Abstract
Manipulating the mouse genome is a widespread technology with important applications in many biological fields ranging from cancer research to developmental biology. Likewise, correlations between copy number variations (CNVs) and human diseases are emerging. We have developed the reference-query pyrosequencing (RQPS) method, which is based on quantitative pyrosequencing and uniquely designed probes containing single nucleotide variations (SNVs), to offer a simple and affordable genotyping solution capable of identifying homologous recombinants independent of the homology arm size, determining the micro-amplification status of endogenous human loci, and quantifying virus/transgene copy number in experimental or commercial species. In addition, we also present a simple pyrosequencing-based protocol that could be used for the enrichment of homologous recombinant embryonic stem (ES) cells.
Chromosome engineering by random integration (to create transgenic animals) or by homologous recombination (to replace/remove specific genes) has been widely used to decipher gene functions and demonstrate disease causality. However, these techniques have limitations. The concatenation of transgenes integrating into the genome makes it difficult to determine copy number and distinguish homozygotes from heterozygotes. Additionally, gene-targeting events are infrequent, and identification of correctly targeted embryonic stem (ES) cells can be laborious. The advent of the bacterial artificial chromosome (BAC) recombineering method makes the use of extremely long homology arms (>10 kb) possible, increases homologous recombination efficiency, and reduces the need of isogenicity between targeting constructs and recipient ES cells (Muyrers et al. 1999; Cotta-de-Almeida et al. 2003; Testa et al. 2003; Valenzuela et al. 2003; Yang and Seed 2003; Warming et al. 2005). However, traditional screening methods, including PCR and Southern blot, are not compatible with homology arms >10 kb. Alternative screening methods aiming to solve this problem attempt to either quantify a twofold difference in the copy number of a targeted allele via quantitative PCR (Valenzuela et al. 2003) and multiplex ligation-dependent probe amplification (MLPA) (Langerak et al. 2005) or to visualize integration sites by fluorescence in situ hybridization (FISH) (Yang and Seed 2003). These technologies remain technically demanding (Gomez-Rodriguez et al. 2008). (Refer to Supplemental Table S1 for the comparison of current screening methods.)
In addition to investigator-initiated genome manipulation, recent studies reveal that the human genome contains copy number variations (CNVs), and such gains or losses of genomic DNA contribute to disease susceptibility (Gonzalez et al. 2005; Aitman et al. 2006; Fanciulli et al. 2007; Hollox et al. 2008). Various methods have been developed to investigate CNVs at either genome-wide or locus-specific levels (Aten et al. 2008; Lee and Jeon 2008). By labeling test and reference DNAs with different fluorescence dyes and hybridizing them to millions of printed probes on a single array, the comparative genome hybridization (CGH) method is suitable for whole-genome CNV scanning and has since revolutionized our understanding of CNV frequency in mammalian genomes (Kallioniemi et al. 1992; Pinkel et al. 1998; Carter 2007). Recently, array-based technologies once specialized for SNP (single nucleotide polymorphism) genotyping have been modified for genome-wide CNV studies (Carter 2007; Lee and Jeon 2008). In addition, the advent of the next-generation sequencing methods, including 454 Life Sciences (Roche)/FLX, Illumina/Genome Analyzer, and Applied Biosystems SOLiD, offers even more powerful means to detect CNVs at the whole-genome level (Mardis 2008). However, all of these methods are still relatively expensive and are, therefore, not optimal for the diagnostic analysis of a large number of samples (Aten et al. 2008). Moreover, CNV data obtained from genome-wide studies often require further validation at the single locus level.
Current methods for studying CNV at the locus-specific level include real-time PCR (Fanciulli et al. 2007), multiplex quantitative PCR (Charbonnier et al. 2000), MLPA (Schouten et al. 2002), and the paralog ratio test (PRT) (Armour et al. 2007). Among these, both real-time and multiplex PCR are customizable but require careful optimization of reaction conditions. MLPA is gaining popularity owing to its multiplex capability, but commercially available probes are currently limited to a small number of human genes, and the difficulty of probe preparation makes customization arduous. The PRT method could accurately measure gene copy number and has been successfully used for the association study between CNVs of the beta-defensin gene and psoriasis (Hollox et al. 2008), yet it depends on the presence of an endogenous pseudogene as a reference (Armour et al. 2007), which limits its utility. Clearly, a simple method that could accurately measure any desired CNVs, identify homologous recombinants in any species, and correctly enumerate the copy number of any integrated DNA fragment of any length will be of value to the academic, commercial, and clinical fields.
