Abstract
The aphid Acyrthosiphon pisum population is composed of different morphs, such as winged and wingless parthenogens, males, and sexual females. The combined effect of reduced photoperiodicity and cold in fall triggers the apparition of sexual morphs. In contrast they reproduce asexually in spring and summer. In our current study, we provide evidence that clonal individuals display phenotypic variability within asexual morph categories. We describe that clones sharing the same morphological features, which arose from the same founder mother, constitute a repertoire of variants with distinct behavioral and physiological traits. Our results suggest that the prevailing environmental conditions influence the recruitment of adaptive phenotypes from a cohort of clonal individuals exhibiting considerable molecular diversity. However, we observed that the variability might be reduced or enhanced by external factors, but is never abolished in accordance with a model of stochastically produced phenotypes. This overall mechanism allows the renewal of colonies from a few adapted individuals that survive drastic episodic changes in a fluctuating environment.
Aphids exhibit a complex mode of reproduction, combining parthenogenesis (spring/summer) and sexual activity (fall/winter) in species such as Acyrthociphon pisum (Dixon 1973; Blackman and Eastop 1984). Aphids thus constitute an excellent model system to investigate how this double reproductive system generates polyphenism, a generic concept used to describe the emergence of distinct morphs, such as winged, wingless, sexual female, and male (Blackman and Eastop 1984; Blackman 1987; Muller et al. 2001). Aphid morph distribution, particularly wing dimorphism, is influenced by environmental conditions, such as population density (crowding effects) (Sutherland 1969) and/or host plant vitality, as well as physical parameters including humidity, temperature, and photoperiodicity (Walters and Dixon 1982; Dixon 1998). This raises fascinating questions regarding the outcomes of alternative developmental mechanisms that cause morph switching in a predictable way (Stearns 1989; Nijhout 1999).
Some aphid species are “sexual” lineages committed exclusively to sexual reproduction, others are “facultative asexual” lineages, which alternate between sexual and parthenogenetic modes depending on the season, while some others are obligate parthenogens (Delmotte et al. 2002; Le Trionnaire et al. 2008). This combined double system of reproduction is shared with many other species like Daphnia (Little and Ebert 2000; Pfrender and Lynch 2000) reef coral Pocillopora damicornis (Miller and Ayre 2004) and many plants (Eckert et al. 2003; Ronsheim and Bever 2000; Balloux et al. 2003). These two reproductive strategies (parthenogenesis and sexuality) have been extensively analyzed to determine whether either one or the other, and/or both, combined modes create more variability of genotypes in aphids (Hebert 1987; Delmotte et al. 2002; Halkett et al. 2008) in the bdelloid rotifers (Fontaneto et al. 2007), in Daphnia (Little and Ebert 2000; Pfrender and Lynch 2000) and/or more phenotypic plasticity in Allium vineale (Liliaceae) (Ronsheim and Bever 2000).
Despite some divergent reports, most authors seem to agree that sexual populations in aphids present a high allelic polymorphism of many genes and predominance of homozygous loci within individuals. In contrast, asexual populations seem to present less allelic polymorphism, but strong heterozygosity at most loci (Delmotte et al. 2002; Kanbe and Akimoto, 2009). It is largely assumed that organisms reproducing asexually should maintain lower genotypic diversity than organisms reproducing sexually. A high rate of clonal reproduction seems to drastically increase heterozygosity and, on the opposite hand, decreases the genotypic diversity (Vorburger et al. 2003; Kanbe and Akimoto 2009).
Little is known about the behavioral and physiological mechanisms allowing asexual aphids to survive in a fluctuating environment for 200 millions years with these complex genetic principles. Adult aphids orient the morphological traits of their progeny by integrating signals from the combined action of photoperiodicity, amplitude of temperature between seasons, thermal difference between day and night, state of resources, and diverse physical stresses like drought and plant desiccation (Brisson and Stern 2006; Le Trionnaire et al. 2007). The exoskeleton, but not the eyes, directly capture the photoperiodic signal by measuring the length of daylight through the cuticle and this suggests that the associated carotene molecules are a key player in absorbing light and transmitting electrons (Le Trionnaire et al. 2007; Wittkopp and Beldade 2009). Moreover microarray analysis has revealed that photoperiodic signals regulate essentially the transcripts of cuticular proteins and kinases involved in the cascade of cellular signaling and signal transduction (Le Trionnaire et al. 2007). In aphids, the control of cuticular permeability is essential to resist the stress of desiccation and to contribute to maintaining the high internal osmotic pressure generated by osmolytes, such as sugars and polyols (Kaersgaard et al. 2004; Brisson and Stern 2006). We observed that the cuticle of adult aphids can be soft and flexible or alternatively rigid depending partly on the degree of humidity and temperature (Neville 1975). Our preliminary work indicates that no major modification in protein expression patterns occur shortly after drastic changes of environmental conditions (temperature, photoperiodicity), which prompts us to think that immediate gene expression and/or enzymatic induction, in other words, phenotypic plasticity, is not the key mechanism for aphid adaptation. In contrast, we observed that later individual offspring appear to exhibit distinct protein patterns as though each one had a defined molecular identity for its lifespan.
Therefore, in this letter we tried to address the question of how much diversity exists in clonal aphids likely sharing the same genome and identical morphological traits. We investigated the range of variabilities in biochemical, physiological, and behavioral traits between clonal individuals of the dominant morph category, the wingless parthenogens. The phenotypic variability in this context was approached by exploring the capacity of a single mother to generate an entire collection of possible phenotypic variants presenting distinct molecular profiles. Therefore, clonally obtained individuals from the same founder mother were propagated separately and their progeny were individually compared to investigate the scale of their molecular diversity. The extent of the repertoire of molecular profiles generated by a single parthenogenetic founder was analyzed in optimal environmental conditions in terms of temperature, photoperiodicity and food resources. This variability was also examined in extreme conditions to evaluate whether the repertoire is stochastically produced, or are conditional and/or oriented when it appears that both scenarios contribute to shaping the morphs distribution. Therefore, we focused on the expression of cuticular proteins due to the fact they appear highly sensitive to many environmental stimuli, acting as a coincidental sensor of diverse changes in the environment, such as photoperiodicity, humidity, and temperature (Le Trionnaire et al. 2007). We reasoned that this protein family constitutes a sort of active barrier to transfer messages from the environment to the germarium/ovariole system in which the embryo fate is decided. Also as they constitute a large family of proteins, we thought that they might be an ideal marker for a refined analysis of the extent of aphid molecular variability.
If aphid asexual progenies are a repertoire of molecularly distinct profiles whose distribution is influenced by the environment, we reasoned that the best way to approach the phenomenon should be to identify some epigenetic markers with the potential to influence the specific expression of alleles and/or the exclusion of some others. Our preliminary results have shown that the aphid genome is heavily methylated, which lead us to investigate whether the clonal descendants of a founder mother raised in different conditions might exhibit specific patterns of covalently modified DNA targets. Epigenetic modifications are induced by CpG island methylation, associated with covalent modifications of histones. Methylation of lysine 9 of histone H3 contributes to silencing genes by the consecutive recruitment of transcription repressor proteins on methylated DNA regions (Chinnusamy and Kang Zhu 2009). In plants, the methylation of CpG islands within specific gene promoters produces distinct phenotypes and, surprisingly, these traits are heritable over a limited number of generations (Kakutani 2002; Zilberman and Henikoff 2005; Richards 2006). Moreover, the methylation of these genes can be reversed and this new status can be heritable again (Kakutani 2002; Kalisz and Purugganan 2004; Zilberman and Henikoff 2005; Richards 2006). Although, of great current interest, epiallelic genes and the extent of epiallelic variation are poorly understood with regards to the mechanisms that sense the fluctuating environmental conditions and consequently drive the best fitness. In our current report, the methylation of cytosine within the CpG islands was probed in the aphid genome as the most probable epigenetic signaling mechanism through which the environment influences the repertoire of phenotypes.
