_1, H19_56, and H6_17), from sequences of non-LTR retrotransposons or endogenous retroviruses whose source loci could be identified (H6_2, H8_F116, H9_2, H22_17, H3_5, and H12_3), or they represent nonrepetitive genomic DNA fragments ranging from 21–107 bp (H1_F_67, H12_2, H1_F_163, H8_2). TSD-flanked intact Alu elements in sense orientation are part of H10_1, H19_56, and H22_17. H3_5 displays secondary (AluY) and tertiary (NPEPPS exon sequences) transduction events. Numbers indicate extensions of different modules of 5′-transduced heterologous (pink boxes) and MAST2 (yellow boxes) sequences and VNTRs. TSDs flanking AluSc and AluSp elements (red arrowheads), as well as all remaining Alu sequences (blue bars) are shown. Open boxes indicate genomic DNA flanking the SVA insertion. (C) Members of transduction group 3 include 3′ transductions. SVA_F1 members with 3′ transductions and their source elements are shown. Primary 3′ transductions of HX_4 and H9_1 originate from SVA-free source loci (data not shown). The source element of H6_6 is H1_F_160 (in brackets), a member of transduction group 1 (Fig. 7A). MER57A represents internal portions of the nonautonomous endogenous retrovirus 1 (ERV1) family. Yellow boxes depict 5′-transduced MAST2 sequences and/or 3′ transductions, respectively. Numbers denote the length of 5′- and 3′-transduced sequences, their components, and VNTR regions in nucleotides. Red ovals, TSDs flanking SVA elements; blue ovals, TSD sequence of the respective source element that became part of 3′ or 5′ transductions. (D) Members of transduction group 4 are derivatives of source element H10_1. Structures of the 13 identified members of transduction group 4 and their source element H10_1 are shown. Two consecutive transcriptional termination signals (asterisks) in the 3′-flanking sequence of H10_1 resulted in 3′ transductions of 400 and 479 bp, respectively. The 3′ TSD of the source element is overlapping with the 5′ TSD of AluSp. Lengths of 5′-truncated VNTRs of H10_7, H15_2, H15_3, and H6_5 range from 241–2098 bp. H19_47 is a source element of H1_F_163 (Fig. 7B); Black bars, extension of query sequences 1 and 2.
Overall, we thus identified 84 SVA_F elements that are members of or derived from members of this particular MAST2 5′ transduction group (Fig. 7; Supplemental Table 3). It is to be expected that numerous additional genomic SVA copies originated from a MAST2 5′-transducing source element but cannot be identified as such because 5′ truncations might have caused the loss of any distinguishing 5′-transduced MAST2 sequence. Thus, members of the MAST2 5′ transduction group represent at least 32% of the roughly 260 human-specific subfamily F members and 3% or more of all genomic SVA copies. Due to their significant number, we assigned all members of the MAST2 5′ transduction group to a separate subfamily termed SVA_F1. They most likely originated from one single source element that was formed and propagated after the separation of human and chimpanzee lineages about 4–6 Mya (Fig. 8).
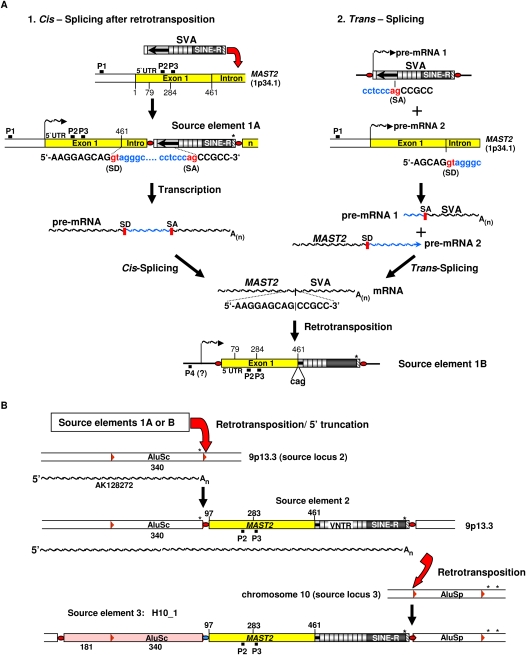
Figure 8.
Evolutionary scenarios for the emergence of SVA subfamily F1. (A) Schematic diagram of two alternative scenarios giving rise to hypothetical precursor source elements 1A or B of the SVA_F1 subfamily, respectively. Alternative 1: Cis-splicing after retrotransposition; after SVA retrotransposition into the MAST2 gene downstream of exon 1, transcription of source element 1A is controlled by the MAST2 promoter P1 or by potential regulatory sequences in exon 1 (P2, P3) resulting in a chimeric MAST2-SVA pre-mRNA. Cis-splicing of the pre-mRNA removes the intronic sequence flanked by the splice donor (SD) site at the MAST2 exon 1–intron junction and the cryptic splice acceptor (SA) site localized within the Alu-like region. Retrotransposition of the spliced, chimeric MAST2-SVA transcript results in the 5′-transducing source element 1B. Transcription of this element could have been controlled by internal promoter sequences located within the MAST2 sequence (P2, P3) or by an external promoter localized upstream of the SVA_F1 insertion (P4). Nucleotide position 79 indicates the 5′-end of the MAST2 sequence of SVA H15_5 (Fig. 7A) that harbors the most extensive MAST2 5′-transduction. Position 284 denotes the 5′-end of the putative cryptic promoter sequence P3; asterisks indicate transcription termination signals. Alternative 2: Trans-splicing; two separate pre-mRNA molecules encoding an SVA element (pre-mRNA 1) and the MAST2 protein (pre-mRNA 2), respectively, are synthesized. Pre-mRNA 2 provides the 5′ splice site (splice donor [SD]), pre-mRNA 1 harbors branchpoint sequence, poly-pyrimidine tract, and a cryptic 3′ splice site (splice acceptor [SA]). The exons of the two separate pre-mRNA molecules are joined by trans-splicing to create a single chimeric RNA. Retrotransposition of this RNA would result in source element 1B. The 5′-transduced MAST2 sequence of source element 1B could either be full-length or 5′-truncated. In theory, members of transduction group 1 (Fig. 7A) could be direct descendants of source elements 1A or B. Splice donor and splice acceptor sequences are highlighted in red. P1, MAST2 promoter; P4, hypothetical external promoter controlling transcription of source element 1B. (B) Evolutionary scenario giving rise to source element H10_1. Retrotransposition of source element 1A or B into the 3′ TSD of an AluSc element on chromosome 9p13.3 is setting the newly integrated 5′-truncated MAST2-SVA element (source element 2) under control of the AK128272 promoter. Transcriptional read-through causes a chimeric AK128272-MAST2-SVA transcript. 5′ Truncation in the process of retrotransposition of the chimeric RNA into the 5′ TSD of an AluSp element in source locus 3 results in the 5′-transducing source element H10_1. The SVA-free source locus 3 is still present on chromosome 10 of both rhesus macaque and chimpanzee genomes but absent from humans. H10_1 served as source element for all members of SVA_F1 transduction group 4.
