Abstract
DNA methylation is a vital epigenetic mark that participates in establishing and maintaining chromatin structures and regulates gene transcription during mammalian development and cellular differentiation. Differences in epigenetic patterns between individuals may contribute to phenotypic variation and disease susceptibility; however, little is known about the extent of such variation or how different epigenetic patterns are established. Here we have compared DNA methylation profiles of macrophages from two inbred mouse strains (C57BL/6 and BALB/c) at 181 large genomic intervals that were selected based on differential gene expression patterns. Using a DNA methylation-dependent fractionation approach based on a combination of methyl-CpG immunoprecipitation and locus-wide tiling arrays, we identified several hundred differentially methylated regions, and simultaneously uncovered previously unrecognized genetic variability between both mouse strains at the studied loci. DNA sequence and methylation differences were validated by DNA sequencing and mass spectrometry analysis of bisulfite-treated DNA for a subset of regions. Importantly, we show that in F1 hybrids, the majority of strain-specific methylation patterns in somatic cells were maintained on the parental allele, regardless of their status in the male germ line. The common association of differentially methylated regions with sequence polymorphisms suggests that the genomic context determines the developmentally regulated epigenetic status at most nonimprinted regions of mammalian genomes.
Epigenetic processes are fundamental to the differentiation and development of multicellular organisms by controlling chromatin accessibility and transcription (Bird 2002). Although phenotypic variation is mainly driven by genetic traits, there is also evidence that epigenetic mechanisms may contribute to interindividual phenotypic differences in mammals (Rakyan et al. 2002). Examples for epigenetic differences between individuals are comparatively rare and mostly, but not exclusively, confined to the level of DNA methylation, which represents a relatively stable epigenetic modification that can often be measured in an allelic context. There is well-documented evidence that epigenetic states can be inherited across generations, e.g., at the Agouti viable yellow (Avy) allele in mice (Morgan et al. 1999; Blewitt et al. 2006), and that allelic variation at certain epigenetic modifier genes in mice, like DNA methyl transferases or chromatin remodeling factors may influence the inheritance of CpG methylation patterns in trans (Chong et al. 2007; Blewitt et al. 2008). It has also been demonstrated that supposedly genetically identical, monozygotic twins can show differences at the level of DNA methylation that are acquired during the lifetime of each individual (Fraga et al. 2005; Kaminsky et al. 2009) through largely unknown mechanisms.
In addition to trans regulation, there are several reports demonstrating that DNA sequence variants associate with specific epigenetic states (Murrell et al. 2004; Flanagan et al. 2006; Heijmans et al. 2007). Along this line, a recent study in humans identified several cases of allele-specific DNA methylation at nonimprinted gene loci (Kerkel et al. 2008), where the methylation status of each allele was likely controlled in cis by the local DNA sequence. The published data suggest that three types of inheritance of CpG methylation patterns may exist in vivo: Methylation patterns at nonimprinted loci may be inherited across generations based on genetic mechanisms (in cis and in trans) or based on epigenetic mechanisms. The extent of contribution of each type of mechanism in shaping individual epigenetic or phenotypic differences is currently unknown.
Inbred mice are ideally suited to study the inheritance of an epigenetic mark like DNA methylation, because the sameness of genetic backgrounds within a strain allows for reproducible mating conditions between two inbred mouse strains. To study the inheritance of an epigenetic mark in the F1 generation, one first needs to define regions that are epigenetically different in cells of both strains. However, systematic screens for epigenetic differences in mice have not been performed and only a handful of epigenetically variable regions have been studied so far (Rakyan et al. 2002). In the present study, we performed a combined genetic and epigenetic profiling of strain-specific differences in inbred mice and uncovered more than 400 differentially methylated regions (DMR). The methylation patterns of DMR in F1 hybrids clearly suggest that the majority of allelic differences between individual mammals are determined by cis-acting sequences.