Results
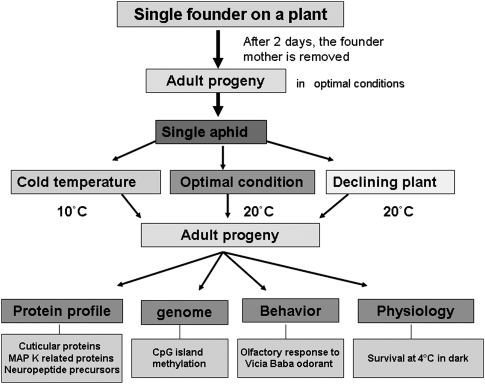
The scheme and the rationale behind our experimental design are summarized in Figure 1. A single founder parthenogenetic mother was used to propagate clonal progenies and several individuals from the successive generations were placed in different environmental conditions to perpetuate clones. A series of molecular analyses were then performed on individuals collected from the same plant. Each series differed by the fact that the experiments were carried out with different generations of adult aphids (from the same founder mother) raised on different plants and environmental conditions.
Figure 1.
General scheme of the experimental steps that were carried out to analyze the clonal progenies of a single founder aphid.
Variations in the cuticle protein pattern between clonal adult individuals
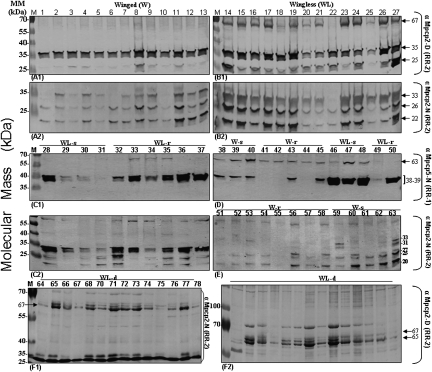
Cuticular proteins (CP) were initially used for the comparison of individual aphids as morphs are predicted to have an external CP skeleton composition that is adapted to their respective behavioral tasks (such as flight for winged aphids), as well as physiological responses to physical stress like desiccation. Insect cuticles comprise chitin fibers (poly N-acetyl-D glucosamine) embedded in a matrix of proteins (Neville 1975). The relative proportion of chitin and CP determines the rigidity and flexibility of the cuticle (Neville 1975). Insect cuticular proteins were divided into two major groups, RR-1 and RR-2 (Andersen 1998), which were later defined by the presence of consensus chitin binding domains (RR1 [Togawa et al. 2004] and RR2 [Rebers and Willis 2001]). Several cuticular genes (and proteins) have been identified and isolated from several aphid species (Dombrovsky et al. 2003, 2007a). In our current study, total cuticular proteins were extracted from individual aphids, and one-half of the individual CP extract (Fig. 2A1,A2,B1,B2,C1,C2,F1,F2) was tested in parallel using antibodies against the native and denatured forms of the RR-2 or RR-1 conserved motifs (Dombrovsky et al. 2007a,b). To assure a comparative analysis only gels showing equal protein staining in each track were kept for analysis. The unchanged band, like the 35-kDa band in Figure 2, A1 and B1, obtained with one antibody (against denatured RR-2) was used as an internal standard for evaluating differences in bands obtained with the other antibody (against native RR-2) in Figure 2, A2 and B2. Variations of molecular profiles are convincingly illustrated in Figure 2E, where bands are missing or expressed depending on the tested aphids. Strong differences in intensity of bands were also observed between aphids (winged or wingless) using the antibody against RR-1 (see Fig. 2C1,D). Finally, we found that unchanged bands in one series can be highly variable in another: for instance the 25-kDa band obtained for white aphids generated in declining plants (Fig. 2F1) is stable (used as internal control), but highly variable in pink aphids raised under optimal plant conditions at 20°C (in Fig. 2A2,B2,C2,E). In contrast the 35-kDa band stable in Figure 2A1,B1 highlights the variability in the 25- and 67-kDa bands in the same gels. Interestingly, our data show evidence of variability within the winged or wingless categories and/or within walker (r) or sedentary (s) wingless behavioral groups, although correlation between molecular profiles and subcategories seems very weak (Fig. 2C1,C2,D,E).
Figure 2.
Cuticle protein profiles of individual A. pisum adults. (A–F) Extracted proteins from single aphid samples were separated by SDS-PAGE (A–E, 15%; F, 12%). Western blot analysis was then performed using four different antibodies raised against M. persicae cuticle proteins MpCPAb (1:1500). The extracts from each single aphid were resolved by equal loading onto duplicate gels. Therefore, each track within the paired gels (A1,A2,B1,B2,C1,C2,F1,F2) corresponds to one-half of the extracted protein sample from the same individual aphid. Only gels showing identical protein loading, as determined by densitometry after red Ponceau staining, were used in the analysis. Moreover, aliquots were analyzed by blue coomassie staining in parallel to assess the exact amount of protein loading in each track. The paired gels were then subjected to Western blot analysis using either the (N) or (D) αMpcp2 antibody (produced in rabbit against the native (N) or denatured (D) forms of related cuticle proteins). The same procedure was carried out using the (N) or (D) αMpcp5 antibodies. RR-2/RR-1 are the chitin binding domains of cuticle proteins used to produce the antibodies Mpcp2 and Mpcp5, respectively. Seventy-eight aphids were tested individually and gels without numbers correspond to half of numbered samples (A1,A2,B1,B2,F1,F2). W-s/WL-s, sedentary aphids (winged and wingless); W-r/WL-r, rover aphids (winged and wingless) that show exploratory behavior. The profiles A–E were obtained from pink aphids (using optimal plant conditions for propagation) and (F1,F2) from white aphids, WL-d, wingless raised on declining plants. MM, molecular mass markers.
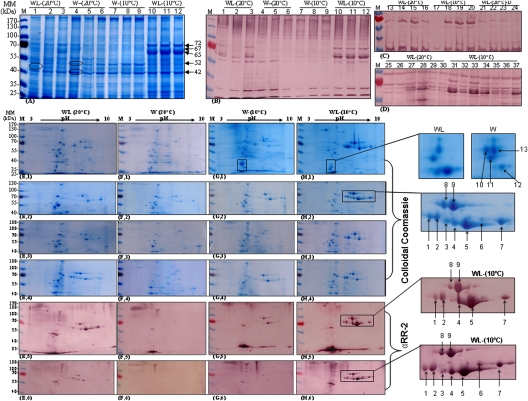
CP profiles of each individual aphid raised at 10°C or 20°C were examined further by two-dimensional (2D) gel electrophoresis. The wingless aphids raised at 20°C manifested strong variations related to the expression levels of three major protein bands with the estimated molecular masses of 65, 67, and 72 kDa, whereas their sister siblings raised at 10°C showed high expression of all of these protein species (Fig. 3A,B). In contrast, these three CP isoforms were found at low abundance in aphids maintained on declining plants (Fig. 3C,D). Significant differences were again observed between the groups and strong variability was evident within groups. The most variable region at 20°C was located on the gel in the range of 60–75 kDa at a pI from 7 to 9 (Fig. 3E–H). In contrast, the wingless aphids propagated at 10°C exhibited a constant pattern of high protein expression in this area of the gel (Fig. 3[E,1–E,4]–[H,1–H,4]). The highly variable proteins numbered one to nine in the higher magnification insets in Figure 3 were characterized as CP proteins based on their positive detection with MpCPAb, (Fig. 3[E,5–E,6]–[H,5–H,6]) and by amino acid sequencing via LC-MS-MS. Interestingly, the analyses consistently confirmed higher variation in the wingless morphs rather than in the winged ones, regardless of the environmental conditions in which they were raised. However, variability was more evident under optimal conditions than in extreme conditions.
