Abstract
Genome-wide association studies (GWAS) have now successfully identified important genetic variants associated with many human traits and diseases. The high cost of genotyping arrays in large data sets remains the major barrier to wider utilization of GWAS. We have developed a novel method in which whole blood from cases and controls, respectively, is pooled prior to DNA extraction for genotyping. We demonstrate proof of principle by clearly identifying the associated variants for eye color, age-related macular degeneration, and pseudoexfoliation syndrome in cohorts not previously studied. Blood pooling has the potential to reduce GWAS cost by several orders of magnitude and dramatically shorten gene discovery time. This method has profound implications for translation of modern genetic approaches to a multitude of diseases and traits yet to be analyzed by GWAS, and will enable developing nations to participate in GWAS.
Genome-wide association studies (GWAS) have successfully identified single nucleotide polymorphisms (SNPs) associated with many human diseases (McCarthy et al. 2008). GWAS have broken a bottleneck in the understanding of common complex diseases as no a priori knowledge of underlying molecular mechanisms is required (McCarthy et al. 2008). The high cost of SNP arrays remains a major barrier to widespread utilization of such studies. Typically, arrays capable of genotyping 3 × 105–1 × 106 SNPs are analyzed for every case and control individually. GWAS generally utilize large data sets with DNA extraction followed by SNP array genotyping costs running to >US$1 million, accompanied by long-time requirements for genotyping. Uncertainty about the genetic architecture of a given trait or disease means there is no guarantee of a successful outcome despite the large investment required. Many groups researching diseases in developing nations, or traits of perceived lesser clinical significance, have been unable to pursue GWAS methodology due to the high budgets required. One strategy for reducing cost is equimolar pooling of separately extracted DNA samples (Macgregor et al. 2006; Pearson et al. 2007). Rather than individually genotyping each subject in the case and control cohorts, stringently quantitated equimolar DNA pools derived from all cases and all controls are constructed separately and then genotyped. Pool construction for this genotyping strategy is labor-intensive and requires sophisticated DNA quantitation procedures (Macgregor et al. 2008).
We hypothesized that equal volume, but unquantitated whole blood aliquots, from case and control cohorts could be pooled prior to DNA extraction. Although this clearly introduces a potential error relative to pooling of DNA, by deriving a formula for the degree of error expected given typical cell count values, we show that the additional error is very small. The “pooling of blood” (POB) method imparts large savings in time, labor, arrays, and reagents. We provide strong proof of principle data indicating that POB-based GWAS can identify known genes for eye color, pseudoexfoliation syndrome, and for age-related macular degeneration (AMD), the commonest cause of blindness in the Western world. The POB approach is easily applied to array-based GWAS, which aim to identify common variants of small effect. However, with full-genome sequencing becoming an accessible commodity, the identification of rare causal variants of larger effect may also prove tenable using our novel POB method.
Results
By pooling blood instead of DNA, individuals will not be equally represented in the pool. In the Supplemental material we show that the decrease in effective sample size of the pool is ∼1 + c2, where c is the coefficient of variation of the contribution of each individual to the pool. The value of c is typically around 0.25 (Bain 1996). This implies that the decrease in effective sample size for blood pooling, relative to DNA pooling, will only be ∼5%–6% and that the results from POB will be broadly similar to those obtained using DNA pooling and individual genotyping.