Results
Comparative methyl-CpG immunoprecipitation (MCIp) to dissect genetic and epigenetic differences
We utilized MCIp (Gebhard et al. 2006; Schilling and Rehli 2007; Schmidl et al. 2009) to compare methylation profiles of a defined cell type (bone-marrow-derived macrophages) from two inbred mouse strains (C57BL/6 and BALB/c) that represent prototypic models for Th1- or Th2-dominated immune responses (Locksley et al. 1987; Gessner et al. 1993). A schematic overview of the methodical approach is shown in Figure 1. We chose to analyze macrophages, because they may actually contribute to some immune-related phenotypic differences in these strains, and because they can be grown as a homogenous cell population (usually >98% macrophages) from mouse bone marrow. We expected to find functionally relevant, strain-specific methylation differences with greater probability in regions with differential transcriptional activity, and therefore limited our analysis to whole gene loci that showed strain-specific gene expression in C57BL/6 versus BALB/c macrophages, plus a handful of control regions that were equally expressed (or not expressed) in both cell types. The expression profiling is described in detail in the Supplemental material and examples of qRT-PCR validation of strain-specific differences in gene expression are given in Supplemental Figure S1. Based on the transcriptome analysis, a tiling microarray was designed that covered 28 megabases of the mouse genome and contained 181 genomic segments (with a median size of 110 kb), more than 550 proximal promoter regions and at least 800 genes, which included a number of well-known and immunologically relevant genes like Plau, Marco, Ltb, Il1b, etc. (a complete list of regions is given in Supplemental Table S1).
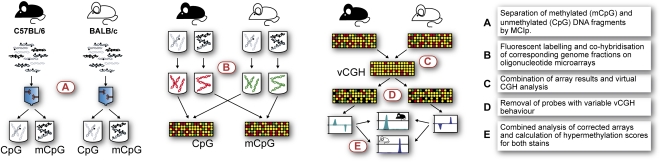
Figure 1.
Simultaneous detection of epigenetic and genetic differences using methyl-CpG immunoprecipitation (MCIp). The experimental workflow is presented schematically. (A) Fragmented genomic DNA from bone marrow-derived macrophages of either mouse strain was fractionated using a MBD-Fc column and separated into methylated (mCpG) and unmethylated (CpG) DNA pools. (B) Both DNA pools are fluorescently labeled and compared between mouse strains by cohybridization on a locus-specific microarray using stringent conditions. (C) Array results are combined in a virtual CGH analysis to detect CNVs and sequence polymorphic regions, that are removed (D) from further analysis. (E) Differentially methylated regions are detected by analyzing remaining array probes for diametrically opposed enrichment behavior between both hybridizations (e.g., a region that is relatively enriched in the unmethylated pool of BALB/c and shows reverse enrichment behavior in the methylated pool is considered hypomethylated in BALB/c).
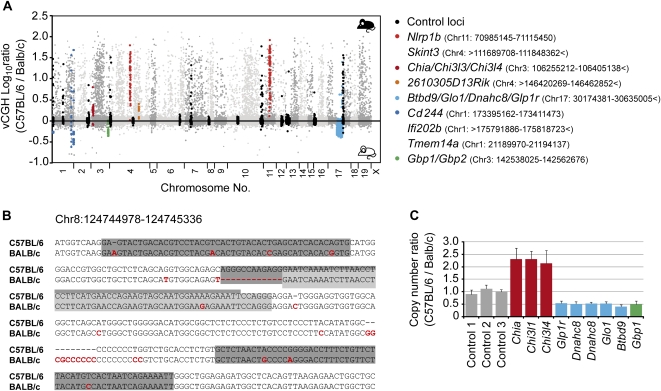
We separated gDNA of macrophages from both strains into unmethylated (CpG) and methylated pools (mCpG) using MCIp and cohybridized the two unmethylated or the two methylated DNA pools to the locus-wide tiling arrays. (Each hybridization was performed twice using independent sample preparations.) As enriched DNA fragments from one strain in the methylated fraction should be depleted in the unmethylated fraction, the signal intensities in CpG pool and mCpG pool hybridizations should complement themselves and thereby allow the identification of differentially methylated regions (DMR). However, the analysis is complicated by the fact that the sequence of microarray probes corresponds to only one strain's genome (the reference genome of C57BL/6) and sequence variations between mouse strains can additionally influence the hybridization efficiency of microarray probes. Since the extent of genetic variation between both strains at the investigated loci was largely unknown, we utilized the fact that we had essentially analyzed the entire genome of both strains in two halves, and recombined the two independent microarray results of each of the two independent biological replicates to obtain a virtual comparative genome hybridization (vCGH) of both strains. As shown in Figure 2A, a large fraction of microarray probes (15.1%) demonstrated preferential hybridization with gDNA from C57BL/6 macrophages, which is most likely due to the presence of sequence variants between C57BL/6 and BALB/c. In line with this, the