Figure 3.
Variability of CP expression in individual A. pisum adults and its correlation with temperature. (A–D) A. pisum cuticle protein extracts (ApCPE) were separated by 1D 12% SDS-PAGE. (A) Coomassie staining. (B–D) Western blot analysis using antibodies raised against M. persicae CP (1:1500); (B,C) αMpcp2(D) antibody; (D) αMpcp2(N) antibody. (E,F) 2D separation of ApCPE by 15% SDS-PAGE. pH values are shown at the top and ranged from 3 to 10. ([E,1–E,4]–[H,1–H,4]) Gels were stained with colloidal coomassie blue (Sigma) and individual protein spots were then selected (numbered 1–9 and denoted by arrows) for subsequent protein sequencing by LC-MS/MS. ([E,5,E,6]–[H,5–H,6]) Western blot analysis using the αMpcp2(D) antibody. In C, WL (20°C)-D indicates aphids raised on declining plants.
These results suggest that there is a high level of molecular variability. Furthermore, some aphids in the wingless category were found to resemble winged aphids. In addition, the molecular profiles resulting from cold conditions were evident in the wingless collection propagated at 20°C. Winged molecular profiles were also found in the wingless collection at 20°C without the corresponding visible traits (Figs. 2, 3).
The possibility that other protein families beside cuticle proteins are equally variable was investigated. We wished more precisely to investigate if proteins of developmental importance were affected due to the fact that the environment can influence the phenotypic fate of embryos. The expression patterns of the evolutionarily conserved extracellular signal-regulated kinases 1 and 2 (ERK1/2) were examined as a marker of cell signaling in development (see review Nishimoto and Nishida 2006). As indicated in the literature, experiments using microarray technology applied to aphid transcriptome analysis have revealed that kinases are affected by the environment and light (Le Trionnaire et al. 2007). Also strong differences were once again observed between the winged and wingless groups and huge variations were evident mainly for the 57-kDa band (Supplemental Fig. S1A,C,F). Patterns of molecular polymorphism were also observed from one series of wingless aphids to another where the 57-kDa band is present or absent. We noted that some wingless aphids showed the same profile as those of winged aphids (Supplemental Fig. S1, A vs. C). The patterns obtained with adult head extracts or new born progenies are also heterogeneous (Supplemental Fig. S1D,E).
Distinct behavioral and physiological traits between clonal individuals
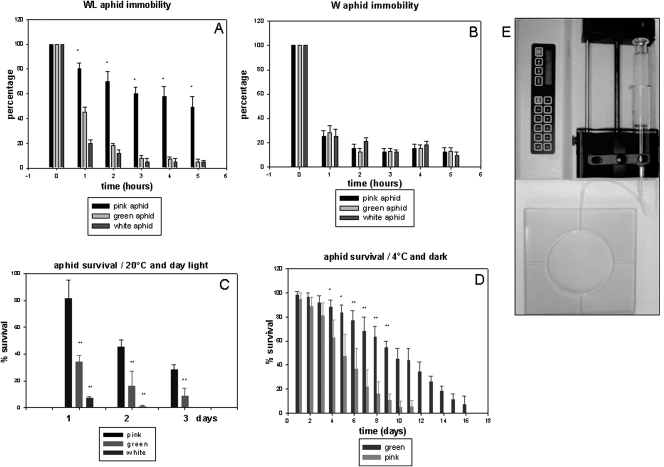
The strong diversity of expressed cuticular proteins between asexual individuals in conditions where the genome is likely unchanged prompted us to investigate whether these differences might be correlated with a large range of life history traits. To investigate this hypothesis further we took advantage of progenies born from a single founder mother and raised them in three sets of distinct environmental conditions. In Acyrthosiphon pisum colonies, the cuticle is usually associated with variable pigmentation between individuals depending on population density, temperature, and state of the plants. The fact that a predominant pigmentation takes over in extreme conditions, like 10°C or declining plants at 20°C after a few generations, was an unexpected opportunity to unravel the physiological and behavioral differences associated with cuticle colors. We constantly observed variability in cuticle pigmentation in aphids raised under optimal conditions (mixture of pink and yellow individuals on the same plant stem with rare green phenotypes). White aphids were mainly obtained on declining plants i.e., poor nutritional conditions, and green aphids, characterized by a green abdomen and a pink head were selected and propagated at 10°C. These phenotypic variants were tested in a trajectometry device using olfactory stimuli to measure the exploration/walking behavior trait. Significant differences between aphid categories were observed (Fig. 4A–E). These results show that a long-lasting immobilization of pink wingless aphids was obtained by air saturation with plant odorants (Vicia faba) and that the opposite exploration and/or mobility traits were triggered in green and/or white wingless aphids by the same odorants (Fig. 4A). The winged aphid categories (white, pink, and green) show indistinguishable levels of induced mobility (Fig. 4B).
Figure 4.
Behavioral studies of A. pisum aphid subclones. (A) Ninety-nine adult wingless aphids (33 white, 33 green, and 33 pink) were placed in an arena and exposed to saturating odors generated by Vicia faba plant extracts as described in Supplemental material. When animals were immobilized, the arena was opened and the time course of the percentage of insects that remained immobile was calculated. Experiments were repeated eight times and the bars indicate the SD. *P < 0.001. (B) The same experiment was performed as described in A except that the three phenotypes analyzed (pink, green, and white) are winged. (C) Aphid survival under conditions of starvation at room temperature. The adult green aphid (induced and propagated at 10°C), the adult pink aphid (propagated at 20°C), and the white aphid (propagated on declining plants at 20°C) were placed at room temperature without a host plant. The mortality rates were then measured every 24 h. This experiment was carried out with 60 aphids and repeated 10 times. The results are presented as a percentage of surviving animals. SD, standard deviation. **P < 0.001. (D) Aphid survival in starving and cold conditions. Adult aphids (pink, green, and white) were placed in an incubator at 5°C in the dark without a host plant and mortality was again measured every 24 h. This experiment was carried out using a group of 60 aphids and repeated 10 times. The results are presented as the percentage of surviving animals. SD, standard deviation. *P < 0.01; **P < 0.001. (E) Photograph of the apparatus and system used for behavioral studies (pusher, syringe, and arena).
To investigate further the physiological differences that might occur between these cuticular variants, life expectancy in extreme conditions was tested. A simple experiment was carried out that consisted of measuring the survival rate of aphids placed at 4°C and in the dark without food. The green wingless variants (raised at 10°C) could survive a long period (LD50, 10 d) whereas the pink variety (raised at 20°C) died much sooner (LD50, 5 d) (Fig. 4D). On the other hand, comparisons of the survival ability revealed that green wingless variants died quickly without food at 20°C under seasonal photoperiodicity conditions (LD50, 14 h) and their pink counterparts survived for longer (LD50, 48 h) (Fig. 4C). Therefore, we conclude that the differences within the cuticular pigmentations are linked to specific behavioral and physiological traits.