Based on their structural organization, SVA_F1 elements can be subdivided into four major groups: Group 1 includes those elements that carry exclusively MAST2 sequences as 5′ transduction events (Fig. 7A). In group 2, 5′ transductions contain nonhomologous sequences in addition to the MAST2 sequence (Fig. 7B); Group 3 covers SVA elements with both MAST2 5′ transductions and variable 3′ transduction events (Fig. 7C). 3′-Transducing derivatives of the source element H10_1, a group 2 SVA_F1 element, constitute group 4 (Fig. 7D).
The origin of the SVA_F1 subfamily
By applying an “alternative splice site prediction (ASSP) program” (Wang and Marin 2006; http://www.es.embnet.org/∼mwang/assp.html), we found that in SVA_F1 elements, the splice donor site of MAST2 exon 1 is fused precisely to a cryptic splice acceptor site in the Alu-like region. Therefore, it is likely that the ancestral MAST2 SVA source element was generated by pre-mRNA splicing in cis or trans (Fig. 8A) (for trans-splicing, see Maniatis and Tasic 2002; Horiuchi and Aigaki 2006). Insertion polymorphism studies on a diverse human population panel suggest that this ancestral SVA_F1 source element is not present in any of the tested populations (data not shown), although sample size may be critical in the ascertainment of the element. However, the structural similarity of the MAST2 exon 1–SVA junction to the RHOT1 exon–SVA junction of the RHOT1 transduction group (Fig. 5E; Table 2) whose source element is still present in the genome (Hancks et al. 2009) argues for the involvement of pre-mRNA splicing in the formation of the ancestral SVA_F1 source element. Splicing in cis presupposes the existence of an SVA_F element that is localized downstream of MAST2 exon 1 within the transcriptional unit. Although such an element could not be identified in the human genome reference sequence, it might have been polymorphic for insertion presence/absence in the population at the time of initial propagation of the SVA_F1 lineage and has been lost subsequently through drift or selection. Alternatively, an SVA_F1 source element could have been generated by splicing in trans (Fig. 8A), as recently suggested to explain the existence of zebrafish L1–RNA fusion transcripts for which no source element could be detected (Tamura et al. 2007). Although there is also the formal possibility that retrotransposition of an SVA_F element into the exon 1–intron junction of the MAST2 gene put the SVA element under control of the MAST2 promoter, it is highly unlikely.
Depending on the pathway that led to the assembly of the SVA_F1 ancestral source element (Fig. 8A), it was associated either with MAST2 exon 1 and transcribed from an external MAST2 promoter (source element 1A) or with at least 382 bp of the 5′-transduced MAST2 sequence and transcribed from either a nearby external promoter or from an internal promoter localized in MAST2 exon 1 (source element 1B). As the largest MAST2 5′ transduction identified in the human genome encompasses 382 bp (H15_5) (Fig. 7A), a source element generated via cis- or trans-splicing must have covered at least this fraction of MAST2 exon 1 (Fig. 8A).
The MAST2 promoter that was most likely recruited by the ancestral SVA source element 1A for its mobilization is reported to be highly active in mammalian testicular tissue, although the promoter sequence has not been localized yet (Walden and Cowan 1993; Walden and Millette 1996). Testis-specific MAST2 RNA increases in abundance during prepuberal testis development, peaking at the spermatid stage, and it has been suggested that the protein serine/threonine kinase activity of MAST2 plays a role in spermatid maturation (Walden and Millette 1996). Therefore, recruitment of the MAST2 promoter would assure that the 5′-transducing SVA is coexpressed with the L1-protein machinery, which is known to be present in germ cells (Branciforte and Martin 1994; Ergun et al. 2004) and assumed to trans-mobilize SVA elements (Ostertag et al. 2003).
To localize any potential external or internal MAST2 promoter that was recruited by an SVA source element, we analyzed 1000 bp of genomic sequence upstream of the MAST2 transcriptional start site as well as the MAST2 exon 1 sequence applying different promoter prediction programs (see Methods). We found a strong external promoter candidate that was predicted to initiate transcription at position 1 of the MAST2 gene as well as putative cryptic promoter sequences at positions 205–256 and 284–333 relative to the transcription start site (P2, P3) (Fig. 8A). The presence of such regulatory sequences in MAST2 exon 1 is supported by analyses performed with the ESPERR regulatory potential track of the UCSC Genome Browser (King et al. 2005). These potentially regulating motifs (P2, P3) (Fig. 8A) are present in 25% of all SVA_F1 subfamily members.