Here we describe a novel approach based on converting these quantitative problems into a determination of single nucleotide variation (SNV) frequencies by pyrosequencing. Pyrosequencing sequences single-strand DNA by coupling nucleotide incorporation onto a sequencing primer with the production of bioluminescence signals, producing readouts called “pyrograms” (Ronaghi et al. 1998) (http://www.pyrosequencing.com/). Each peak in a pyrogram corresponds to a specific nucleotide, and the height is proportional to the amount of nucleotide incorporated during each synthesis step (see Fig. 2; Ronaghi et al. 1998). Pyrosequencing has gained enormous popularity in DNA methylation analysis (Uhlmann et al. 2002; Tost and Gut 2007), microbial identification (Clarke 2005), detection of allelic expression imbalance (Wang and Elbein 2007), and next-generation sequencing (454 Life Sciences (Roche)/FLX; http://www.454.com/). In particular, pyrosequencing has been widely used for SNP detection (Fakhrai-Rad et al. 2002; Langaee and Ronaghi 2005; Ronaghi et al. 2007) because it offers an easy, quantitative, and accurate method to detect the relative abundance of alleles in a mixture, provided they differ by a single nucleotide and that their relative fraction is between 10% and 90% (Ronaghi et al. 1998; Wasson et al. 2002). We realized that with two simple modifications, pyrosequencing could be used to effectively identify changes in copy number at any locus: First, insertion of SNVs into the probes (a reference probe and a query probe) eliminates the reliance on natural variation among alleles. Second, if the reference and query probes are ligated into a single reference/query pyrosequencing (RQPS) probe, their molar ratio will be fixed at 1:1, allowing elucidation of the copy number by pyrosequencing these two SNVs in mixtures of the test DNA and the RQPS probe. We show here that RQPS could easily determine the copy number of any query gene (or DNA fragment) in a genome, be it a man-made transgene, the result of a natural CNV (including both gain and loss), or gene loss as a result of homologous recombination. Unlike previous methods (Pielberg et al. 2003; Söderbäck et al. 2005), RQPS does not require any prior knowledge of the duplication break point or an endogenous reference fragment. It thus offers a flexible and universal CNV determination tool that can be applied to any individual from any species. In addition, it is ideal for the detection of targeted genes in ES cells. The simplicity, automation-friendly protocols, and low cost make it suitable for processing a large number of samples for any specific locus, which could be particularly useful for clinical disease-related CNV screening and patient diagnosis.
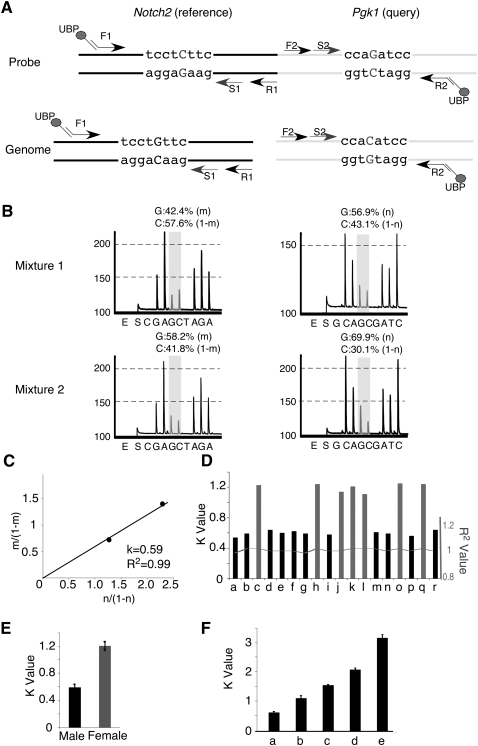
Figure 2.
RQPS accurately determines gene copy number. (A) Illustration of the Notch2-Pgk1 probe with SNVs introduced to differentiate it from the genomic counterparts. In the RQPS probe, the reference Notch2 fragment has a G-to-C SNV while Pgk1 has a C-to-G SNV. The fragment containing the Notch2 SNV was amplified with a three-primer system: (1) the forward tailed primer F1 with a complementary region to (2) the universal biotinylated primer (UBP), and (3) the untailed reverse primer R1. The fragment containing the Pgk1 SNV was amplified with the same UBP primer and two gene-specific primers (untailed forward primer F2 and a tailed reverse primer R2). The biotinylated PCR product was purified with streptavidin-coated Sepharose beads and sequenced with a sequence primer (S1 for Notch2 and S2 for Pgk1, respectively) hybridizing near the desired SNV. (B) Example of a readout (pyrogram) from a Biotage PSQ96-MA machine. Two probe/genomic DNA mixtures were prepared in this example. Reference and query SNVs were quantified by pyrosequencing of each mixture; the relative ratio is shown. Note that a G, instead of a C, determines the ratio of the RQPS probe in each mixture because the sequencing primer for the Notch2 reference gene binds to the minus strand. (C) The plot of m/(1 − m) against n/(1 − n) gives a line that passes (0, 0) with slope k = 0.59, which suggests that there is one copy of Pgk1 in this mouse and hence it is a male. (D) RQPS accurately assigns gender by determining the copy number of the X-linked Pgk1 gene. All male mice show a k-value of ∼0.6 and all females ∼1.2. (E) Statistical analysis of k-values from 18 animals tested demonstrates the robustness of RQPS. (F) RQPS accurately measures the copy number of exon 30 from the Notch1 gene, encoding a part of the Notch1 intracellular domain, in five different lines of mice that are known to have (a) one, (b) two, (c,e) three, and (d) four copies of the NICD1, respectively. When the N1 + /▵1;Gt(ROSA)26SorNotch/Notch mouse is used, owing to a deletion removing exon 30 (query) and exon 23 (reference) from one allele (Conlon et al. 1995), three copies of N1 ICD produced k = 3.1 since a haploid reference gene was used (e).