Strong variability in CpG island methylation patterns within the aphid genome
We reasoned that if variants are generated stochastically and/or in a partially oriented mode by environmental conditions, early alternative mechanistic switches might dictate the developmental fate of embryos. The CpG island methylation pattern was probed to test whether asexual reproduction involves active enzymes capable of generating epigenetic modifications on DNA. The methyl-dependent endonuclease McrBC was used to generate DNA fragments and in parallel, pull down experiments of MseI genome fragments were carried out using an engineered hybrid protein with the transcription motif recognizing methyl CpG Island and a tag for precipitation (see Methods). A version of this later procedure allowed us to retrieve numerous sequences restricted to heavily methylated CpG islands (at least six methyl groups per fragment). The comparison of the extensive collection of heavily methylated fragments from green and pink adult aphids is shown in Supplemental Table S1.1. The associated genes were identified by Blast searches of sequences, first against the aphid genome database in order to obtain the extended sequences, then against the NCBI database (Supplemental Table S1.1). By comparing the heavily methylated fragment collection, which was retrieved in these conditions, we observed that the list of identified sequences did not overlap between the two categories of aphids (pink and green) (Supplemental Table S1.1).
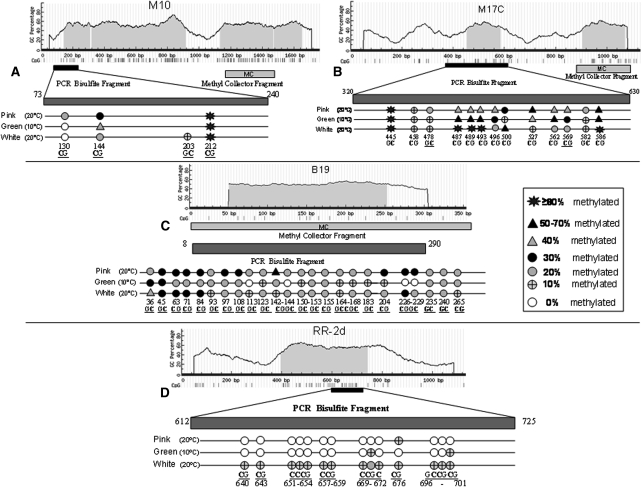
To investigate this phenomenon further, genome bisulfite treatment was carried out and a few putative heavily methylated sequences retrieved by software prediction were sequenced in order to localize the methyl groups. The methylated/unmethylated ratio on each site was also determined (see Methods; the primers listed in Supplemental Table S1.2). The intensity of methylation on each CpG island was found to vary drastically between the three categories: pink, green, and white aphids. Variability was also observed within each category when analyzing a few methylated fragments (Fig. 5A–C; Supplemental Figs. S2–S5). The cuticular genes showed variable degrees of methylation within their coding regions and the methylated form of one member of the RR-2 family (RR-2D) was found to correlate with the white phenotype (Fig. 5D). Methylation of cuticular protein genes at a low level (which is compatible with high degree of methylation in few cells) is shown in Supplemental Figure S5.
Figure 5.
Bisulfite sequencing of selected genomic fragments. Bisulfite sequencing of the methylated region of the affinity precipitated M10 fragment gi|171789667: (A) 1315–1713 Contig56007; (B) the affinity precipitated M17C fragment gi|171824612: 8609–8756 Contig21050; (C) the affinity precipitated B19 fragment; (D) the selected fragment encoding for RR-2 cuticular protein located at contig124 position 21,493–22,627. The prediction of the CpG Island (shown as light gray shading) was performed by software analysis and primers were designed for bisulfite sequencing analysis using the MethPrimer program. Quantification of methylation levels was performed by direct bisulfite sequencing of 10 independent clones for A,C, and D, and 20 independent clones for B.
Phenotypes induced by methyl transferase inhibitors
To further investigate methylation in the aphid genome, the effects of methyltransferase inhibitors were tested in terms of the growth rate of larva stages and adult population, morph distribution, and phenotypic traits. Two types of inhibitors were used: (1) Zebularine (Sigma) which has long-lasting effects due to the fact that it is an analog of cytosine, becoming incorporated into the genome, and inactivating methyl transferase molecules by direct covalent binding (Cheng et al. 2004) and (2) RG108 (Calbiochem), a competitive inhibitor acting at the level of the catalytic site of the methyl transferases, which generates brief periods of inhibition (Brueckner et al. 2005). These inhibitors were injected into pink, green, and white adult aphid mothers and the induced phenotypes/morphotypes were analyzed in the successive clonal generations.
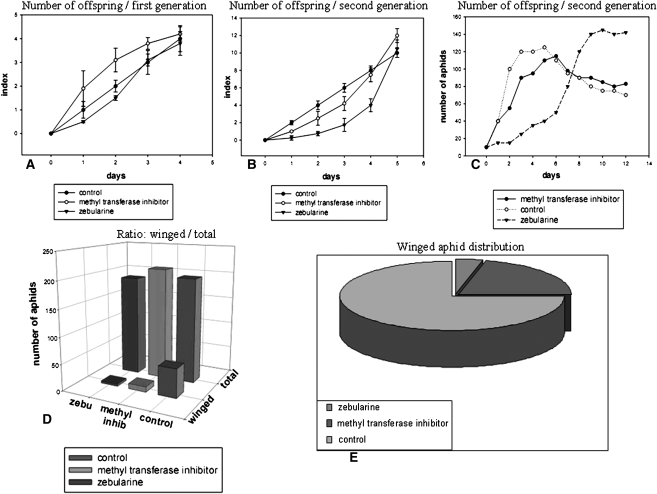
Regarding pink aphids, we observed that Zebularine had a strong effect upon the growth rate, which was found to be delayed in the second generation (Fig. 6). However, after a 10-d time course of the aphid cycle, the growth rate of the population was consistently higher and the pigmentation became pink/red (Fig. 6). The winged morphs significantly disappeared after treatment. Moreover, this inhibitor synchronized the populations of first-generation progenies after 12 d with an inversion of the population ratio of the four developmental stages (Fig. 7). In contrast, the RG108 inhibitor induced moderate effects with the same orientation. Interestingly, when performed on green aphids (raised at 10°C) these pharmacological tests showed no significant differences in terms of growth rate of population, pigmentation, and ratio wing/wingless versus the control (data not shown). Surprisingly, when an adult white aphid (raised on declining plants) was injected, these inhibitors caused a high mortality in the first generation, which was not seen in either pink or green aphids (data not shown). These results argue in favor of a strong impact of CpG island methylation upon growth rate, morph distribution, pigmentation, and consequently, physiological and behavioral traits.
Figure 6.
DNA methyl transferase inhibitors on polyphenism and number of offspring. Mature parthenogenetic wingless adults were injected separately with two different inhibitors of DNA methyl transferases. The first, zebularine, is a cytosine analog that becomes integrated into the genome. The second, RG108, blocks the active site of these enzymes. Wingless adult aphids were injected with 10 μL of zebularine solution (50 μM) and/or 10 μL of RG108 (10 μM). (A) After 24 h the progenies of injected mothers were counted over a period of 5 d. Ten mothers for each category (control, RG108, and zebularine) were analyzed in parallel and the experiment was repeated three times. Index represents comparative ratio of offspring growth. Values are the mean ± SE. (B) After reaching adulthood the progenies of the first generation were placed on a new plant and “granddaughters” were counted over a period of 5 d. Ten aphids were used in parallel and the experiment was repeated three times. Index represents comparative ratio of offspring growth. Values are the mean ± SE. (C) Five adult aphids of the first generation (daughters of control, zebularine, and RG108-injected aphids) were placed on a new plant in parallel and the progenies were counted each day. The experiment was repeated four times and the graph represents the cumulative numbers. (D) Winged progenies of the second generation of drug-injected aphids were counted versus the total. (E) Ratio of the winged animals in control and drug-injected aphids. Ten drug-injected aphids were placed on plants and the cumulative number of winged animals was counted for the next three generations. The ratio is drawn from three cumulative separate experiments.