majority of known SNPs (13,535) overlapped with probe sequences showing unequal hybridization behavior, whereas all other probes associated with relatively few SNPs (2801). We sequenced a representative set of regions that were indicative of sequence variations (the complete list of alignments is given in Supplemental Fig. S2) and found that every affected probe contained at least one, but more often two or three, sequence variations, including SNP, microdeletions, or insertions. The number of probes demonstrating unbalanced hybridization behavior (34,096) was substantially larger than the number of known variations, suggesting that sequences of both strains are far more variable at the studied genomic regions than previously published SNP data (Frazer et al. 2007) suggests. This is in line with an earlier study, suggesting that the published SNP data only capture a fraction of the variations found in the laboratory mouse (Yang et al. 2007). The distribution of known SNP markedly varied across all interrogated regions with 15%–20% of all regions having comparably low or high SNP densities (less than 2/10,000 or more than 15/10,000, respectively; a diagram is provided in Supplemental Fig. S3A). The average SNP density (1 SNP every 1200 bp) on our array was approximately two times higher as compared to the whole genome (1 SNP every 2500 bp), suggesting that differentially expressed genes tend to contain more polymorphisms. Accordingly, the variation detected in our vCGH approach was generally higher in genes that showed differential expression in macrophages of both strains (Fig. 2A, gray dots) than in equally expressed/nonexpressed control genes (Fig. 2A, black dots). In addition, we also identified a number of regions where vCGH signals indicated copy number variations (CNVs) and validated gene duplications for three locations using qPCR (see Fig. 2C). Some of the larger CNVs were previously identified using standard CGH approaches (She et al. 2008), suggesting that the vCGH analysis correctly identified genetic alterations between both mouse strains.
Figure 2.
Detection of genetic variation. (A) The histogram shows a CGH-like presentation of combined signal intensities from the separate hybridizations of methylated and unmethylated DNA pools. Control loci (in black) show relatively few C57BL/6-enriched signals. Two regions (Nlrp1b and Skint, in red) are deleted in BALB/c, two regions (Chia/Chi3l3/Chi3l4 and 2610305D13Rik; in dark red and orange, respectively) are duplicated in C57BL/6, and five regions (Btbd9/Glo1/Dnahc8/Glp1r, Cd244, Ifi202b, Tmem14a, and Gbp1/Gbp2; in blue or green) appear (at least) duplicated in BALB/c. Genomic locations are provided for individual regions. The signs > and < indicate that the affected regions extended over the analyzed area. Data represent mean values of two biological replicates. (B) Sequences of an exemplary region where several probes (boxed in gray) showed C57BL/6-enriched signals. Compared to the reference strain, BALB/c contains several nucleotide exchanges (highlighted in red). (C) QPCR validation of three CNVs (Chia/Chi3l3/Chi3l4, three amplicons; Btbd9/Glo1/Dnahc8/Glp1r, five amplicons; Gbp1/Gbp2, one amplicon) detected by vCGH.
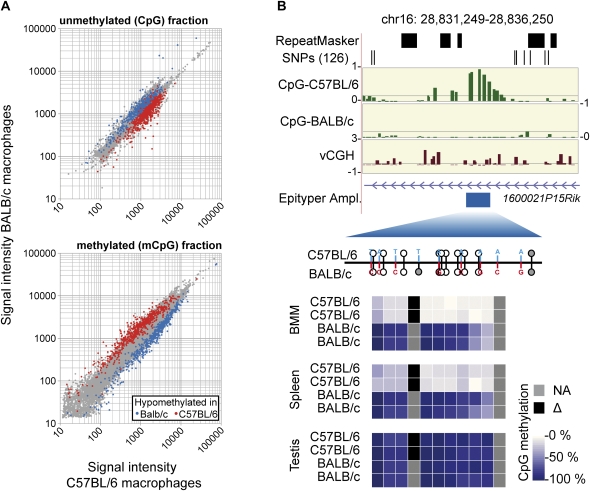
All probes indicating sequence variation were excluded and the residual set was used to identify DMR. A representative scatter plot of vCGH-corrected comparative microarray hybridizations from unmethylated (CpG) and methylated pools (mCpG) is shown in Figure 3A, where microarray probes showing the expected complementary behavior are colored in red (hypomethylated in C57BL/6 BMM [bone marrow-derived macrophages]) and blue (hypomethylated in BALB/c BMM). In total, we identified 435 regions with lineage-specific CpG methylation that were associated with 171 genes. A complete list of DMR, together with expression data of associated genes, is given in Supplemental Table S2. As expected, DNA methylation at proximal promoter regions correlated with lower gene expression levels in all cases, whereas no significant correlation between the methylation and the transcription state was evident at promoter distal (intergenic), or intragenic sites. At the current stage it is unclear, whether promoter distal DMR are functionally relevant and whether they contribute to the observed phenotypic differences between both mouse strains.