In the course of developing RQPS, we established new protocols that enable fast and economic enrichment for homologous recombinant ES cells by simple modifications of the targeting vectors. This is achieved by introducing one SNV into a mouse phosphoglycerate kinase 1 (Pgk1) promoter, which drives the expression of the antibiotic selective cassette, and another into one of the homology arms of the targeted gene. Only male ES cells experiencing homologous recombination would display an equal ratio between the introduced SNVs residing in the targeting construct and the respective endogenous loci for both Pgk1 and the targeted allele (for details, see Fig. 5). A simple pyrosequencing screen (pyroscreening) could reduce the screening task by removing up to 80% of nonhomologous ES cell clones from the candidate pool. This significantly reduces the effort and increases the number of clones that can be screened effectively for hard-to-target regions. While we demonstrate the utility of RQPS for identification of homologous recombinants in ES cells and show that RQPS, either alone or in combination with pyroscreening, provides an easy, safe, cheap, BAC and conventional targeting vector compatible ES cell screening method, its real strength is that it offers a powerful and flexible tool for gene copy enumeration in any DNA-containing organism.
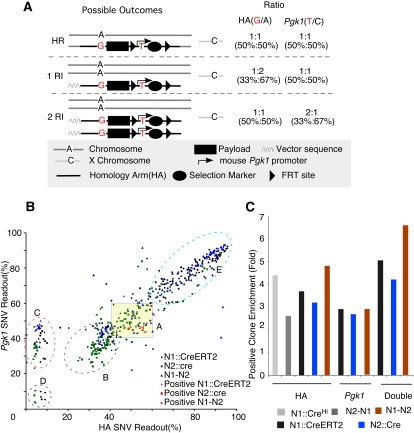
Figure 5.
Pyroscreening of artificially introduced SNVs in targeting constructs dramatically enriched for positive HR clones. (A) Schematic illustration of pyroscreening. Artificially designed SNVs are introduced into targeting constructs: one in the HA (∼50 bp away from payload) and another one in the mouse Pgk1 promoter driving the expression of the selection marker. Different fates of the targeting construct in ES cells (HR, homologous recombination; RI, random integration) yield different ratios of the two linked SNVs to their endogenous counterparts. See text for details. (B) Pyroscreening of ES cells electroporated with (black dots) N1∷CreERT2, (green squares) N2∷cre, and (blue triangles) N1–N2. ES cells cluster into different groups. (Group A) Both HA and PGK SNV ratios fall between 40% and 60%, which suggests these are possible HR candidates. (Group B) HA SNV ratio ∼33% and PGK ∼40% suggest one copy of random integration. (Group C) The ratio suggests that these clones retained only one copy of the neomycin selection gene. (Group D) Poorly growing ES cells with heavy, G418-resistant MEF contamination. (Group E) ES cells that may have experienced multiple integrations. Note that nine out of 10 positive clones from (red dots) N1∷CreERT2, one out of one from (red square) N2∷Cre, and four of four from (red triangles) N1-N2 fall within group A. (C) When only one SNV is used for pyroscreening, the enrichment for positive clones is roughly threefold. This is increased to more than fourfold when two SNVs are combined.