No matter which pathway led to the formation of source element 1, the ancestral element and/or its offspring spread extensively afterward, picking up additional sequences at both their 5′- and 3′-ends and dispersing them throughout the genome.
Transcriptional control by multiple external promoters causes consecutive SVA_F1-mediated 5′ transduction events
In addition to MAST2 sequences, transductions of members of SVA_F1 5′ transduction group 2 (Fig. 7B) encompass heterologous sequences. Therefore, the (now extinct) source elements of group 2 members must have been transcribed from external promoters. In the case of the most successful SVA_F1 source element H10_1, the 5′ transduction could be traced back to an annotated transcription unit (AK128272) (Fig. 8B). The AK128272 promoter which was recruited by the source element confers high level expression in testis. Members of transduction group 2 (Fig. 7B) encompass heterologous sequences that are derived from (1) a common source locus 2 (Fig. 8B) on chromosome 9p13.3 (e.g., H10_1), (2) source loci covering exclusively sequences from non-LTR retrotransposons (L1, L2, Alu) and endogenous retroviruses like MER21A (e.g., H6_2), (3) source loci representing short fragments of nonrepetitive DNA (e.g., H8_2), or (4) those containing both nonrepetitive and Alu sequences (e.g., H3_5). A striking feature of the subsequent transductions is the large fraction of Alu sequences that are found to be comobilized.
SVA_F1 elements transduce 3′-flanking sequences
The genome also harbors at least 18 SVA_F1-mediated 3′ transduction events (Fig. 7C,D). Source loci of 3′ transductions of HX_4 and H9_1 (Fig. 7C) are SVA-free. Both 3′-transducing SVA_F1 subfamily members served as source elements for H3_8 and H20_17 that carry 571 and 17 bp of secondary 3′ transduction events, respectively. The source element of H6_6 (H1_F_160) is a member of SVA_F1 transduction group 1 (Fig. 7A) that is flanked by MER57A sequences (Kohany et al. 2006). Retrotransposition of an H1_F_160 transcript generated 5′ as well as 3′ transductions in SVA H6_6.
Thirteen 3′-transducing SVA_F1 members can be assigned to one single 3′ transduction group whose members are derived from source element H10_1 and are subsumed in transduction group 4 (Fig. 7D). H10_1 is one of the most active source elements identified so far, given that it generated 13 or more copies with 3′ transductions despite the fact that it belongs to the evolutionarily younger SVA subfamily F. Transcriptional termination and polyadenylation at two alternative sites in the 3′-flanking sequence of H10_1 resulted in 3′ transductions covering 400 and 479 nucleotides of the source locus. In each case, an intact AluSp element is part of the transduced sequence.
Six members of transduction group 4 are devoid of any MAST2 fragments or additional heterologous 5′-transduced sequences because their 5′ truncation sites are localized either within the VNTR region or at the 3′-end of the SINE-R component (Fig. 7D). The rearranged H4_2 element consists of two SVA segments of which the 5′ segment covering the VNTRs and the 5′ part of SINE-R is inverted. A similar 5′-truncated and inverted SVA structure has been shown to be the cause for a case of hereditary elliptocytosis (Ostertag et al. 2003). Analogous rearrangements have been reported for L1 elements, and it was suggested that such structures are the consequence of a twin-priming mechanism by which 5′-inverted elements are formed (Ostertag and Kazazian 2001). Clearly, members of transduction group 4 are a consequence of 3′-transducing transcription of source element H10_1 and subsequent 5′ truncation occurring during reverse transcription and/or integration.
The SVA_F1 subfamily is currently transcribed in the human genome
Because transcription of potential SVA source elements is an essential prerequisite for their retrotransposition, we investigated the current transcriptional activity of SVA subfamily F1. In order to identify ESTs from SVA subfamily F1 that include MAST2 sequences, we performed a BLAST search (Altschul et al. 1990) against the human EST database using a 401-bp fragment of H10_1 (Fig. 9A) as a query. The sequence covers the 364-bp MAST2 sequence and the adjacent truncated Alu-like region. ESTs matching source element H10_1 or its derivatives of SVA_F1 transduction group 4 were identified by applying query sequence 2 (Fig. 9A). ESTs aligning to the source element HX_4 and its derivative H3_8 were identified from the annotations of both loci in the UCSC Genome Browser. In total, we extracted 17 ESTs that originated from transcription of members of the SVA_F1 subfamily (Fig. 9A–C; Table 3). The identification of their most likely elements of origin suggested that at least 19 out of the 78 SVA_F1 elements are currently transcribed. However, we cannot draw any conclusion on the general expression level and/or retrotransposition rate of SVA subfamily members because (1) tissues that express SVAs or support retrotransposition might not be adequately represented in the databases, and (2) transcription of different members might be variably regulated because they are controlled by different external promoters. EST data suggest that SVA_F1 members are expressed not only in reproductive tissues, stem cells, and tumor cells but also in a variety of somatic cell types and tissues.
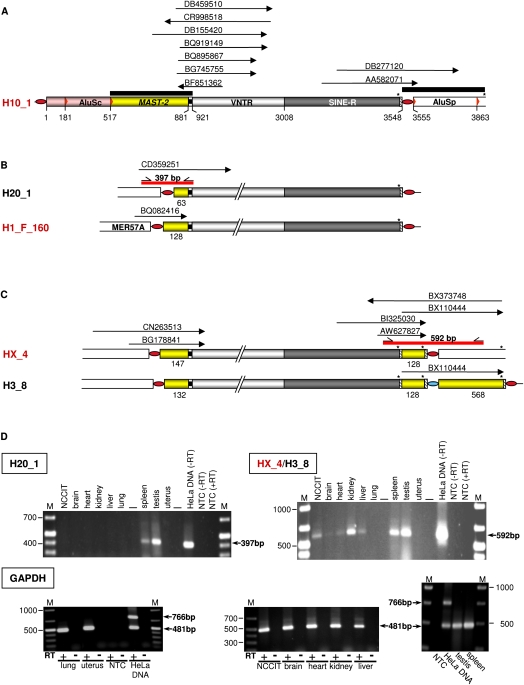
Figure 9.