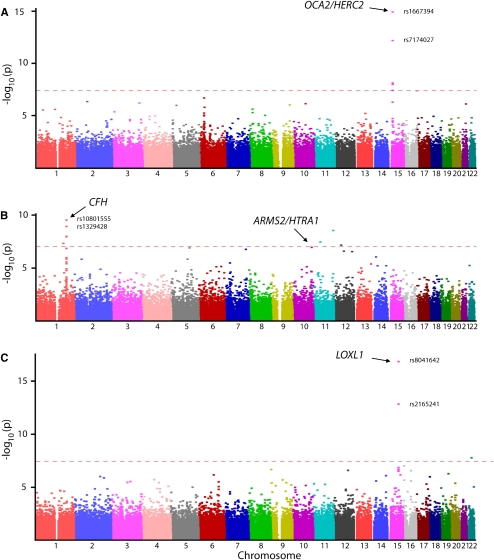
To test if our theoretical prediction that POB would be effective holds up in practice, 100 μL aliquots of fresh whole blood were pooled from 100 individuals with blue eyes and 100 people with brown eyes, respectively. DNA was then extracted (two extractions in total, ∼US$40) and array hybridization performed using Illumina HumanHap550 SNP arrays. Concordance between POB allele frequency estimates and individual reference frequencies was excellent, and even better between case and control pools (Fig. 1). Tests of association revealed seven SNPs in the eye color study that achieved genome-wide significance (P < 5 × 10−8), all at the OCA2/HERC2 locus (chromosome 15q11.2-q12), previously implicated in eye color determination (Fig. 2A; Duffy et al. 2007; Sulem et al. 2007; Sturm et al. 2008). Multiple hits were also found in other known pigmentation genes in the top 1000 associated SNPs: TYR, TYRP1 and top 10,000: SLC24A4, SLC24A5 (see Supplemental Table S1; Graf et al. 2007; Stokowski et al. 2007; Sulem et al. 2007).
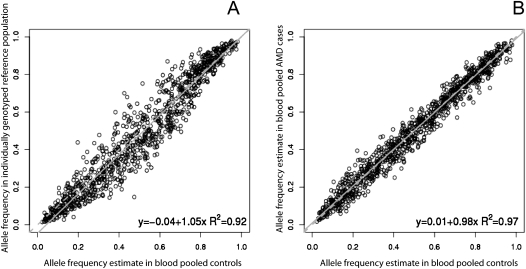
Figure 1.
(A) Comparison in allele frequency between 540 individually genotyped Caucasian reference samples compared to the 216 control POB samples. (B) Correlation between estimated allele frequencies in the POB AMD case and control subjects. (A,B) A subset of 1000 randomly chosen SNPs are plotted, with the broken gray line being y = x and the solid gray line the regression line for the full set of SNPs. The reduced correlation in A is due to the unequal amplification of alleles (see main text for discussion).
Figure 2.
(A) Identification of the OCA2/HERC2 eye color locus (15q11.2-q12) using POB GWAS. Horizontal line at P = 5 × 10−8 indicates genome-wide significance. Multiple significant SNP associations were also found in other known pigmentation loci in the top 1% of genotyped SNPs (TYR, TYRP1, SLC24A4, and SLC24A5). (B) Age-related macular degeneration POB GWAS. SNPs at the CFH locus (1q32) clearly reached genome-wide significance. SNP rs10922153 in CFHR5 adjacent to CFH was also strongly associated P = 6.9 × 10−9. SNP rs3750847 in ARMS2/HTRA1 (10q25-q26) yielded P = 6.4 × 10−8. (C) Pseudoexfoliation syndrome POB GWAS. Strong association of SNPs at the LOXL1 locus (15q22), where two SNPs (rs8041642, rs2165241) reached genome-wide significance.
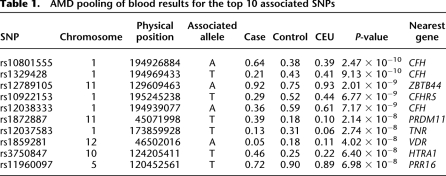
To test whether fresh blood is a requirement for successful POB, a second case-control cohort was studied in which whole blood had been stored at 4°C for >12 mo. Previous studies of AMD have identified two major susceptibility loci; CFH (chromosome 1q32) and ARMS2/HTRA1 (chromosome 10q25-q26) (Edwards et al. 2005; Hageman et al. 2005; Klein et al. 2005; Dewan et al. 2006; Yang et al. 2006; Fritsche et al. 2008). We used a modified POB protocol (Supplemental material) to construct a pool from 131 cases of exudative AMD and another from 216 examined normal controls. These samples were hybridized on Illumina 1M arrays. The association of exudative AMD with the CFH gene was easily detected (Fig. 2B) with multiple SNPs in the most strongly associated region. The ARMS2/HTRA1 locus also appeared in the top 10 associated SNPs (Table 1; Supplemental material).