Results
Principle of RQPS
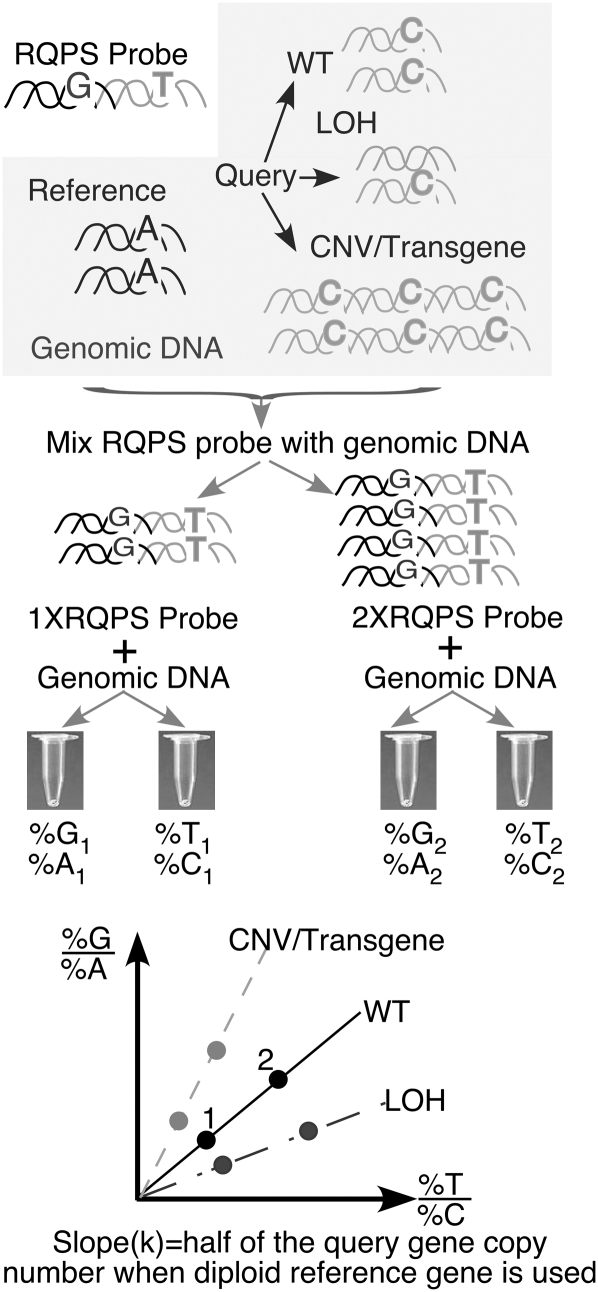
Unambiguous identification of positive homologous recombinants (HR), independent of homology arm length, is desired to facilitate the use of BAC targeting vectors. To achieve this, we developed the RQPS method (Fig. 1). The key to this method is the creation of a DNA probe consisting of two fused DNA fragments. The first fragment is designed from a reference gene with known copy number, preferentially one that produces a haploinsufficient phenotype (to ensure both copies are present), and the second from the query gene. Importantly, each of the fragments could be as short as 50 bp and designed to contain a novel SNV to differentiate the probe from the wild-type allele. Such probes can be created by cloning and mutagenesis, by synthesis of long oligos, or by PCR amplification with primers containing desired SNVs. This strategy ensures that the two artificial SNVs in the mixture are always at a 1:1 molar ratio.
Figure 1.
Schematic illustration of RQPS. A RQPS probe, which consists of a DNA fragment from the query gene physically linked to a DNA fragment from a reference gene, both containing artificially designed SNVs, is mixed with genomic DNA samples to attain a molar ratio between 1:9 and 9:1. The molar ratio of R-SNV, Q-SNV between probe and sample genomic DNA in each mixture is accurately determined by quantitative pyrosequencing. If the query gene is diploid, %G/%A = T%/C% (wild type, WT in the diagram). If one copy of the query gene is lost, %G/%A < T%/C% (loss of homozygosity, LOH in the diagram). If multiple copies of the query gene are present, then %G/%A > T%/C%. See text for details.
Figure 1 describes how to determine the copy number of any query gene (endogenous, integrated, or episomal). First, the RQPS probe is added to genomic DNA samples of known concentration. Since pyrosequencing is most accurate in mixtures where the percentage of SNVs is between 10% and 90% (Wasson et al. 2002), two mixtures are generated for each sample. In the first, the molar ratio of the RQPS probe to sample genomic DNA is ∼1:1, and in the second it is higher (accuracy is not important; more probes may be added when the query copy number is very high). We define the reference SNV as R-SNV and the query SNV as Q-SNV. In this example, assume that the probe R-SNV is a guanine (G), the probe Q-SNV is thymidine (T), the sample reference (R) (genomic or other) is adenine (A), and the sample query (Q) is cytidine (C); then the total (100%) of (R-SNV) + (R) in the mixture (%G + %A) is equal to the total (Q-SNV) + (Q) or (%T + %C). When an autosomal gene with two copies per genome is used as a reference, m is defined as the fraction of R-SNV and n is defined as the fraction of Q-SNV in a mixture, the fraction of R (%A) is (1 − %G) or (1 − m) and the fraction of Q (%C) is (1 − %T) or (1 − n). While the sum (R-SNV) + (R) is always equal to (Q-SNV) + (Q), the T/C ratio is equal to the G/A ratio only if the query, like the reference gene, is a diploid locus. T/C would be twice G/A if one query allele is lost (as expected if a homologous recombination has occurred and one copy of the wild-type allele is deleted or replaced with the payload). If many copies of a query transgene (or virus) were integrated, T/C would have a smaller value than G/A. In a more abstract form, the molar ratio of probe to sample in the mixture (G/A above) could be expressed as 2m/(1 − m) for a diploid locus. This ratio is equal to tn/(1 − n), where t is the copy number of the query gene in the genome. (If the query is also a diploid locus, t = 2; if more copies of the query are present, then t > 2. See Methods for details.) To derive the value (t), we plotted the G/A values against the T/C values to produce a line with zero intercept and calculate its slope (k). The value (t) is equal to 2k (t = 2k) when the diploid reference locus is used. To increase the accuracy, we plotted at least two m/(1 − m) values obtained in different reaction mixtures against n/(1 − n) values of the corresponding mixture. A graphic representation of these calculations is shown in Figure 1.