Ongoing transcription of SVA_F1 subfamily members is demonstrated by ESTs and RT-PCR analyses. (A) ESTs (arrows) aligning specifically to the source element H10_1 and its descendants (DB277120, AA582071) and to MAST2–SVA junctions of a subset of SVA_F1 subfamily members. Extensions of the queries used to identify ESTs (black bars) are shown. (B) Transcriptional control of transduction group 1 members H20_1 and H1_F_160 is effected by external promoters. Identified ESTs match 5′ junctions of the SVA elements with primary 5′ transductions. MER57A is an internal portion of the nonautonomous retrovirus 1 (ERV1). (C) ESTs indicating ongoing transcription of source element HX_4. Those ESTs that correspond to the 3′-end (BI325030, AW627827) and the 3′ junction (BX110444) of HX_4 are also consistent with transcription of the HX_4 derivative H3_8 (Fig. 7C). 3′-Ends of sense ESTs BI325030 and BX110444 as well as the 5′-end of antisense EST BX373748 are directly at the polyadenylation sites of HX_4 and/or H3_8. SVA copies highlighted in red were verified to be source elements. (D) RT-PCR analyses confirm transcription of distinct SVA_F1 subfamily members. Agarose gel electrophoresis of H20_1 and HX_4/H3_8-specific RT-PCR products derived from total RNA from human tissues and NCCIT cells. Primer binding sites and extension of the RT-PCR products are indicated as short arrows and red bars, respectively, in B and C. To control for both successful cDNA synthesis and DNA-free RNA preparations, PCRs with GAPDH-specific, intron-spanning primers (see Methods) were performed on RNA preparations from each tissue before (−RT) and after (+RT) cDNA synthesis. PCR on genomic HeLa DNA served as a positive control. The 766- and 481-bp products originate from the unspliced GAPDH gene and a processed GAPDH pseudogene, respectively (Harper et al. 2003). Sizes of obtained PCR products are indicated and correspond to expected fragment sizes. RT, reverse transcriptase; M, size marker; NTC, no template control.
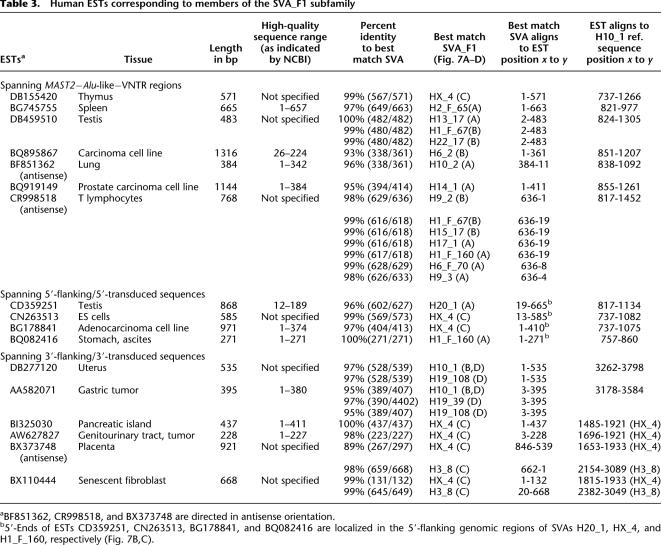
Table 3.
Human ESTs corresponding to members of the SVA_F1 subfamily
Two ESTs matching the SINE-R–AluSp junction specific for the H10_1 source element and its descendants of transduction group 4 argue for ongoing transcription of these elements (Fig. 9A). ESTs matching the 5′ junctions of H20_1, and the source elements H1_F160 and HX_4 (Fig. 9B,C) demonstrate current transcriptional control of these SVA elements by external promoters. We confirmed transcriptional control of H20_1 by external promoter sequences by RT-PCR on total RNA isolated from a variety of human tissues with a primer pair specifically amplifying a 397-bp fragment of the 5′ junction between H20_1 5′-end and adjacent genomic sequence (Fig. 9D). H20_1-specific RT-PCR products could be detected in spleen and testis only. The identification of three ESTs (AW627827, BI325030, BX110444) mapping the junction between the 3′-end of SINE-R and 3′ transductions of both HX_4 and its derivative H3_8 in sense orientation (Fig. 9C) is consistent with transcription of both elements. This demonstrates transcriptional read-through into genomic sequences beyond the 3′-end of the SINE-R region. Transcriptional read through into the 3′-flanking genomic sequence of HX_4 that gave rise to its derivative H3_8 is documented by EST BX110444 (Fig. 9C). This was confirmed by RT-PCRs using a primer pair specifically amplifying the 3′ junction between the 3′-end of the SINE-R region of the source element HX_4 (or its derivative H3_8) and its 3′-flanking genomic sequence, which becomes a secondary transduction in H3_8 (Fig. 9C,D). HX_4/H3_8-encoded transcripts could be identified in all tissues tested except for lung and uterus. Taken together, EST data and RT-PCR demonstrate that SVA_F1 elements are transcribed in reproductive as well as in somatic tissues. Our expression data confirm both that SVA transcription is initiated from upstream promoters and that RNA polymerase can bypass the SVA element's transcriptional termination signal.