Table 1.
AMD pooling of blood results for the top 10 associated SNPs
In the third POB GWAS, which utilized data generated from the same control cohort, and a fresh POB sample from 100 patients with pseudoexfoliation syndrome, we easily identified the LOXL1 gene, which has been recently associated with this disease (Fig. 2C; Thorleifsson et al. 2007; Hewitt et al. 2008).
Estimation of pooling error
Pooling allele frequency estimates are known to be most useful in estimating differences in frequency between pools rather than absolute frequencies (Macgregor et al. 2006), on account of the potential unequal amplification of alleles at some SNPs. To a close approximation, any unequal amplification effects cancel when frequency differences are used, partly explaining the higher concordance seen in Figure 1A compared with Figure 1B.
Pooling error was assessed for the AMD pools (Illumina 1M arrays). The variation in the difference in pooling allele frequency estimates between the case and control pools was compared with what would be expected if pools of this size were individually genotyped. If either the number of individuals contributing to the pooling allele frequency estimate was lower than that specified/expected (e.g., due to some individuals contributing substantially more DNA than other individuals) or the array specific error was high, then the variation in the difference in pooling allele frequency would be unusually high. Using the previously described method (Macgregor et al. 2006), an explicit estimate of the pooling variance resulted in an estimate of 6.8 × 10−5. This estimate comprises both error due to pool construction (here attributable to unequal contributions of cells from different individuals and to pipetting or measurement error) and due to array probe. This estimate is similar to that observed in previous DNA pool studies using Illumina HumanHap arrays, validating our theoretical calculation comparing POB and DNA pooling. Although our theoretical calculation focused on variation due to unequal WBC counts, other variation in total DNA yield (including that caused by potential degradation of samples in our refrigerator stored AMD data set) per person must have been small for us to have obtained this small estimate. The estimate of 6.8 × 10−5 is 5–10 times smaller than the variance due to random sampling of individuals from the population (for fuller discussion, see Macgregor 2007; Macgregor et al. 2008). The arrays used here (1M) differed from those we have reported previously (Macgregor et al. 2008; HumanHap300), but HumanHap300 and 1M arrays have similar arrangements of beadscores on the arrays so similar performance for different array types is expected. In Macgregor et al. (2008), three arrays were used for each (equal sized) pool, which should give similar array specific variance as the arrangement used here (two and six arrays for N = 131 AMD case and N = 216 control pool, respectively). Different numbers of case and control pool arrays were used because there were different numbers of individuals in the two pools. The low estimates of pooling error using this methodology implies that disease associated variants of small effect size will be identifiable using adequately powered large cohorts of cases and controls. Such estimates of pooling error imply an effective relative sample size (Macgregor et al. 2006) of ∼90% for the AMD case-control sample. This suggests that, consistent with DNA pooling results from larger pools (Brown et al. 2008; Macgregor et al. 2008), POB will prove useful for detecting smaller effect sizes in larger pools. In contrast to up-scaling individual or equimolar DNA pooling strategies to much larger data sets, with blood pooling the budget remains very low irrespective of the sample size.
Discussion
Using the novel POB method we identified the associations between previously identified genes in all three separate GWAS. Importantly, the individual allelic architecture of these three case-control cohorts had not been previously determined. Despite the extreme simplicity of the underlying hypothesis and method, estimates of pooling error in these cohorts were remarkably small relative to random sampling error. A plethora of human diseases and traits warrant genome-wide investigation, and it is foreseen that POB prior to DNA extraction will both broaden access to this cutting edge modality and expedite disease-gene discovery. For novel findings, additional replication cohorts will be required as for any properly conducted GWAS.