Gene copy number determination with RQPS
To demonstrate the utility of this method, we determined the gender of 18 animals in a blind experiment by applying RQPS to assess the copy number of the X-linked Pgk1 gene (males have one X chromosome, females have two). We made a plasmid containing a ligated fragment from the Pgk1 promoter (as the query probe) and a fragment from a Notch2 exon (as the reference probe) (Fig. 2A). Each was mutated to include a SNV. Two mixtures with different ratios of Notch2-Pgk1 RQPS probe to genomic DNA were prepared and quantitatively pyrosequenced in four reactions per individual, one for each SNV in each mixture (Fig. 2B). The m/(1 − m) value was plotted against n/(1 − n) using Microsoft Excel (see example in Fig. 2C). The animals were segregated into two groups: one with the slope k = 0.6 and another with the slope k = 1.2, consistent with t = 2k values of 1.2 for males and 2.4 for females (Fig. 2D,E). Importantly, the gender of all of the animals was correctly determined.
To further assess the accuracy of RQPS, we next measured the copy number of exon 30 in the Notch1 intracellular domain (N1ICD) in four different transgenic lines with clearly defined copy numbers (see Methods) (Murtaugh et al. 2003; Vooijs et al. 2007). The RQPS probe used for this experiment consisted of two short DNA fragments, one from Notch1 exon 23 and another from exon 30, mapping to Notch1 extracellular and intracellular domains, respectively, and fused together to function as reference and query probes (Methods; Supplemental Table S2). The slopes (k) obtained were ∼0.6, 1.1, 1.6, and 2.1 (Fig. 2F), correctly identifying animals with t = 1, 2, 3, and 4 copies of exon 30. This analysis was successfully repeated with additional Notch1 alleles under characterization in our laboratory, correctly identifying the number of N1ICD in various genomes.
Importantly, in mice containing the N1 + /Δ1 allele, which harbors a deletion removing exons 23–30 (Conlon et al. 1995), only one copy of exon 23 is present in the genome. When analyzing N1 + /Δ1; Gt(ROSA)26SorNotch/Notch DNA, we obtained a slope of k = 3.1 because three copies of exon 30 were present as expected and the reference allele is haploid (where t = k) (Fig. 2F,e). This highlights the importance of selecting a reference gene with a known copy number; selection of a haploinsufficient locus as a reference will ensure that the reference DNA is present in two copies per genome in healthy individuals.
Transgene copy number and zygosity determination with RQPS
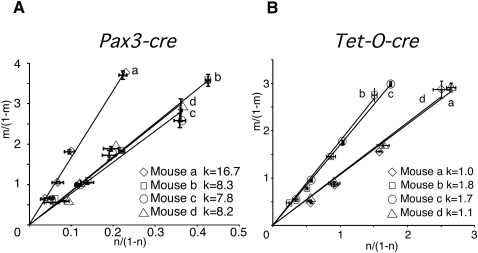
The frequent use of transgenic Cre lines and the difficulty in determining transgene copy number motivated us to apply RQPS to differentiate homozygous from heterozygous pups, a task that previously required knowledge of the transgene integration site or a test breeding. For this purpose, a general RQPS probe (Notch2-Cre RQPS) was prepared. Pax3-Cre transgenic mice (Li et al. 2000) were chosen for proof-of-principle analysis since the copy number and integration site of this transgene remained unknown. As shown in Figure 3A, we calculated the value (t) for four male pups by plotting the results from different probe dilutions for each genomic DNA sample. We observed three lines with the slope k = ∼8 and one with the slope k = ∼16. Multiple rounds of mating these studs with wild-type females confirmed that RQPS identified the lone homozygous male (Fig. 3A). Simultaneously, RQPS estimated the Pax3-Cre transgene copy number in hemizygotes to be roughly 16 (Fig. 3A). To determine if the Notch2-Cre RQPS probe could be used as a universal reagent for all Cre transgenic mice, we tested the Tet-O-Cre (Perl et al. 2002) and Six2-Gfp∷Cre (Humphreys et al. 2008) lines and determined that both contained two copies in hemizygotes (Fig. 3B; Supplemental Fig. S1). These results validated RQPS as a rapid, simple solution for the simultaneous determination of transgenic breeder zygosity and copy number. (The Notch2-Cre RQPS probe is available upon request.)
Figure 3.