Discussion
We set out to thoroughly analyze the structural variants of SVA elements on a single chromosome because, first, we expected to find evidence for the assembly process leading to the composite retrotransposon SVA and, second, we argued that the organization of 5′-ends of SVA insertions and their genomic environment would allow conclusions for the localization of the promoter sequences controlling SVA transcription. In this study, we found SVA elements with 3′ truncations, both 5′ and 3′ truncations, and both 5′ and 3′ transductions in combination or alone. The identification of 5′-transducing SVAs on chromosome 19 led to the first comprehensive genome-wide analysis of 5′ transduction groups of a non-LTR retrotransposon. Overall we identified 220 SVA elements with 5′ transductions comprising 8% or more of all SVA elements. Our findings show that SVA elements can use external promoters not only for their own transcriptional regulation but also for attaching new genomic sequences to their 5′-ends by means of the retrotransposition pathway leading to structural diversity of SVA elements. Furthermore, SVA-mediated 5′ transduction represents a novel mechanism contributing to structural human genomic diversity.
SVA-mediated 5′ transduction indicates transcriptional control by external promoters
5′ Transduction events were discussed for the first time in order to explain the variability observed among 5′-end sequences in different L1 subtypes (Boeke and Pickeral 1999; Lander et al. 2001). 5′ Transduction involves retrotransposon transcription from an external promoter followed by retrotransposition of the transcript containing additional heterologous sequences at its 5′-end. Following a first description of two 5′-transducing L1 copies in the human genome (Lander et al. 2001), Symer et al. (2002) provided the first experimental evidence for retrotransposition-mediated 5′ transduction events using an L1 retrotransposition reporter assay in cell culture. More recently a disease-linked L1-mediated 5′ transduction event has been reported in mice (Chen et al. 2006).
The fact that more than 8% of human SVA elements carry 5′ transductions, together with our RT-PCR results and 5′-RACE experiments performed in the Kazazian laboratory (Hancks et al. 2009), indicates that SVA elements frequently parasitize adjacent cellular promoters. The most likely explanation for this process is the intrinsic weakness of a still elusive SVA-specific internal promoter. This reasoning is supported by previous attempts to identify any SVA-internal promoter sequence using reporter gene assays which led to ambiguous results (A Damert et al., unpubl.; Hancks et al. 2009).
The identification of 93 different primary and 26 diverse secondary 5′ transduction events (Supplemental Tables 2, 3) strongly suggests that during hominid evolution there were or still are more than 100 different cellular promoters regulating transcription of downstream SVA elements in germ cells.
It is very likely that the actual number of SVA elements that resulted from retrotransposition of an SVA source element controlled by an external promoter exceeds the number of genomic SVA elements with 5′ transductions. The reason is that those 5′-truncated SVA retrotransposition events that are derived from 5′-transducing source elements will not be identified as such if the 5′ truncation site is localized downstream the 5′-transduced sequence. This is supported by the identification of six 5′-truncated SVA_F1 subfamily members that were devoid of any MAST2 sequence but could be unambiguously assigned to this subfamily due to a specific 3′-transduced sequence (Fig. 7D).
5′ Transductions involve a variety of genomic sequences ranging from 14–2161 bp, and it is obvious that they significantly contribute to SVA and overall genomic structural diversity. Modification of the SVA RNA secondary structure by 5′-transduced cellular sequences might result in a better target for any putative trans-mobilizing protein machinery. Alternatively, splicing can bring an SVA element in close vicinity of transcribed, gene-internal, host-encoded RNA polymerase II promoter sequences or transcriptional regulatory sequences thereby generating a novel SVA subtype that might be characterized by a superior expression rate. Promoter acquisition is exemplified by the dominant A-type L1 elements in mouse which evolved from F-elements by adopting the A-type promoter (Adey et al. 1994). In both cases, 5′ transductions would be evolutionarily advantageous for the SVA element.
It is conceivable that 5′ transduction events played a role in the assembly process that led to the current organization of SVA elements. This would be supported by the structure of the “GA repeat” 5′ transduction group (Table 1; Supplemental Table 2), which is characterized by variable length additions of simple GA repeats. The SVA hexamer repeat region might have been acquired by 5′ transduction as well. One example for the impact of structural diversity brought about by 5′ transduction events is the SVA_F1 subfamily: Transcription of a single ancestral SVA source element by the MAST2 promoter gave rise to a whole new subfamily—with properties obviously favoring its rapid expansion in the human genome.
Exon shuffling mediated by SVA 5′ transduction
To date, SVA elements have been reported to contribute to human genome evolution through insertional mutagenesis (Kobayashi et al. 1998; Rohrer et al. 1999; Wilund et al. 2002; Ostertag et al. 2003; Conley et al. 2005; Makino et al. 2007; Takasu et al. 2007), by constituting alternative exons (Damert et al. 2004) and by exon and gene shuffling mediated by 3′ transduction (Xing et al. 2006). Similar to 3′ transduction events, the process of 5′ transduction entails the juxtaposition of two previously unlinked host genomic segments. It thus represents another mechanism of diversification of the genome via increasing genome plasticity and facilitating novel combinations of coding and regulatory sequences. Given that retrotransposition of SVA and other transduction-capable retroelements (e.g., L1) is an ongoing process in the genome, transduction events generated in this manner afford the possibility for rapid lineage-specific evolution of gene families, the proteome, and the transcriptome.
SVA-mediated 5′ transductions can be composed of genomic fragments or spliced cellular RNA sequences with functional splice acceptor sites being located either within intronic sequences or in the 5′-transducing SVA element (Fig. 6; Table 2). If RNA splicing is involved, exons of protein-coding genes or noncoding RNAs are translocated into a new regulatory and potentially also coding context. Thus, SVA-mediated 5′ transduction resembles exon shuffling by L1-mediated 3′ transduction (for review, see Boeke and Pickeral 1999). In contrast, SVA-mediated 3′ transduction has been shown to be capable of translocating entire transcriptional units, thus retaining the original regulatory context for the transduced genomic copies (Xing et al. 2006).