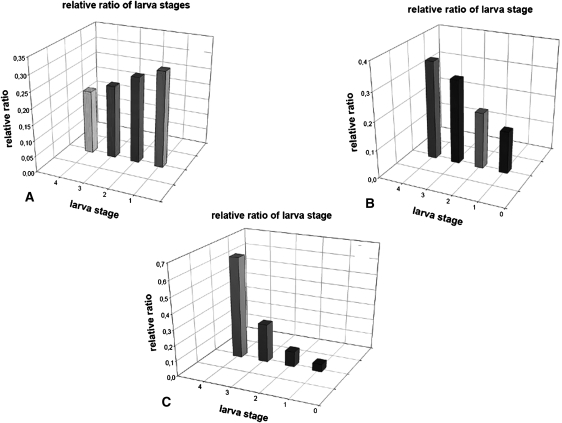
Figure 7.
DNA methyl transferase inhibitors effects on developmental stages distribution. Aphid larval development is characterized by four stages between birth and adulthood. The graphs represent the relative proportion of the number of individuals in each larva stage category. Counting was carried out from day 12 to 15 after injection of 10 aphids placed on the same plant. The values are the mean of three separate experiments. For stage 4, mean ± SE: (A) 0.15 ± 0.05; (B) 0.3 ± 0.1; (C) 0.58 ± 0.1. P < 0.005 for B and C versus the control A (Student's t-test). (A) Control; (B) RG108 inhibitor; (C) zebularine.
Neuropeptides as putative sensors of DNA methylation status
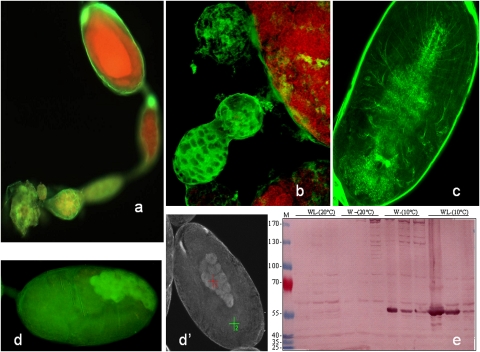
Due to the fact that our data point to methylation as a mechanism for the molecular variability in genetically identical aphids, we looked at possible candidates that trigger the fate deciding mechanism. Previous work has shown that the fate of aphid morphs seems to be decided in the stem cells of old embryos inside the ovariole of the mother (Blackman 1987; Miura et al. 2003). This means that the neurosensorial systems of the mother guide the phenotypic fate two generations after their exposure to particular environmental cues. The positive labeling of the ovariole envelope with anti-HRP antibodies against the insect neuronal marker (Jan and Jan 1982), especially at the embryo junction connections, might suggest its neuroectodermal origin, as this antibody labels the neuronal network within the old embryos in the ovariole (Fig. 8a–c). Interestingly, the allatostatin antibody labels a distinct cluster of cells in older embryos (Fig. 8d,d′). Western blot analysis showed strong variations in the processing of allatostatin precursors (major differences in the levels of intermediate by-products with neuropeptide motifs could be seen) between the green (selected at 10°C) and pink (optimal conditions at 20°C) aphid (Fig. 8e). This is likely to have played a role in determining the final contour of our observed repertoire. Although, to test the neuropeptide hypothesis in more detail, the regulation of the transcription of these genes will require the investigation of DNA methylation in their promoters.
Figure 8.
Immunohistological analysis of neuropeptides in aphid ovarioles. Dissected wingless adult ovarioles and embryos were fixed in 70% ethanol and overlaid with one of the following antibodies: anti-HRP or allatostatin followed by FITC second antibody labeling (see Supplemental material). (a) An intense fluorescent signal is evident in the ovariole envelope when stained with anti-HRP, and this pattern highlights the structural cells of the germarium (yellow). (b) Confocal microscopy shows strong fluorescent labeling of the germarium and the individualization of the first embryo. (c) Neuronal network inside an embryo labeled with an antibody anti-HRP recognizing insect neurons. (d,d′) Labeling with anti-allatostatin. The FITC fluorescent probe was confirmed by absorption spectrum in spectrometry. (e) The extracted A. pisum proteins were analyzed by a Western blot with antibodies against allatostatin (1:1000). (W) Winged aphid; (WL) wingless aphid; (M) molecular mass markers.
Discussion
In cyclical parthenogenic aphids, the genetic variability in a colony of clonal individuals found on a single plant appears extremely low. The genetic identity seems to be the consequence of founder effects involving a short number of generations. Genetically different clones are found mostly in large geographical areas and appear to be generated by the previous cyclic sexual phases (Dixon 1973, 1998). The embryos resulting from fertilized eggs are expected to exhibit genetic variability by independent sorting of parental chromosomes and recombination at the meiosis stage, which will be transferred to the asexual lineage next spring. It is largely accepted that at this stage any modification of the genome seems very low (Hebert 1987; Brisson et al. 2007). Asexual populations have an intrinsic property to grow at high rates. In theory, the asexual lineage exponentially doubles the number of its population each generation compared to the sexual lineage that produces only half of this number (De Jong 1980; Maynard Smith 1990; Haday and Becker 2007). This explains why strong clonal population growth occurs mainly when resource conditions are optimal, increasing the chance of outcome of rare phenotypic variants if these are stochastically produced.
Therefore, the clonal individuals obtained over a limited number of generations from a founder mother, offers the opportunity to address the question of variability within the same colony, based on protein expression profiling and behavioral/physiological analysis. The reported experiments herein allowed us to investigate the range of stochastic variation and the environmental influences that restrict variability in selecting the profiles presenting the best fitness. We found that variation goes far beyond the morph categories and is in accordance with a repertoire of molecularly distinct aphids, even when the morphological traits are identical. This diversity, highlighted by our experiments, is obviously advantageous in a fluctuating environment.
Our data suggest strongly that the external conditions manipulate the ratio of phenotypes within the repertoire without ever abolishing any of them. This does not exclude alternative mechanisms that could generate some particular variants. For example, some of them might be stochastically produced at the same rate, but extreme conditions would recruit one phenotype, while the others would be eliminated after birth. On the other hand, the role of environmental cues that influence embryonic fate commitment to increase the rate of a given morph is well documented (Braendle et al. 2006; Brisson and Stern 2006). However, our data also suggest that multiple mechanisms might be intertwined due to the fact that the fate of embryos is likely decided in the grandmother (two generations after environmental cue exposure) and the window of plasticity in the development process of A. pisum is still unknown (Dixon 1998; Brisson et al. 2007) (emerging embryos are present in old embryos inside adult aphid, which inspires the term of telescopic generations; Dixon 1998). However, in other aphid species (Myzus persicae, Aphis gossypii) post-natal plasticity regarding morph fate commitment has been reported (Dixon 1998). Whatever the external conditions in which our aphids were raised, the variants constitute a repertoire that is never abolished, although some profiles turned out to be statistically better represented and others arose only rarely. Our data strongly suggest that a single genotype reproduced clonally is a powerful system for generating distinct individuals in terms of their physiological, behavioral and molecular identities. The complexity of such a repertoire appears enhanced under optimal environmental conditions and reduced in extreme conditions, such as declining resources, high population density, or cold temperatures.