Figure 3.
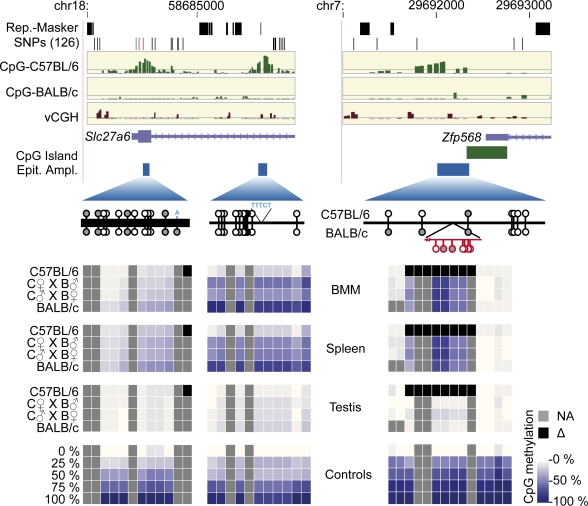
Validation of strain-specific CpG methylation by MALDI-TOF MS of bisulfite treated DNA. (A) Scatter plots of normalized signal intensities from independent hybridizations of methylated (mCpG) and unmethylated (CpG) DNA pools. Probes in differentially methylated regions (colored in red and blue) show the expected intensity distribution (enriched in one pool and depleted in the other one). (B) One example of a DMR detected by the MCIp-microarray approach and validation using MALDI-TOF MS of bisulfite treated DNA (three additional examples are presented in Supplemental Fig. S4). MCIp results are presented in the upper panels. Shown are the following tracks (from top to bottom) that were generated using the UCSC Genome Browser (http://genome.ucsc.edu/): repetitive regions as identified by the RepeatMasker program, single nucleotide polymorphisms from the dbSNP (NCBI database for genomic variation) build 126 (both in black), hypomethylation scores for BMM of both mouse strains (defined as the difference product of log10 signal intensity ratios of both hybridizations; in green), vCGH signals indicating the presence of genetic variation at probe level (in brown), as well as gene structures (in purple) and the position of amplicons (Epityper Ampl., in blue) that were designed for MALDI-TOF MS analysis of bisulfite treated DNA. The relative position of CpGs within amplicons is indicated below by small lollipops (with the upward orientation representing C57BL/6, and the downward orientation representing BALB/c). Sequence variations are highlighted in red and blue, black bars mark the position of exons, and gray lollipops are not analyzed by the MS. Methylation levels of individual CpGs in the indicated cell types (two individuals for each strain) are shown color-coded. The scale ranges from pale yellow (0% methylation) to dark blue (100% methylation), strain-specifically absent CpGs are colored black, nondetectable CpGs are marked in gray.
We also studied correlations between genetic variability and differential methylation status. According to the classifications provided by the Perlegen study (Frazer et al. 2007), 80 of our genomic intervals contained DMR of intersubspecific origin and 66 intervals contained DMR of intrasubspecific origin. Thirty-two regions were predicted to be of the same haplotype. However, the majority of DMR in the latter class were also associated with probes showing unbalanced hybridization behavior in the vCGH, suggesting that most of them actually are of intrasubspecific origin (Supplemental Fig. S3B). In total, about 90% of the identified DMR contained probes that showed unbalanced hybridization behavior in the vCGH. The DMR and SNP count per region correlated (r2 = 0.59) suggesting a link between DMR occurrence and genetic variation between both strains. A corresponding diagram and a box plot on DMR/SNP correlation are provided in Supplemental Figure S3C,D.