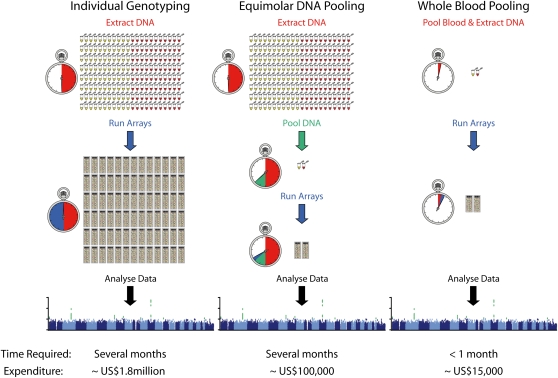
The dramatic reduction in costs and time required for GWAS using the POB technique has profound implications for genetics research. It is especially noteworthy that application of this methodology would also allow larger GWAS to be cheaply performed (Fig. 3). A conventional individual genotyping experiment on 2000 cases and 2000 controls would cost ∼US$1.8 million and take several months to perform. This GWAS would typically be the expensive first stage in a multiple stage design examining two or more data sets. In contrast, a POB experiment could yield similar first-stage results in a few weeks for ∼US$15,000. Since POB performance was broadly similar to DNA pooling performance, previous cost benefit calculations for the allocation of resources in pooling-based multistage designs (Macgregor et al. 2008) remain valid.
Figure 3.
Flowchart comparing available options for performing genome-wide association studies. Costs and time calculations were based on experiments with 2000 cases and 2000 controls.
The simplicity and robustness of our method will expedite scientific discovery, laying open the field of genome-wide association to smaller research groups, and to answer biological questions that would otherwise not be asked. Our described methodology is directly applicable to researchers in the developing world, as well as investigators not previously involved in genetics work. We also foresee that the pooling of blood samples could enable the discovery phase of initial GWAS to be performed rapidly and cheaply with positive results followed up in previously established cohorts. A relative disadvantage of POB is that since it is not necessary to extract DNA from each individual sample, then DNA would not be available for subsequent individual genotyping. Clearly, the lack of stored DNA from each individual included in the blood pools does not permit individual genotyping in stage two of analysis. However, given the low pooling error we demonstrate here with POB, most of the available information is extracted from the POB samples (e.g., expressed in terms of effective relative sample size; Macgregor et al. 2008) and further resources would be best spent on genotyping the implicated SNPs in independent replication samples. Furthermore, in the event of positive results, DNA can be extracted later from stored blood samples if deemed worthwhile.
There are several established analytical restrictions with equimolar DNA pools that are also applicable to POB. It must be noted that any quality control step or analysis that requires individual-level genotypes cannot be performed using current pooling methods, including POB. This includes haplotype construction, SNP imputation and analysis of case-control status, while adjusting for covariates (Homer et al. 2008a; Jawaid and Sham 2008). Additionally, poorly performing individual samples and bias due to population stratification are not easily controlled; however, they may be overcome by pooling design (Sham et al. 2002). Nonetheless, novel statistical methods are continually being developed for genome-wide imputation and haplotype-based analysis of pooled data (Kirkpatrick et al. 2007; Homer et al. 2008b; Zhang et al. 2008). It is also worthwhile noting that given such a small number of arrays are required it is possible to use more expensive state-of-the-art arrays with a larger number of SNPs in pooling studies. In such cases (e.g., when using the 1 million SNP Illumina arrays on Caucasian samples), the need for imputation is diminished because a larger number of SNPs are genotyped directly. Another important limitation of this methodology is that the lack of individual-level data limits the study to a single phenotype for each constructed pool, even if many more phenotypes were recorded. The lack of individual genotype data and, in turn, the inability for covariate adjustment, decreases the power of pooling-based GWA studies. Additionally, copy number variants are difficult to quantify, and cannot be currently analyzed.