To gain more information regarding the observed molecular variability in protein expression, the epigenetic modification of CpG islands in the aphid genome was investigated. We predicted that this was the most probable link between environmental cues and the numerous observed cuticular phenotypes. Therefore, we reasoned that phenotypic variability might come from covalent modifications of cytosine by methylation. The presence of the gene encoding for DNA methyl transferase-2 (Dnmt-2) within the aphid genome and EST data bank, as well as an isolated clone from our laboratory raised A. pisum (A Dombrovsky, TN Ledger, and A Robichon, unpubl.) was consistent with this hypothesis. Our current analyses show that some identified CpG islands are heavily methylated and that the statistical presence of the methyl group on each site is partially controlled by the environment. Furthermore, the inhibition of DNA methyl transferase by injection of drugs into pink aphid mothers induces changes in distribution patterns of the granddaughter generation, mostly eliminating the winged component and modifying the pigmentation of the aphids. These data strongly suggest that variant selection by environmental conditions is the result of recruitment of DNA sites for methylation. Consistently, some DNA fragments were found to be heavily methylated in white aphids and poorly methylated in green aphids. Interestingly, a strong methyl transferase inhibitor-induced phenotype was obtained in the pink aphids, which correlates with a higher degree of molecular diversity in this strain.
To further investigate these epigenetic events, we designed experiments in order to assess how environmental cues might influence and/or decide embryonic fates downstream of DNA methylation events. We postulated that the neurosensory circuitry might act as the link between external stimuli and aphid embryogenesis. Allatostatin is known to inhibit the biosynthesis of the juvenile hormone (JH) in the cockroach Diploptera punctata corpora allata (Pratt et al. 1991). Corazonin, which at the moment has not been found in the aphid genome, has been demonstrated to induce the passage from a solitary to gregarious phase in the locusts S. gregaria and L. migratoria and to stimulate ecdysis triggering hormones (ecdysone) (Tanaka 2001). We present evidence of drastic changes in allatostatin, induced by temperature shift, suggesting that the level of neuropeptides might well be the trigger through which external stimuli, via DNA methylation, influence alternative phenotypic outcomes in embryogenesis.
Gene recombination is not likely a process through which phenotypic diversity is produced in clonal reproduction. Although, the phenomenon of recombination in asexual reproduction has been described in eukaryotes like Candida albicans (Gräser et al. 1996), it is a rare event and not likely to occur here. The level of genetic variation in such cyclic parthenogenetic populations is enhanced by the sexual phase, but the existence and persistence of clonal diversity in local populations of obligate parthenogens proves that the coexistence of clones is not necessarily the consequence of such sexual recruitment (Hebert 1987). The general acceptance that asexual reproduction leads to an unchanged genome lacking efficient DNA repair mechanisms, whereas sexual reproduction acts as a powerful tool driving genomic diversity and concomitantly repairs DNA, might need to be reevaluated. However, in support of this contention, a previous study has reported that the asexual lineages of the microcrustacean Daphnia pulex accumulate deleterious amino acid substitutions in proteins at a fourfold higher level than in the sexual lineage (Paland and Lynch 2006). In the aphid, clonality might also generate diversity partly due to “mistakes” like DNA mutations and/or a high level of errors in protein synthesis, which should allow the environment to eliminate the unfit profiles and select others. One of the functions of the sexual phase might be to reset the genome integrity.
On the other hand, comparative analysis of the variability of morphometric traits has been documented in other species like the grasshopper Warramaba virgo in closely related parthenogenetic and bisexual species (Atchley 1977; Vorburger 2006). The parthenogenetic taxa in this species appears to be more often variable than the sexual species regarding the morphology (Atchley 1977; Vorburger 2006). In contrast, no significant variability in morphometric traits within A. pisum morph categories was observed through our observations in our laboratory. In connection with our work, morphological variability in the cyclical and obligate parthenogenetic aphid M. persicae was not observed as a temperature effect despite the phenotypic differences found in relation with the cold climate tolerance (Vorburger 2004). At this stage of our knowledge only few reports deal with the genetic and molecular basis of aphid diversity. However, some transcriptomic differences between morph categories, but never between individuals of the same category, have been reported using microarray technology (Lushai and Loxdale 2002; Braendle et al. 2006; Ghanim et al. 2006; Brisson et al. 2007).
In this study, our data reveal the extent of molecular diversity in clonal individuals presenting definitively acquired traits that appear to constitute an identity for their lifetime. The asexual phase in this bimodal system of reproduction seems to generate a surprisingly high level of diversity. This might act like a powerful mechanism to recruit rare profiles in harsh conditions and to restart a heterogeneous population, which should constitute an immense advantage in a fluctuating environment. Studies in aphids, to date, have highlighted their impressive adaptability and survival capability in unstable and unpredictable environments, which probably explains the persistence of this species for more than 200 million years (Brisson and Stern 2006).
Methods
Maintenance and propagation of aphids
The “Pea Aphid” Acyrthosiphon pisum belongs to the order Homoptera Family (Aphididae). Aphids were maintained on Vicia faba plants in cages in an incubation room at 20°C and a photoperiodicity of 16/8 h light/dark, respectively. Green and pink aphids are presented in Supplemental Figure S6.
Extraction of cuticular proteins from a single aphid
The extraction of cuticle proteins (CP) was based on a procedure developed for other insects and adopted for the aphid (see Supplemental material). Several modifications were introduced to adjust this CP extraction for a single aphid as follows: Each single aphid (adult) was starved for 2 h, then ground separately in an individual Eppendorf tube with 500 μL of a buffer (a low-salt mix, designated TBS-L) containing 20 mM Tris-HCl, 0.15 M NaCl pH 7.5, 3 mM PMSF (Sigma), and 10 mM β-mercaptoethanol (Sigma).
Western blotting analysis and two-dimensional gel electrophoresis
Antibodies against Myzus persicae cuticular proteins were generated as follows: two genes encoding M. persicae CP, Mpcp5 with an RR-1 consensus site (accession no. DQ108939) and Mpcp2 with an RR-2 consensus site (accession no. DQ108935) were expressed in Escherichia coli (see details in Supplemental material). These antibodies were used in Western blot analysis after performing one- or two-dimensional gel electrophoresis on aphid protein extracts.
For amino acid sequencing, the spots corresponding to cuticle proteins were cut from 2D gels after identification. Cuticle protein samples were prepared as indicated above except that the final extraction step was carried out with 150 μL ReadyPrep 2D starter kit (Bio-Rad) for 2D gel electrophoresis analysis (see details in Supplemental material).
Behavioral analysis
An arena was designed (30 cm diameter and 7 mm in depth) as indicated in Figure 4. This system was set up to generate a gradient of odorants in a controlled manner inside the arena. The plastic structure of the system contains four channels (2 mm/diameter), one of which is connected to the odorant source. This structure is placed in a sandwich between two glass plates. Plants (Vicia faba) were ground up in a glass homogenizer and the extracted liquid (5 mL) was loaded into a 30-mL glass syringe and used to generate the odor. The air in the syringe, connected to the arena by a capillary, was injected using an automatic syringe pusher at a speed of 5 mL/min to first create a gradient and then to saturate the arena atmosphere.