Fine mapping of strain-specific DNA methylation differences in macrophages, spleen, and male germ line
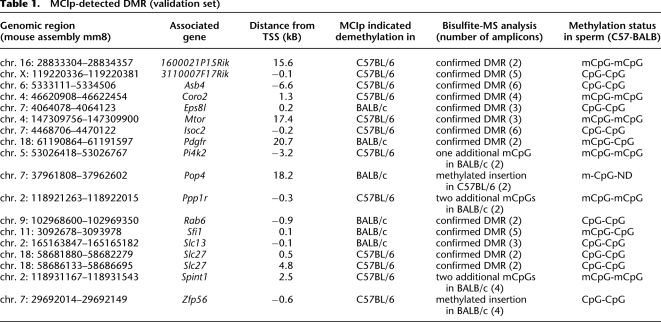
To validate and quantify methylation differences, a representative set of DMR was sequenced in both strains before subsequent analysis by MALDI-TOF MS of bisulfite treated DNA. (For information on sequences and primers see Supplemental Fig. S2 and Supplemental Table S3, respectively.) DNA sequencing indicated that DMR were often associated with strain-specific insertions of CpG-containing repetitive sequences (not overlapping with microarray probes), which were likely methylated and responsible for the observed methylation pattern. We primarily focused our validation panel on sequences showing no repeat variations and performed an initial round of MALDI-TOF MS analysis at 18 selected DMR. (For information on amplicons and complete MALDI-TOF MS results for the initial test set, see Supplemental Tables S4, S5). With one exception (Eps8l1), the selected and validated DMR contained sequence variations between strains. Data for the validation set are summarized in Table 1 and results of representative DMR are shown in Figure 3B and Supplemental Figure S4. Thirteen out of 18 regions showed “true” methylation differences. In the remaining five cases, the differential MCIp enrichment behavior resulted from an increased number of methylated CpGs in one strain, either due to the presence of SNPs (Pi4k2b, Ppp1r14d, Spint1) or due to the insertion of methylated sequences in one strain (Pop4, Zfp568). One DMR-associated gene (Asb4) was previously shown to be paternally imprinted (Mizuno et al. 2002), however, the observed methylation pattern at the DMR was clearly strain-specific.
Table 1.
MCIp-detected DMR (validation set)
To check whether the observed differences related to the artificial BMM culture system and how methylation patterns differed between somatic and germ line cells, we additionally analyzed the two tissues, spleen and testis. The latter was prepared to mainly contain germ line cells (70%–90%) as judged by the predominantly unmethylated status (95%–85%) of the maternally imprinted Snrpn and Mest alleles in these preparations (data not shown). The two biological replicates of each cell or tissue type were highly similar (median r2 > 0.95) at the analyzed regions. The two somatic samples, spleen and BMM, also correlated well within the same strain (median r2 > 0.93), suggesting that the observed patterns are not a cell culture-induced artifact.
Interestingly, the majority (15/18) of the validated DMR only acquired differential DNA methylation in somatic cells (BMM, spleen), but showed similar methylation states in the testis/germ line preparations of both strains. Thus, the differential patterns are likely established after fertilization, and DNA methylation itself cannot be the inherited mark.
Inheritance of DNA methylation differences in F1 hybrids
How, then, are the observed methylation patterns established during development? To address this question, we generated F1 hybrids in both possible mating combinations and compared the degree of CpG methylation at selected loci of F1 hybrids with their wild-type parents.
The initial screen included only male mice. Bisulfite/MALDI-TOF MS analysis of methylation patterns in females suggested that the DMR are equally detected on autosomes of both genders. Methylation patterns in F1 animals were highly similar between samples from the somatic tissues suggesting a minor impact of mating combinations or offspring gender. A complete correlation matrix is given in Supplemental Figure S5. Methylation ratios for all analyzed amplicons and samples are given in Supplemental Tables S6–S8 (BMM, spleen, and testis, respectively). A comparison of microarray and MALDI-TOF MS analysis for representative regions is given in Figure 4 and Supplemental Figure S6.
Figure 4.
Inheritance of stain-specific methylation patterns in F1 hybrids. Two examples (Slc27a6 and Zfp568) for allelic inheritance of stain-specific methylation patterns are shown. In the top panels, hypomethylation scores for BMM of both strains are displayed as described in Figure 3. Averaged methylation levels of individual CpGs were determined by MALDI-TOF analysis at the indicated DMR in BMM, spleen, and testis and are shown color-coded (as in Fig. 3) for parental strains and F1 hybrids. Data are mean values of two to four individual samples.