Further development of this POB methodology could be directly applied to diverse fields, such as animal trait mapping, or be transferable to other array-based technologies, such as those emerging in the fields of proteomic and epigenetics research (Kendziorski et al. 2005; Docherty et al. 2009), or even newer generation sequencing techniques (Shendure and Ji 2008). Perhaps the greatest benefit to flow from the described methodology will be the opportunity for clinicians and researchers in the developing world to participate in the GWAS biological revolution.
Methods
Subject recruitment and DNA extraction
All participants provided written informed consent for this study, which was approved by The Flinders Medical Center Human Research Ethics Committee. Venous blood samples from Caucasian individuals with blue eyes (n = 100), brown eyes (n = 100), and pseudoexfoliation (n = 100), were collected in 9-mL EDTA tubes. One-hundred-microliter aliquots of whole blood from each of the 100 individuals in each group were combined after gentle agitation into three 10-mL pools of blood (POB). Whole blood samples, which had been stored at 4°C for between 12 and 24 mo, from 131 Caucasian patients with exudative AMD and 216 examined disease-free Caucasian controls, were also pooled following lysis of each individual sample to improve homogeneity of the stored samples. Genomic DNA was extracted from each blood pool according to manufacturer guidelines using the QIA-AMP DNA blood maxi kit (QIAGEN). The extracted DNA was quantified by spectrophotometry.
Generation and analysis of array data
The blue and brown eye color pools were hybridized to two Illumina HumanHap550 duo arrays (Illumina), as reference samples are readily available for these arrays (Schymick et al. 2007). The pools were configured to each appear once on each of the two duo arrays, resulting in raw data in the same format as if two replicate single-sample arrays had been used on each pool. The AMD case pools were genotyped with two (single sample) Illumina 1M arrays, the pseudoexfoliation case pools with four Illumina 1M arrays, and the larger control pool genotyped with six Illumina 1M arrays.
Bead-level intensity data were analyzed. SNPs with beadscore values <−50 were excluded. Copy number variants and SNPs from the sex chromosomes were not analyzed. SNPs were also removed in the initial quality control if they had a minor allele frequency <1% in the HapMap CEU reference samples.
Beadscores required calibration because green beadscores tended to be larger than red beadscores. Illumina HumanHap550 arrays had 10 stripes (previously denoted as “strands” [Macgregor et al. 2008]) per array, each with ∼55,000 SNPs. Illumina 1M arrays had 20 stripes per array, each with ∼50,000 SNPs. Within each stripe, half of the SNPs were from the Illumina “TOP” strand (A/C and A/G SNPs) and half were from the Illumina “BOTTOM” strand (T/C and T/G SNPs). The pooling allele frequency (PAF) was computed as the corrected red intensity divided by the total (corrected red plus green) intensity. Normalization was performed within stripe, separately for each strand by rescaling the red beadscore to make the mean PAF value = 0.5 (for all SNPs on that stripe/strand). A very small number of SNPs had <5 PAF estimates available and were dropped. SNPs were removed from the analysis if they had a −log10(P) quality control value >6. SNPs from different strands exhibited different levels of pooling error; hence, 261,887 (HumanHap550) and 375,246 (1M) SNPs from the BOTTOM strand are presented.
After correction for green/red ratio variability, a linear model was used to test for allelic association in which the PAF estimate for each SNP was the response variable. A test statistic that corrected for the pooling error was used to rank SNPs, with P-values based on a χ2 distribution with 1 degree of freedom (Macgregor et al. 2006). Since this method is based upon contrasting case and control pools, the effect of unequal amplification of alleles is minimal as such effects cancel out (Macgregor et al. 2006, 2008). Using a previously described method (Macgregor et al. 2008), the explicit estimate of pooling variance in the AMD experiment was 6.8 × 10−5, similar to that observed with previous equimolar DNA pools.