RQPS differentiates homozygous from heterozygous transgenic animals. (A) RQPS on four Pax3-Cre animals. In this experiment, four different dilutions of RQPS probe were made with genomic DNA samples from each mouse; this was done to demonstrate that any two concentrations would have worked as well. Note the very small standard deviations for each data point, indicating that triplicates or duplicates are not necessary. Pyrosequencing of each mixture was performed in triplicate, and the average values were plotted; the standard deviation of each data point is shown. (B) RQPS of Tet-O-Cre mice. The copy number of the Tet-O-Cre transgene was measured in four animals. Four different mixtures were pyrosequenced in duplicate for each sample. RQPS detected two homozygotes and two heterozygotes, with about two and four copies of the transgene, respectively.
Detection of copy number variation in human patients using RQPS
We next tested if RQPS could be used to measure the gene copy number in seven human patient samples previously found to have duplication or loss of specific loci with array-CGH. Without knowing if loss of homozygosity (LOH) or duplication had occurred, the samples were subjected, in a semi-blind manner, to RQPS using probes consisting of small DNA fragments from genes located in the aberrant region, as query, and NOTCH2, as reference. The results obtained from five samples by RQPS were consistent with array-CGH data, including all four LOH cases and one duplication (Table 1). However, RQPS results conflicted with array-CGH for two samples. In both cases, the readout from RQPS was smaller (two copies by RQPS vs. three copies by array-CGH). Since the RQPS probes that we designed did not overlap with array-CGH probes, this could reflect a scenario wherein the region covered by the 60-bp-long array probe is duplicated, whereas the region detected by our RQPS probe is not. Indeed, when RQPS probes were redesigned to match the array-CGH probes, the two methods produced consistent results, identifying duplication in both loci. The discrepancy between the exact copy numbers (three copies vs. four copies) may not be surprising since probe response to ratio change in oligonucleotide arrays tends to be suppressed (Carter 2007). These data demonstrate that RQPS offers an economic alternative for confirmation of array-CGH results and is particularly effective in detecting defined deletions/amplifications. RQPS could be also used to map the boundaries of CNV.
Table 1.
CNV analysis in human patient samples by RQPS
For each sample, an RQPS probe, consisting of an exon fragment (∼50 bp) from within a randomly selected query gene residing within the CNV region fused to a NOTCH2 fragment, was prepared. Both fragments were mutated to contain an SNV. These probes were then used to estimate the copy number of seven loci in a semi-blind test. After RQPS, the results were compared to previously determined array-CGH data. Two discrepancies were found, which were resolved when RQPS was repeated with query sequences matching the CGH probe.
aRepeat reaction with RQPS probe matching the CGH probe.
bInconsistent result between RQPS probe to a region not covered by a CGH probe and the CGH array.
cBoth RQPS and array-CGH data detected amplification; they differ in estimating copy number.
Screening for homologous recombinants in ES cells
Next, we used RQPS to determine if we could correctly identify homologous recombination in ES cells by detecting the loss of one allele at the targeted locus. We tried this with ES cells that were electroporated with the N1∷CreERT2 construct. Homologous recombination will replace the Notch1 intracellular domain with DNA encoding CreERT2 and a FRT flanked selective marker; hence, only one wild-type allele remains in HR clones. We tested 32 ES cell clones enriched for HR candidates using a RQPS probe containing the Notch2 reference fragment and a query DNA fragment corresponding to the intracellular domain of Notch1 (Notch2–Notch1 RQPS) (see enrichment protocol below; Supplemental Table S3). Most clones had k-values of ∼1 (diploid), but nine clones had k-values near 0.5, as expected if one Notch1 allele is lost (Fig. 4A). A Southern blot of all 32 clones confirmed the identity of the nine clones (Fig. 4B), and RQPS correctly identified all the homologous recombinants in this pool. Therefore, the RQPS screening produced an accurate, simple, BAC-compatible HR identification protocol.
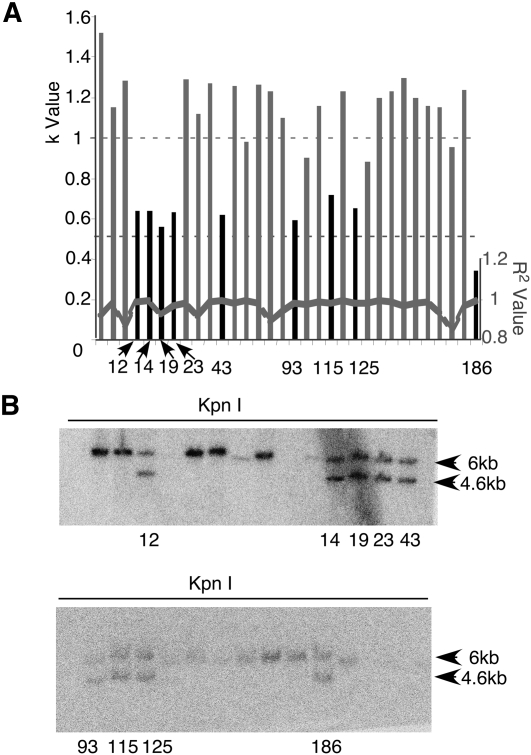
Figure 4.