At most DMR regions analyzed by MALDI-TOF MS, the overall methylation level of individual CpGs in F1 hybrids equaled the mean methylation level of their parents (see Fig. 5A for BMM; Supplemental Fig. S7 for spleen). The simplest and most plausible explanation for this pattern is that the methylation level of each allele is inherited independently and that the methylation level is controlled in cis during development. DMR that show a different distribution between parents and offspring (like the Sfi1 pseudogene, Eps8l1, and Isoc2b) may have a trans-regulating component in their inheritance.
Figure 5.
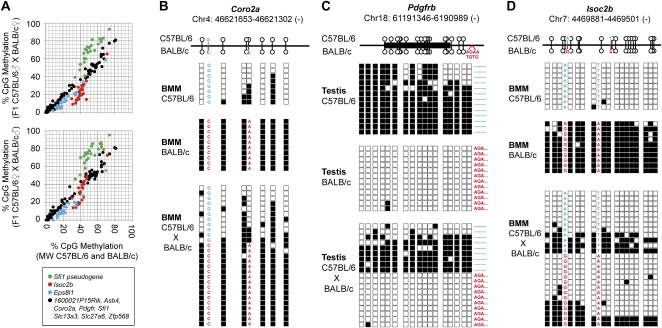
Strain-specific methylation patterns are mainly controlled in cis. (A) Averaged CpG methylation ratios of parental BMM (n = 3 for each strain) are plotted against averaged CpG methylation ratios of F1 hybrids derived from C57BL/6 (top, n = 5) or BALB/c (bottom, n = 3) sires. In eight out of 11 DMR analyzed by MALDI-TOF (marked in black), methylation patterns in F1 hybrids are almost identical to the average methylation level in parental strains (r2 > 0.97). Three DMR (Sfi1 pseudogene, Isoc2b, and Eps8l1, marked in red, green, and blue, respectively) either acquire (Sfi1 pseudogene) or lose methylation (Isoc2b, Eps8l1) in F1 hybrids relative to parental strains. (B–D) Allele-specific bisulfite sequencing of DMR in Coro2a (B, controlled in cis), Pdgfrb (C, controlled in cis), and Isoc2b (D, controlled in trans). The genomic position of CpGs within the amplicons is shown at the top. Sequence variations used to distinguish the different parental alleles are marked in blue for C57BL/6 and in red for BALB/c. Individual CpGs are represented by either white (unmethylated) or black (methylated) squares. Lines of squares represent independently sequenced clones derived from two independent sample preparations derived from reciprocal crosses. Three additional examples of DMR controlled in cis are given in Supplemental Figure S8.
Because the MS analysis does not discriminate between the two different parental alleles, we performed traditional bisulfite sequencing on independently cloned amplicons that maintained a characteristic sequence difference after bisulfite treatment. As shown in Figure 5B for Coro2a, in Figure 5C for Pdgfrb (in Supplemental Fig. S8 for Asb4, 1600021P15Rik, and Slc27a6), allelic methylation patterns in F1 hybrids were established essentially as observed on the parental alleles, suggesting that DNA methylation at these sites is indeed largely controlled in cis. Only at the Isoc2b promoter (see Fig. 5D), a mixed DNA methylation pattern was observed with some sequences being unmethylated and others being methylated, independent of their origin. Here the differential pattern must be controlled in trans, e.g., through a yet unknown strain-specific epigenetic modifier. Isoc2b methylation levels in F1 hybrids were considerably more variable (ranging from 15% to 45% in individual F1 mice) as compared to cis-controlled loci, likely representing a novel example of a metastable epiallele in inbred mice.
Discussion
In this study we performed a broad analysis of differential DNA methylation in two inbred mouse strains using DNA methylation-dependent genome fractionation and locus-wide tiling arrays. By using stringent hybridization conditions for microarray analyses of CpG-methylated and unmethylated genome fractions, we could simultaneously identify several hundred DMRs, as well as previously unrecognized strain-specific sequence variations that track with the large majority of DMRs. The microarray results were validated by DNA sequencing (to confirm sequence variations) and a combination of bisulfite treatment of genomic DNA and mass spectrometry (to confirm CpG methylation differences) suggesting that our approach is highly reproducible and correctly identifies both types of differences. To our knowledge, our study describes the first extensive identification of allele-specific DNA methylation in mice, and provides a resource for further mechanistic studies.