Acknowledgments
The Ophthalmic Research Institute of Australia, the Glaucoma Research Foundation, the American Health Assistance Foundation, and the Australian National Health and Medical Research Council provided financial assistance. We thank Jozef Gécz and Nicholas Hayward for their helpful comments.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to Gene Expression Omnibus ( http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE17557.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.094680.109.
References
- Bain BJ. Ethnic and sex differences in the total and differential white cell count and platelet count. J Clin Pathol. 1996;49:664–666. doi: 10.1136/jcp.49.8.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- Docherty SJ, Davis OS, Haworth CM, Plomin R, Mill J. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenetics Chromatin. 2009;2:3. doi: 10.1186/1756-8935-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Montgomery GW, Chen W, Zhao ZZ, Le L, James MR, Hayward NK, Martin NG, Sturm RA. A three-single-nucleotide polymorphism haplotype in intron 1 of OCA2 explains most human eye-color variation. Am J Hum Genet. 2007;80:241–252. doi: 10.1086/510885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- Graf J, Voisey J, Hughes I, van Daal A. Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat. 2007;28:710–717. doi: 10.1002/humu.20504. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt AW, Sharma S, Burdon KP, Wang JJ, Baird PN, Dimasi DP, Mackey DA, Mitchell P, Craig JE. Ancestral LOXL1 variants are associated with pseudoexfoliation in Caucasian Australians but with markedly lower penetrance than in Nordic people. Hum Mol Genet. 2008;17:710–716. doi: 10.1093/hmg/ddm342. [DOI] [PubMed] [Google Scholar]
- Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson JV, Stephan DA, Nelson SF, Craig DW. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008a;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N, Tembe WD, Szelinger S, Redman M, Stephan DA, Pearson JV, Nelson SF, Craig D. Multimarker analysis and imputation of multiple platform pooling-based genome-wide association studies. Bioinformatics. 2008b;24:1896–1902. doi: 10.1093/bioinformatics/btn333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawaid A, Sham P. Impact and quantification of the sources of error in DNA pooling designs. Ann Hum.Genet. 2008;73:118–124. doi: 10.1111/j.1469-1809.2008.00486.x. [DOI] [PubMed] [Google Scholar]
- Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick B, Armendariz CS, Karp RM, Halperin E. HAPLOPOOL: Improving haplotype frequency estimation through DNA pools and phylogenetic modeling. Bioinformatics. 2007;23:3048–3055. doi: 10.1093/bioinformatics/btm435. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S. Most pooling variation in array-based DNA pooling is attributable to array error rather than pool construction error. Eur J Hum Genet. 2007;15:501–504. doi: 10.1038/sj.ejhg.5201768. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Visscher PM, Montgomery G. Analysis of pooled DNA samples on high density arrays without prior knowledge of differential hybridization rates. Nucleic Acids Res. 2006;34:e55. doi: 10.1093/nar/gkl136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Zhao ZZ, Henders A, Nicholas MG, Montgomery GW, Visscher PM. Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res. 2008;36:e35. doi: 10.1093/nar/gkm1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N, Brun M, Szelinger S, Coon KD, Zismann VL, et al. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am J Hum Genet. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, Kasperaviciute D, Hernandez DG, et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: First stage analysis and public release of data. Lancet Neurol. 2007;6:322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- Sham P, Bader JS, Craig I, O'Donovan M, Owen M. DNA Pooling: A tool for large-scale association studies. Nat Rev Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, Jarman C, Filsell W, Ginger RS, Green MR, van der Ouderaa FJ, et al. A genomewide association study of skin pigmentation in a South Asian population. Am J Hum Genet. 2007;81:1119–1132. doi: 10.1086/522235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, Martin NG, Montgomery GW. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Jonasdottir A, Stefansdottir G, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang HC, Yang Y. PoooL: An efficient method for estimating haplotype frequencies from large DNA pools. Bioinformatics. 2008;24:1942–1948. doi: 10.1093/bioinformatics/btn324. [DOI] [PubMed] [Google Scholar]