5′ Transduction events involving splicing at alternative/cryptic splice acceptor sites result in the modification of the 3′-end of the transduced ORF. Provided that integration of the 5′-transducing element occurs downstream of a suitable regulatory region, this could lead to expression of mutant proteins that are truncated and/or modified at their C termini. Splice acceptor sites used during SVA-mediated 5′ transduction events can be located within SVA sequences (Fig. 6A, H6_1088; Table 2), which is reminiscent of Alu (for review, see Kreahling and Graveley 2004) and L1 (Belancio et al. 2006; Tamura et al. 2007) elements, which have been shown to provide functional splice sites to host RNAs. However, while splicing to SVA sequences was only observed in the context of SVA-mediated exon shuffling so far, splicing of/to Alu (Sorek et al. 2002) and L1 sequences (Mulhardt et al. 1994; Meischl et al. 2000) was exclusively reported to lead to exonization.
The alternative mechanism resulting in the modification of the 3′-end of any SVA-transduced ORF sequence involves activation of upstream intronic cryptic splice sites (Fig. 6A, e.g., H1_108). Recent reports demonstrated activation of an upstream cryptic splice acceptor site by an Alu insertion into dystrophin intron 11 (Ferlini et al. 1998) and by an L1 insertion into an intron of the porcine KPL2 gene (Sironen et al. 2006, 2007). Also in these cases, alternative splicing induced by Alu and L1 insertions led to exonization and not to exon shuffling as observed for SVA 5′ transductions.
Taken together, splicing into and upstream of Alu and L1 elements on one side and SVA elements on the other side have profoundly different consequences: Alu and L1 sequences are exonized within a given regulatory and coding context. In contrast, splicing of 5′ transducing SVA RNAs followed by retrotransposition leads to exon shuffling, which generates new combinations of modified coding and/or regulatory sequences at genomic integration sites.
What is the basis for the exceptional success of the SVA_F1 subfamily?
With at least 84 members, the SVA_F1 subfamily is the largest 5′ transduction group identified to date. The fact that it originated from the youngest human-specific SVA subfamily F implies that it must have reached its copy number within a relatively short period of time. There are two possible explanations for the outstanding success of the SVA_F1 subfamily:
SVA_F1 source elements recruited external promoters that are highly active in the germ line. The ancestral source element 1A (Fig. 8A) recruited the promoter of the MAST2 gene whose expression peaks in germ cells at the spermatid stage in mice (Walden and Cowan 1993). Transcriptional control of a potential source element by the MAST2 promoter assures that the 5′-transducing SVA element is coexpressed with the L1 protein machinery, which is known to be present in germ cells (Branciforte and Martin 1994; Ergun et al. 2004) and suspected to trans-mobilize SVA elements (Ostertag et al. 2003). Additional heterologous sequences that are part of SVA_F1 5′ transductions (Fig. 7B–D) indicate that their source elements were subject to transcriptional control by external promoters other than the MAST2 promoter. Identification of SVA_F1-specific ESTs and RT-PCR products extending into genomic flanks upstream of specific SVA_F1 elements indicate ongoing transcriptional regulation by external promoters in both reproductive and somatic tissues (Fig. 9).
Newly acquired 5′- and 3′-transduced sequences made SVA_F1 subfamily members a better target for trans-mobilization by the L1 protein machinery. A striking feature of numerous SVA_F1 secondary transductions is the presence of intact Alu elements (Fig. 7B,D). The most successful source element H10_1, in particular, can give rise to transcripts containing intact AluSc and AluSp RNAs at their 5′- and 3′-ends, respectively (Fig. 7D). The same mechanism hypothesized recently to be responsible for the preferential trans-mobilization of Alu elements by the L1-encoded protein machinery (Boeke 1997) could thus explain the favored mobilization of SVA RNAs harboring intact Alu elements in their transduced sequences. In this model, the Alu sequence is docked on ribosomes via the SRP9/14 complex and captures the L1 ORF2 protein as it is translated from an active L1 element mRNA. By capturing ORF2p at the ribosome, Alu and, possibly, Alu-containing SVA RNAs can efficiently substitute its RNA for the normal L1 mRNA during the process of target-site primed reverse transcription (TPRT). Functionally intact Alu core elements were recently identified from four of the six AluS subfamilies, and binding of the Alu RNA to SRP9/14 was shown to be essential for Alu retrotransposition activity (Bennett et al. 2008). Provided that sequences flanking Alu elements in SVA transcripts allow formation of the three-dimensional structure required for SRP9/14 interaction (Weichenrieder et al. 2000), Alu sequences could mediate docking of the respective RNA to the ribosome via SRP9/14 and thus facilitate efficient capture of ORF2 proteins (Boeke 1997; Dewannieux et al. 2003; Mills et al. 2007).
In a different scenario, 5′-transduced Alu sequences in antisense orientation (Fig. 7B) might substitute for the truncated Alu-like region in SVA_F1 subfamily members. The SRP9/14 model of Alu trans-mobilization suggests that hybridization of the SVA Alu-like region that is complementary to intact ribosome-bound Alu elements could bring SVA transcripts into close proximity to the nascent L1 ORF2 polypeptide chain and may, in this way, enhance the probability of ORF2p capture (Mills et al. 2007).
The presence of both sense and antisense Alu sequences on the same transducing RNA (Fig. 7B) might increase the trans-mobilization rate of such an SVA mRNA even more. Clearly, all three of these scenarios remain to be tested experimentally.