Having defined a large number of allele-specific methylation events, we were able to ask how methylation patterns are established at a representative set of genomic regions. Interestingly, most DMR (15/18) that were studied in detail were only differentially methylated within the somatic tissues studied (BMM and spleen) and showed equal levels of methylation in the male germ line, suggesting that the observed differences are established post-fertilization and DNA methylation itself cannot be inherited. Furthermore, the exemplary mass spectrometry validation of DMR and allele-specific examination of DNA methylation in F1 hybrids, using traditional sequencing of bisulfite treated genomic DNA, suggest that the propensity for acquiring DNA methylation during development mainly depended on the local sequence context in cis. It is formally possible that another, yet uncharacterized epigenetic mark at these sites determines the allelic methylation status in heritable fashion. However, since we found a common association of DMR with sequence variations, it is much more likely that the establishment of differential methylation patterns during embryonic development may involve cis-acting regulators, including sequence-specific DNA-binding proteins or other sequence-dependent regulatory mechanisms. The genomic intervals investigated in this study were enriched for genetic variation as compared to the whole genome, which could produce a bias toward cis-dependent DMR. However, the regions studied do contain balanced numbers of all SNP densities that clearly correlate with the density of DMR. Thus, only a relatively small proportion of strain-specific DMR (like the ones identified in Isoc2b, Sfi1 pseudogene, or Eps8l1) are expected to depend dominantly on trans-acting mechanisms. In fact, this is not surprising, since the absence or functional inactivation of the few known epigenetic modifiers that mediate DNA methylation in trans show profound developmental abnormalities (Ashe et al. 2008).
Our findings are in line with a number of previous studies linking allelic DNA methylation and genetic variation (Murrell et al. 2004; Flanagan et al. 2006; Heijmans et al. 2007; Kerkel et al. 2008) and share an obvious corollary with the recent observation that the genetic sequence is largely responsible for directing species-specific transcription in mice carrying the human chromosome 21 (Wilson et al. 2008). Our analysis contrasts a recently published study that did not find significant variation in DNA methylation between inbred or outbred mice and concluded that the impact of DNA polymorphisms on DNA methylation would not seem to be common (Kaminsky et al. 2009). The reasons for the deviant findings are not clear, but may relate to the different technologies and readout systems used in each study. For example, the above study used a CpG island microarray covering 2176 unique CpG island regions of the mouse genome that are likely more conserved, frequently contain gene promoters, and are often unmethylated. Based on our findings that differential DNA methylation is associated with genetic variation and promoter-distal sites, we would predict that the microarray platform utilized by Kaminsky et al. (2009) is less likely to detect allele-specific DNA methylation than the considerably larger and locus-wide microarray used in our study.
The presented findings suggest that genetic variability has a major impact on epigenetic variability, likely due to its influence on the propensity of individual alleles to become methylated (or demethylated) during development. This relationship is likely not restricted to early embryonic development in mice, but may also apply to other developmental (including disease- and age-related) processes in mammals. In essence, our results imply that the majority of DNA sequences (at nonimprinted loci) carry intrinsic information that recruits cis-acting factors and determines its DNA methylation status during mammalian development.
Methods
A full description of all experimental methods is provided in the Supplemental material.
Cells and tissues
Wild-type inbred C57BL/6JCrl and BALB/cAnNCrl mice were purchased from Charles River Laboratories. F1 hybrid offspring were produced by natural matings. The generation of bone marrow-derived macrophages (BMM) is described in the Supplemental material. Total cellular RNA was isolated using the RNeasy Kit (Qiagen). Genomic DNA was prepared using the Qiagen Blood & Cell Culture DNA Kit. DNA and RNA concentrations were determined using the NanoDrop ND-1000 Spectrophotometer (PeqLab) and quality was assessed by agarose gel electrophoresis. In addition, spleen and testis DNA were prepared using a TissueLyser (Qiagen) and the DNeasy Blood & Tissue kit (Qiagen).