Identifying HR clones in targeted ES cells using RQPS. (A) Refined screening of 32 ES cells falling within group A (Fig. 5B). Two different dilutions of RQPS probe were made for each ES cell genomic DNA sample and pyrosequenced. The black bar represents HR candidates (clone ID shown) with k-values indicating loss of one Notch1 allele. R2 values for a zero intercept line are also plotted. (B) All HR candidates identified in A were confirmed by Southern blot, each containing the WT allele fragment (6 kb) and the targeted allele fragment (4.6 kb) expected after KpnI digestion.
Enrichment for HR by incorporating SNV(s) into targeting vectors
RQPS can work as a primary screening method to accurately identify homologous recombinants regardless of homology-arm (HA) lengths, but it is still laborious if one is to screen hundreds of ES cells. To simplify the task and enrich for ES cells that experienced homologous recombination events independent of HA size, we introduced another simple innovation: insertion of two SNVs to the targeting vector (Fig. 5A). As mentioned in the introduction, one SNV is placed on the homology arm (the HA-SNV), roughly 50 bp away from the payload; the other was inserted into the neomycin selection cassette, using the same Pgk1 promoter SNV described in Figure 2. Enrichment for putative HR is accomplished by simply examining the ratio of these SNVs to their endogenous counterparts in a single quantitative pyrosequencing reaction (Fig. 5A). Because most facilities use male (XY) ES cells, only ES cell clones that have experienced a single integration, homologous or random, would display an allele ratio of 1:1 for the X-linked Pgk1 SNV. Any additional integration will increase the proportion of the vector Pgk1 SNV (two integration 66%, three 75%, etc.). On the other hand, the HA-SNV will only show a 1:1 ratio in homologous recombinants or in cells that experienced two random integrations, while under all other conditions, the fraction of HA-SNV will fall either around 33% (one random integration) or above 60% (at least three integrations). Therefore, only HR will display a 1:1 ratio for both SNVs. We called this enrichment method “pyroscreening.”
As a proof-of-principle, ES cells were electroporated with one of five different gene targeting constructs (a detailed description of each line will be provided elsewhere). Four of the five vectors were constructed in C57/B6 BAC clones using the BAC recombineering method (Warming et al. 2005), during which the desired SNVs were incorporated (Supplemental Fig. S2). Although pyroscreening is insensitive to HA length and should work equally well with BAC targeting vectors, we subcloned each targeting construct into a plasmid before electroporation into ES cells to facilitate parallel analysis with traditional PCR and Southern blot screening methods. Of these targeting vectors, two contained only the HA-SNV, while the other three also contained Pgk1 SNVs. Colonies were picked from cells selected by G418 resistance, expanded on mouse embryonic fibroblast (MEF) cells, and 808 clones were pyroscreened. As quantitative pyrosequencing has an error rate of ∼5%, we collected all clones with pyrosequencing readout between 40% and 60% (50% ± 2 × 5%) and tested them for enriched HR candidates. Even when only a single SNV was used, PCR analysis confirmed that a threefold enrichment was achieved (Fig. 5C; Supplemental Fig. S3). With two SNVs, a fourfold enrichment for HR was attained by selecting clones with an ∼50% SNV ratio for both Pgk1 and HA (Fig. 5B,C). Comparison with traditional PCR and Southern blot screening of the entire population determined that only two of the 21 positive HRs were missed by pyroscreening (Fig. 5B; Supplemental Figs. S3 and S4). Pyroscreening is thus an effective enrichment protocol, streamlining the analysis process by eliminating ∼80% of all G418-resistant clones prior to RQPS, PCR, or Southern blot analysis with an estimated false-negative rate of ∼9.5% (Supplemental Table S3). The false-positive clones are most likely due to the contamination of MEF cells, which affect the ratios of both HA and Pgk1 SNVs. We expect fewer or no false-positives if ES cells are grown without MEF cells. The SNV-containing selection cassette we have created (eukaryotic Pgk1/prokaryotic EM7 dual promoter-driven neomycin/kanamycin flanked by FRT site) is available upon request and should facilitate pyroscreening.
Discussion
We report here the adaptation of quantitative pyrosequencing to measure the copy number for any gene or DNA fragment in any species and its use in detecting homologous recombinants, transgene copy number, zygosity, and CNV validation. In order to apply pyrosequencing to any sequence, the investigator needs to generate a RQPS probe consisting of two SNV-containing fragments with homology with a query locus and a reference locus, respectively. Query loci could be integrated DNA or endogenous loci that underwent micro duplication or loss of DNA. The design of the SNV-containing RQPS probes renders our method independent of naturally occurring SNPs and reference alleles, distinguishing it from other pyrosequencing-based CNV detection methods reported so far (Pielberg et al. 2003; Söderbäck et al. 2005). This endows RQPS with great flexibility while retaining the simplicity and cost effectiveness of pyrosequencing. Once a probe is made for a specific locus of interest, a large number of samples could be easily and economically processed, which makes RQPS ideal for high-throughput clinical diagnosis of the CNV status at a limited number of loci.