Cloning and sequence analysis
PCR products, McrBC digested genomic DNA fragments, and the DNA fragments obtained by methyl-collector affinity precipitation were cloned into the pGEM-T-easy plasmid (Promega) or into the pUC19 vector (Fermentas). These plasmids were then transformed into E. coli DH10B cells (Invitrogen), extracted with a plasmid extraction kit (Qiagen), and sequenced (at GATC). The nucleotide and predicted amino acid sequences were determined and aligned with BioEdit software. Sequence homologies were compared using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/) and also BLAST searches against the HGSC genome data that are available for the pea aphid genome, A. pisum (http://www.hgsc.bcm.tmc.edu/blast.hgsc?organism=2; R Gibbs and S Richards, pers. comm.).
Isolation of genomic DNA from aphids
Total DNA was extracted from a single aphid or by grinding 100 mg of frozen A. pisum using a genomic DNA extraction kit (Qiagen). The resulting genomic DNA was used for subsequent experiments.
Methyl cytosine detection by McrBC
Total DNA extracted from A. pisum was digested by McrBC endonuclease (New England BioLabs), which cleaves DNA only at the sites containing methylcytosine on one or both strands.
Methyl-collector methods
Total DNA was extracted from A. pisum and was fragmented by MseI digestion (New England BioLabs), which leaves the CpG island intact and enriches for short DNA fragments with the potential to contain several methylated cytosines. The fragmented DNA was then applied to affinity precipitation (based on the recombinant protein containing a methyl DNA binding domain and 6xHis tag) using a methyl-collector kit (Active Motif). A protocol provided by the manufacturer allowed the isolation of specifically heavily methylated fragments. These sequences were then analyzed and checked for CpG island prediction with the MethPrimer tool (http://www.urogene.org/methprimer/) (Li and Dahiya 2002). Candidate genes putatively regulated by methylation were identified before bisulfite conversion and sequencing.
Bisulfite genomic sequencing
In general, 2-μg aliquots of A. pisum gDNA were used for each bisulfite reaction in 10 separate tubes using the methyl detector kit (Active Motif). The reaction conditions were performed to achieve the maximum conversion of unmethylated cytosines to uracil according to the instructions provided by the manufacturer. The 10 samples of bisulfite modified gDNA were then pooled and served as a template for PCR amplification using specific primers. All the PCR products (two to three repeats for each gDNA amplified fragment) were purified (Qiagen) and cloned into pGEM-T-easy plasmids (Promega). The plasmids were then transformed into DH10B competent cells (Invitrogen) and 10–15 independent colonies were used for plasmid purification and sequencing to determine the percentage of methylation on each genomic fragment.
Pharmacological studies using inhibitors of methyl transferase
Zebularine[1-(β-D-ribofuranosyl)-1,2-dihydropyrimidin-2-one 2-pyrimidone-1-β-D-riboside, (Sigma) is a cytidine analog that inactivates methyl transferases by covalent binding to its active site after its incorporation into DNA (Cheng et al. 2004). RG108 [2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-3-(1H-indol-3-yl) propionic acid] (Calbochem) is a cell-permeable, specific DNA methyl transferase inhibitor (IC50, 115 nM for the CpG methylase M.SssI). Unlike the commonly used 5-azacytidine derivative inhibitors of methyl transferases, RG108 directly interacts with the active site of these enzymes (Brueckner et al. 2005). The injections were performed as described in the figure legends.
Acknowledgments
We thank Gilbert Engler for his invaluable technical help and also for his expertise during the immunohistology analyses. We also thank Claude Castella for useful advice during the 2D analysis, and Rene Feyereisen and Yan Veenstra for providing antisera against allatostatin. We thank Rene Feyereisen, Ralph Greenspan, David Stern, Marc Keller, and Benny Raccah for their critical reading and comments on the manuscript. We also thank the Baylor College of Medicine for allowing open access to the A. pisum genome database. This work was supported by an ANR grant “exsidum” and a post-doctoral fellowship from “Ministère de la Recherche et Enseignement Supérieur” to A.D.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.091611.109.
References
- Andersen SO. Amino acid sequence studies on endocuticular proteins from the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 1998;28:421–434. doi: 10.1016/s0965-1748(98)00028-9. [DOI] [PubMed] [Google Scholar]
- Atchley WR. Evolutionary consequences of parthogenesis: Evidence from the Warramaba virgo complex. Proc Natl Acad Sci. 1977;74:1130–1134. doi: 10.1073/pnas.74.3.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balloux F, Lehmann L, de Meeûs T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–1644. doi: 10.1093/genetics/164.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman RL. Reproduction, cytogenetics and development. In: Minks AK, Harrewijn P, editors. Aphids: Their biology, natural enemies & control. Vol. 2A. Elsevier; Amsterdam: 1987. pp. 163–195. [Google Scholar]
- Blackman RL, Eastop VF. An identification and information guide. Wiley; Chichester, UK: 1984. Aphids on the world's crops. [Google Scholar]
- Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity. 2006;97:192–199. doi: 10.1038/sj.hdy.6800863. [DOI] [PubMed] [Google Scholar]
- Brisson JA, Stern DC. The pea aphid Acyrthosiphon pisum: An emerging genome model system for ecological developmental evolutionary studies. BioEssays. 2006;28:747–755. doi: 10.1002/bies.20436. [DOI] [PubMed] [Google Scholar]
- Brisson JA, Davis GK, Stern DL. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid. Evol Dev. 2007;9:338–346. doi: 10.1111/j.1525-142X.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Boy RG, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, Marquez VE, Jones PA. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004;24:1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Kang Zhu J. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12:133–139. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong G. Some numerical aspects of sexuality. Am Nat. 1980;116:712–718. [Google Scholar]
- Delmotte F, Leterme N, Gauthier JP, Rispe C, Simon JC. Genetic architecture of sexual and asexual populations of aphid Rhopalosiphum padi based on allozyme and microsatellite markers. Mol Ecol. 2002;11:711–723. doi: 10.1046/j.1365-294x.2002.01478.x. [DOI] [PubMed] [Google Scholar]
- Dixon AFG. Studies in biology. No. 44, Edward Arnold; London: 1973. Biology of aphids. [Google Scholar]
- Dixon AFG. Aphid ecology. Chapman & Hall; London: 1998. [Google Scholar]
- Dombrovsky A, Huet H, Zhang H, Chejanovsky N, Raccah B. Comparison of newly isolated cuticular protein genes from six aphid species. Insect Biochem Mol Biol. 2003;33:709–715. doi: 10.1016/s0965-1748(03)00065-1. [DOI] [PubMed] [Google Scholar]
- Dombrovsky A, Sobolev I, Chejanovsky N, Raccah B. Characterization of RR-1 and RR-2 cuticular proteins from Myzus persicae. Comp. Biochem Physiol B Biochem Mol Biol. 2007a;146:256–264. doi: 10.1016/j.cbpb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Dombrovsky A, Gollop N, Songbi C, Chejanovsky N, Raccah B. In vitro association between the helper protein of the zucchini yellow mosaic virus and cuticle proteins of Myzus persicae. J Gen Virol. 2007b;88:1602–1610. doi: 10.1099/vir.0.82769-0. [DOI] [PubMed] [Google Scholar]