Methods
Identification of SVA elements on human chromosome 19 and genome-wide screening for 5′-transducing SVA elements
To identify the complete set of SVA elements on chromosome 19 and 5′-transducing SVA elements genome-wide, we started out by using the RepeatMasker premasked genome section at http://www.repeatmasker.org/cgi-bin/AnnotationRequest (hg 17; May 2004; repeat class: other). Each of the identified SVA sequences was extracted and manually inspected for correct annotation. In order to identify TSD sequences, the BLAST-2–sequences program (bl2seq; http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi; Tatusova and Madden 1999) was applied to align 500-bp 5′- and 3′-flanking sequences of each identified SVA against each other. In case TSDs could not be identified in this way, SVA insertions and their flanking sequences were tested for the presence of TSDs individually. In principle, the minimum number of nucleotides required for TSDs to be considered as such was six.
In case of the genome-wide survey for 5′ transductions, only elements containing at least a fraction of the SINE-R region were analyzed. Thus, SVA2 elements and fragments lacking a poly(A) tail were excluded from the analysis. An element was considered candidate for containing a 5′ transduction if it had (1) unambiguous TSDs (≥6 bp) and (2) a ≥20-bp sequence between the TSD and the 5′-end of the SVA sequence. These criteria were relaxed in those few cases where an unambiguous assignment to a particular transduction group was possible based on the respective transduced sequence. Multiple sequence alignments were calculated using the ClustalW (2.0) program (Labarga et al. 2007).
To identify source loci of transduced sequences, we searched the human genome by using BLAT. Repetitive sequence annotations were obtained from the UCSC Genome Browser. The corresponding descriptions were retrieved from Repbase Update at http://www.girinst.org/server/RepBase/index.php.
Prediction of potential promoter sequences
In order to identify sequences that had the potential to serve as promoters controlling either transcription of the MAST2 gene or expression of members of the SVA_F1 subfamily, we used a specific query sequence. The query encompasses the 461 bp of MAST2 exon 1 and 1000 bp of genomic 5′-flanking sequence. Genomic sequences were analyzed applying promoter prediction programs NNPP2.2 (Reese 2001), TSSW (Solovyev and Shahmuradov 2003), and TSSG (Prestridge 1995; http://www.softberry.com/berry.phtml?topic=tssg&group=help&subgroup=promoter). Obtained results were scrutinized by applying the ESPERR regulatory track of the UCSC Genome Browser (King et al. 2005). Data for this track are obtained by comparing frequencies of short alignment patterns between known regulatory elements and neutral DNA. Score values above 0.1 indicate significant resemblance to alignment patterns typical of regulatory elements in the training set.
Identification of ESTs matching members of SVA subfamily F1
To identify ESTs originating from members of the SVA_F1 subfamily, we searched the human division of dbEST, a descriptive catalogue of ESTs in GenBank, by using the BLASTN program (BLASTN at http://blast.ncbi.nlm.nih.gov/Blast.cgi; default settings except for: word length 16, no filter) with a 401-bp DNA fragment of SVA element H19_56, which is also part of the source element H10_1. The fragment comprises 364 bp of MAST2 exon 1 and 37 bp of the 5′-truncated Alu-like region specific for the MAST2 5′ transduction groups (query sequence 1). ESTs specific for members of SVA_F1 transduction group 4 were searched for by using the 394-bp 3′ transduction specific for this transduction group (Fig. 7D). The sequence is flanked by the H10_1-terminating poly(A) sequence and the first transcriptional termination signal following the AluSp 3′ TSD (query sequence 2). ESTs matching the source element HX_4 and its derivative H3_8 were identified from the annotation of the respective loci in the UCSC Genome Browser.
RT-PCR analyses
Total RNA from different human tissues was purchased from BioCat (brain, heart, liver, kidney) or Agilent/Stratagene (uterus, lung). Total RNA was extracted from human teratocarcinoma cell line NCCIT (ATCC CRL-2073) using TRIzol reagent (Invitrogen). First-strand synthesis was performed using random hexamers (Fermentas) and SuperScript III RT (Invitrogen) according to the manufacturer's instructions. cDNAs synthesized from testis and spleen RNAs were purchased from BioCat. For cDNA amplification AmpliTaq DNA polymerase (Applied Biosystems) was used. PCR cycling conditions were as follows: 5 min at 94°C, 30 sec at 94°C, 30 sec at primer-specific annealing temperature, 30 sec at 72°C, 7 min at 72°C; 40 cycles. HX_4/ H3_8 cDNA was amplified using primers SVA-RT-FW2 5′-TGCCTAGGAAAACCAGAGAC-3′ and HX4-REV 5′-CTCTAATAGTGGGTCACCAATGCC-3′ (62°C). For H20_1, the primer combination 20_1 FW 5′-CCTGTCATTGATCTTGACTTACC-3′ and Alu 3′REV 5′-CGGGCAGAGG CTGCAAT-3′ (63°C) was used.
Control PCRs to exclude genomic DNA contamination and to test for successful cDNA synthesis were performed with GAPDH intron-spanning primers hGAPDH-for 5′-CCATGAGAAGTATGACAACAGC-3′ (Exon 6) and hGAPDH-rev 5′-GTCAAAGGTGGAGGAGTGG-3′ (Exon 8).
Acknowledgments
We thank Dustin Hancks, Haig Kazazian, and Sandy Martin for helpful discussions. We thank Matthias Hamdorf for his indispensable contribution to artwork generation. This research was supported by grant DA 545/2-1 of the Deutsche Forschungsgemeinschaft (A.D., G.G.S), National Science Foundation grant BCS-0218338 (M.A.B.), and National Institutes of Health RO1 GM59290 (M.A.B.).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093435.109.