MCIp and comparative microarray hybridization
The recombinant MBD-Fc protein was produced as previously described (Gebhard et al. 2006). MCIp was essentially performed as described with slight modifications (for details see the Supplemental material). The separation of CpG methylation densities of individual MCIp fractions was controlled by qPCR using primers covering an imprinted region of Mest and a genomic region lacking CpGs. Prior to labeling, fractions containing mainly unmethylated DNA (300–400 mM, CpG pool) or methylated DNA (≥500 mM, mCpG pool) were combined. Based on the expression analysis we created a custom 240,000 tiling array (Agilent) for 181 genomic regions covering 28 Mb of the mouse genome using the web-based tool eArray (earray.chem.agilent.com). Regions and corresponding genes are given in Supplemental Table S1. Two sets of independent comparative hybridizations of fluorescently labeled hypo- and hypermethylated fractions were performed on the 240,000 tilling array using a stringent hybridization protocol as detailed in the Supplemental material. Images were scanned using a DNA microarray scanner (Agilent) and processed using Feature Extraction Software 9.5.1 (Agilent) and the standard CGH protocol. Processed signal intensities were normalized depending on the GC content of probes and imported into Microsoft Office Excel 2007 for further analysis. About 23,000 probes were excluded due to low signal intensities. The vCGH was generated by adding up signal intensities of both genome pools for each strain. Unbalanced hybridization behavior in the vCGH (log10 ratio of cumulative signal intensities >0.15 as compared to the surrounding probes) resulted in the removal of another 33,000 probes. Three of the detected CNVs were validated using qPCR as described in the Supplemental material. DMR were identified in the remaining probe set using a sliding window approach (five probes) as described in the Supplemental material.
Analysis of bisulfite-converted DNA
We chose a representative set of 18 regions based on the MCIp microarray results and sequenced underlying gDNA of both strains before MALDI-TOF-MS analysis, which was performed essentially as described (Ehrich et al. 2005) (see the Supplemental material for detailed information).
For allele-specific analyses, bisulfite amplification products of distinguishable alleles were directly cloned into a pCR 2.1-Topo vector using the TOPO TA Cloning kit (Invitrogen). Insert-containing plasmids of single colonies were sequenced (GENEART) and analyzed using GeneRunner (http://www.generunner.net/) and BioEdit (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html) software tools.
Acknowledgments
Work in the laboratory of M.R. was supported by grants from the Deutsche Forschungsgemeinschaft and the Deutsche Krebshilfe.
Author contributions: Study design: E.S., and M.R. Expression analysis: C.E. MCIp, molecular validations and analysis of DMR: E.S. Bioinformatic analyses: C.E. and M.R. Data interpretation and writing the paper: E.S., and M.R.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data and the results from locus-wide comparative hybridizations data from this study have been submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE14644 and GSE14463, respectively. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ751937–FJ752004].
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.095562.109.
References
- Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, et al. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9:R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006;2:e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Chong S, Vickaryous N, Ashe A, Zamudio N, Youngson N, Hemley S, Stopka T, Skoultchi A, Matthews J, Scott HS, et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39:614–622. doi: 10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van Den BD. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, et al. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Gebhard C, Schwarzfischer L, Pham TH, Schilling E, Klug M, Andreesen R, Rehli M. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- Gessner A, Blum H, Röllinghoff M. Differential regulation of IL-9-expression after infection with Leishmania major in susceptible and resistant mice. Immunobiology. 1993;189:419–435. doi: 10.1016/S0171-2985(11)80414-6. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet. 2007;16:547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Locksley RM, Heinzel FP, Sadick MD, Holaday BJ, Gardner KD., Jr Murine cutaneous leishmaniasis: Susceptibility correlates with differential expansion of helper T-cell subsets. Ann Inst Pasteur Immunol. 1987;138:744–749. doi: 10.1016/s0769-2625(87)80030-2. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Sotomaru Y, Katsuzawa Y, Kono T, Meguro M, Oshimura M, Kawai J, Tomaru Y, Kiyosawa H, Nikaido I, et al. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem Biophys Res Commun. 2002;290:1499–1505. doi: 10.1006/bbrc.2002.6370. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Cooper WN, Douglas E, Apostolidou S, Moore GE, Maher ER, Reik W. An association between variants in the IGF2 gene and Beckwith-Wiedemann syndrome: Interaction between genotype and epigenotype. Hum Mol Genet. 2004;13:247–255. doi: 10.1093/hmg/ddh013. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90:314–323. doi: 10.1016/j.ygeno.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Schmidl C, Klug M, Boeld TJ, Andreesen A, Hoffmann P, Edinger M, Rehli M. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]