- Eckert CG, Lui K, Bronson K, Corradini P, Bruneau A. Population genetic consequences of extreme variation in sexual and clonal reproduction in an aquatic plant. Mol Ecol. 2003;12:331–344. doi: 10.1046/j.1365-294x.2003.01737.x. [DOI] [PubMed] [Google Scholar]
- Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 2007;5:e87. doi: 10.1371/journal.pbio.0050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim M, Dombrovsky A, Raccah B, Sherman A. A microarray approach identifies a new ANT and OS-D as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae. Insect Biochem Mol Biol. 2006;36:857–868. doi: 10.1016/j.ibmb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell TG, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida Albicans exhibits both clonamity and recombination. Proc Natl Acad Sci. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadany L, Becker T. Sexual selection and the evolution of obligatory sex. BMC Evol Biol. 2007;7:245. doi: 10.1186/1471-2148-7-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkett F, Plantegenest M, Bonhomme J, Simon JC. Gene flow between sexual and facultatively asexual lineages of an aphid species and the maintenance of reproductive mode variation. Mol Ecol. 2008;17:2998–3007. doi: 10.1111/j.1365-294X.2008.03798.x. [DOI] [PubMed] [Google Scholar]
- Hebert PD. Genotypic characteristics of cyclic parthenogens and their obligately asexual drivatives. Experientia Suppl. 1987;55:175–195. doi: 10.1007/978-3-0348-6273-8_8. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and grasshopper embryos. Proc Natl Acad Sci. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaersgaard CW, Holmstrup M, Malte H, Bayley M. The importance of cuticular permeability, osmolyte production and body size for the desiccation resistance of nine species of Collembola. J Insect Physiol. 2004;50:5–15. doi: 10.1016/j.jinsphys.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Kakutani T. Epi-alleles in plants: Inheritance of epigenetic information over generations. Plant Cell Physiol. 2002;43:1106–1111. doi: 10.1093/pcp/pcf131. [DOI] [PubMed] [Google Scholar]
- Kalisz S, Purugganan MD. Epialleles via DNA methylation: Consequences for plant evolution. Trends Ecol Evol. 2004;19:309–314. doi: 10.1016/j.tree.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Kanbe T, Akimoto SI. Allelic and genotypic diversity in long term asexual populations of the pea aphid Acyrthosiphon pisum in comparsison with sexual populations. Mol Ecol. 2009;18:801–816. doi: 10.1111/j.1365-294X.2008.04077.x. [DOI] [PubMed] [Google Scholar]
- Le Trionnaire G, Jaubert S, Sabater-Muñoz B, Benedetto A, Bonhomme J, Prunier-Leterme N, Martinez-Torres D, Simon JC, Tagu D. Seasonal photoperiodism regulates the expression of cuticular and signalling protein genes in the pea aphid. Insect Biochem Mol Biol. 2007;37:1094–1102. doi: 10.1016/j.ibmb.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Le Trionnaire G, Hardie J, Jaubert-Possamai S, Simon JC, Tagu D. Shifting from clonal to sexual reproduction in aphids: Physiological and developmental aspects. Biol Cell. 2008;100:441–451. doi: 10.1042/BC20070135. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Little TJ, Ebert D. Sex linkage disequilibrium and patterns of parasitism in three species of cyclically parthenogenetic Daphnia (Cladocera: Crustacea) Heredity. 2000;85:257–265. doi: 10.1046/j.1365-2540.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Lushai G, Loxdale HD. The biological improbability of a clone. Genet Res. 2002;79:1–9. doi: 10.1017/s0016672301009582. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Models of a dual inheritance system. J Theor Biol. 1990;143:41–53. doi: 10.1016/s0022-5193(05)80287-5. [DOI] [PubMed] [Google Scholar]
- Miller KJ, Ayre DJ. The role of sexual and asexual reproduction in structuring high latitude populations of the reef coral Pocillopora damicornis. Heredity. 2004;92:557–568. doi: 10.1038/sj.hdy.6800459. [DOI] [PubMed] [Google Scholar]
- Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DA. Comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) J Exp Zool. 2003;295B:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Muller CB, Williams IS, Hardie J. The role of nutrition, crowding and interspecific interactions in wing development in aphids. Ecol. Ent. 2001;26:330–334. [Google Scholar]
- Neville AC. Biology of arthropod cuticle. Springer; New York: 1975. [Google Scholar]
- Nijhout HF. Control mechanisms of polyphenic development. Bioscience. 1999;49:181–192. [Google Scholar]
- Nishimoto S, Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7:782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paland S, Lynch M. Transitions to asexuality result in excess amino acid substitutions. Science. 2006;311:990–992. doi: 10.1126/science.1118152. [DOI] [PubMed] [Google Scholar]
- Pfrender ME, Lynch M. Quantitative genetic variation in Daphnia: Temporal changes in genetic architecture. Evolution. 2000;54:1502–1509. doi: 10.1111/j.0014-3820.2000.tb00696.x. [DOI] [PubMed] [Google Scholar]
- Pratt GE, Farnsworth DE, Fok KF, Siegel NR, McCormack AL, Shabanowitz J, Hunt DF, Feyereisen R. Identity of a second type of allatostatin from cockroach brains: An octadecapeptide amide with tyrosine-rich address sequence. Proc Natl Acad Sci. 1991;88:2412–2416. doi: 10.1073/pnas.88.6.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers JE, Willis JH. A conserved domain in arthropod cuticular protein binds chitin. Insect Biochem Mol Biol. 2001;31:1083–1093. doi: 10.1016/s0965-1748(01)00056-x. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Ronsheim ML, Bever JD. Genetic variation and evolutionary trade-offs for sexual and asexual reproductive modes on allium vineale (liliaceae) Am J Bot. 2000;87:1769–1777. [PubMed] [Google Scholar]
- Stearns SC. The evolutionary significance of phenotypic plasticity. Bioscience. 1989;39:436–445. [Google Scholar]
- Sutherland ORW. The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 1969;15:1385–1410. [Google Scholar]
- Tanaka S. Endocrine mechanisms controlling body-color polymorphism in locusts. Arch Insect Biochem Physiol. 2001;47:139–149. doi: 10.1002/arch.1045. [DOI] [PubMed] [Google Scholar]
- Togawa T, Nakato H, Izumi S. Analysis of the chitin recognition mechanism of cuticle proteins from the soft cuticle of the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2004;34:1059–1067. doi: 10.1016/j.ibmb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Vorburger C. Cold tolerance in obligate and cyclical parthenogens of the pea potato aphid Myzus persicae. Ecol Entomol. 2004;29:498–505. [Google Scholar]
- Vorburger C. Geographic parthenogeneis recurrent patterns down under. Curr Biol. 2006;16:R641–R643. doi: 10.1016/j.cub.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Vorburger C, Lancaster M, Sunnucks P. Environmentally related patterns of reproductive modes in the aphid Myzus persicae and the predominance of two ‘superclones’ in Vectoria, Australia. Mol Ecol. 2003;12:3493–3504. doi: 10.1046/j.1365-294x.2003.01998.x. [DOI] [PubMed] [Google Scholar]
- Walters KEA, Dixon AFG. Migratory urge and reproductive investment in aphids: Variation within clones. Oecologia. 1982;58:70–75. doi: 10.1007/BF00384544. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: Genetic machanisms and the potential consequences of pleiotropy. Semin Cell Dev Biol. 2009;1:65–71. doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Epigenetic inheritance in Arabidopsis: Selective silence. Curr Opin Genet Dev. 2005;15:557–562. doi: 10.1016/j.gde.2005.07.002. [DOI] [PubMed] [Google Scholar]