References
- Adey NB, Schichman SA, Graham DK, Peterson SN, Edgell MH, Hutchison CA., III Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol Biol Evol. 1994;11:778–789. doi: 10.1093/oxfordjournals.molbev.a040158. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Wooding S, Watkins WS, Ostler CT, Batzer MA, Jorde LB. Human population genetic structure and inference of group membership. Am J Hum Genet. 2003;72:578–589. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD. LINEs and Alus—the polyA connection. Nat Genet. 1997;16:6–7. doi: 10.1038/ng0597-6. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Pickeral OK. Retroshuffling the genomic deck. Nature. 1999;398:108–109, 111. doi: 10.1038/18118. [DOI] [PubMed] [Google Scholar]
- Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: Implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: Lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15:2146–2156. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- Conley ME, Partain JD, Norland SM, Shurtleff SA, Kazazian HH., Jr Two independent retrotransposon insertions at the same site within the coding region of BTK. Hum Mutat. 2005;25:324–325. doi: 10.1002/humu.9321. [DOI] [PubMed] [Google Scholar]
- Damert A, Lower J, Lower R. Leptin receptor isoform 219.1: An example of protein evolution by LINE-1-mediated human-specific retrotransposition of a coding SVA element. Mol Biol Evol. 2004;21:647–651. doi: 10.1093/molbev/msh056. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, Schumann GG. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- Ferlini A, Galie N, Merlini L, Sewry C, Branzi A, Muntoni F. A novel Alu-like element rearranged in the dystrophin gene causes a splicing mutation in a family with X-linked dilated cardiomyopathy. Am J Hum Genet. 1998;63:436–446. doi: 10.1086/301952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O, Tran-Gyamfi M, et al. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Han K, Konkel MK, Xing J, Wang H, Lee J, Meyer TJ, Huang CT, Sandifer E, Hebert K, Barnes EW, et al. Mobile DNA in Old World monkeys: A glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Ewing AD, Tokunaga K, Kazazian HH. Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009 doi: 10.1101/gr.093153.109. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper LV, Hilton AC, Jones AF. RT-PCR for the pseudogene-free amplification of the glyceraldehyde-3-phosphate dehydrogenase gene (gapd) Mol Cell Probes. 2003;17:261–265. doi: 10.1016/s0890-8508(03)00063-x. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Aigaki T. Alternative trans-splicing: A novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreahling J, Graveley BR. The origins and implications of Aluternative splicing. Trends Genet. 2004;20:1–4. doi: 10.1016/j.tig.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European Bioinformatics Institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Makino S, Kaji R, Ando S, Tomizawa M, Yasuno K, Goto S, Matsumoto S, Tabuena MD, Maranon E, Dantes M, et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80:393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Meischl C, Boer M, Ahlin A, Roos D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet. 2000;8:697–703. doi: 10.1038/sj.ejhg.5200523. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Mulhardt C, Fischer M, Gass P, Simon-Chazottes D, Guenet JL, Kuhse J, Betz H, Becker CM. The spastic mouse: Aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Ono M, Kawakami M, Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15:8725–8737. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Twin priming: A proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol. 1995;249:923–932. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Minegishi Y, Richter D, Eguiguren J, Conley ME. Unusual mutations in Btk: An insertion, a duplication, an inversion, and four large deletions. Clin Immunol. 1999;90:28–37. doi: 10.1006/clim.1998.4629. [DOI] [PubMed] [Google Scholar]
- Sironen A, Thomsen B, Andersson M, Ahola V, Vilkki J. An intronic insertion in KPL2 results in aberrant splicing and causes the immotile short-tail sperm defect in the pig. Proc Natl Acad Sci. 2006;103:5006–5011. doi: 10.1073/pnas.0506318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironen A, Vilkki J, Bendixen C, Thomsen B. Infertile Finnish Yorkshire boars carry a full-length LINE-1 retrotransposon within the KPL2 gene. Mol Genet Genomics. 2007;278:385–391. doi: 10.1007/s00438-007-0256-7. [DOI] [PubMed] [Google Scholar]
- Solovyev VV, Shahmuradov IA. PromH: Promoters identification using orthologous genomic sequences. Nucleic Acids Res. 2003;31:3540–3545. doi: 10.1093/nar/gkg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu M, Hayashi R, Maruya E, Ota M, Imura K, Kougo K, Kobayashi C, Saji H, Ishikawa Y, Asai T, et al. Deletion of entire HLA-A gene accompanied by an insertion of a retrotransposon. Tissue Antigens. 2007;70:144–150. doi: 10.1111/j.1399-0039.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- Tamura M, Kajikawa M, Okada N. Functional splice sites in a zebrafish LINE and their influence on zebrafish gene expression. Gene. 2007;390:221–231. doi: 10.1016/j.gene.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Walden PD, Cowan NJ. A novel 205-kilodalton testis-specific serine/threonine protein kinase associated with microtubules of the spermatid manchette. Mol Cell Biol. 1993;13:7625–7635. doi: 10.1128/mcb.13.12.7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden PD, Millette CF. Increased activity associated with the MAST205 protein kinase complex during mammalian spermiogenesis. Biol Reprod. 1996;55:1039–1044. doi: 10.1095/biolreprod55.5.1039. [DOI] [PubMed] [Google Scholar]
- Wang M, Marin A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: A hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Watkins WS, Rogers AR, Ostler CT, Wooding S, Bamshad MJ, Brassington AM, Carroll ML, Nguyen SV, Walker JA, Prasad BV, et al. Genetic variation among world populations: Inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–1618. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- Wilund KR, Yi M, Campagna F, Arca M, Zuliani G, Fellin R, Ho YK, Garcia JV, Hobbs HH, Cohen JC. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum Mol Genet. 2002;11:3019–3030. doi: 10.1093/hmg/11.24.3019. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 2007;135(Suppl. 45):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]