Another application we demonstrated is an enrichment protocol to identify potential homologous recombinants. By introducing two artificial SNVs, one into the Pgk1 promoter of the selection cassette and the second into the homology arm, most negative clones can be discarded (Fig. 5). It is important to point out that the MEF feeder cells used in this study are derived from the F1 embryos of a cross between Jackson transgenic C57BL/6J-Tg (pPWL512hyg)1Ems/J homozygotes and C57BL/6J-TG (pPGKneobpa)3Ems/J homozygotes. In such cells, both hygromycin- and neomycin-selective cassettes are driven by mouse Pgk1 promoters. Assuming an equal number of male and female MEF cells, at least one copy from each transgene and 1.5 copies from the endogenous Pgk1 gene are present, and thus each MEF cell contains at least 3.5 copies of Pgk1 promoter. Despite this confounding factor, we achieved an overall threefold enrichment even with a single SNV pyroscreening. Better performance of pyroscreening will be attained when ES cells are grown without feeder cells (Nichols et al. 1990). It is worth noting that placing a homology-arm, silent SNV into an exon in the gene-targeting construct offers the important additional benefit of distinguishing the two transcripts generated from the endogenous and targeted alleles by quantitative pyrosequencing on reverse-transcribed cDNA.
In contrast to pyroscreening, ES cells can be directly screened with RQPS independent of targeting vectors. Like Q-PCR, RQPS screens for positive clones by detecting the loss of one wild-type allele in the targeted locus (Valenzuela et al. 2003). Yet unlike Q-PCR, RQPS does not measure a twofold reduction in the amount, but rather determines the ratio of two different nucleotides. Because the relative ratio of alleles in a mixture is measured within a single tube, system errors are minimized. We have shown that RQPS can correctly identify all positive clones after enrichment by pyroscreening and could thus replace the Southern blot screening method. As BAC recombineering has made the construction of a large number of gene targeting vectors in a short period of time possible (Valenzuela et al. 2003), the automation-friendly RQPS, alone or in conjunction with pyroscreening, will effectively streamline the ES cell screening process.
Methods
Generation of targeting constructs
The five targeting constructs described in the text are: (1) Notch1∷ creHi; (2) Notch1∷CreERT2, in which the Notch1 intracellular domain (ICD) is replaced with Cre recombinase or tamoxifen-inducible CreERT2 fusion protein, respectively; (3) Notch2∷cre, in which the Notch2 ICD is replaced with Cre recombinase; (4) N2-N1, in which the Notch2 ICD is replaced by the Notch1 ICD; and (5) N1-N2 in which the Notch1 ICD is replaced by the Notch2 ICD. To explain how SNVs are simultaneously incorporated into a targeting construct through BAC recombineering, we provide details of constructing the N1∷CreERT2 vector (below; Supplemental Fig. S2). The construction details and rationale for designing the remaining constructs will be described elsewhere.
In brief, a 6Xmyc-tagged CreERT2 fusion gene was first cloned in front of a FRT-flanked neomycin selection cassette, whose expression was under the control of both mouse Pgk1 and Escherichia coli EM7 promoter. An artificial SNV was introduced into the mouse Pgk1 for pyroscreening by site-directed mutagenesis. Next, we amplified a fragment containing the blastidin promoter-driven ampicillin-resistance gene using pBlueScript as a PCR template. Two pairs of flanking mini-HA were added in subsequent rounds of PCR. The outer mini-HAs (magenta) mediated the recombination of the fragment into a Notch1 BAC; the inner mini-HAs (cyan) mediated subsequent replacement of the ampicillin cassette by recombination of a payload cassette (6xMyc-CreERT2FRT; PGKEM7-Neo). In addition, a second SNV was inserted into the outer mini-HAs (Supplemental Fig. S2). This strategy (insertion of the mini-HR) circumvents the need to amplify the payload in order to introduce homology arms into it and thus facilitates the insertion of large DNA fragments into a BAC. For the studies described here, the CreERT2FRTNeo cassette with 4.8 kb upstream and 2.2 kb downstream flanking BAC DNA was recombined into a pBluescript plasmid as described (Liu et al. 2003).
ES cell electroporation
Forty micrograms of linearized targeting construct was electroporated into either 129 derived SSC10 (for N1∷creHi) or C57BL6 derived B6/BLU (all other constructs) ES cells by the Siteman Cancer Center ES Core at Washington University (http://escore.im.wustl.edu). Neomycin-resistant clones were selected by the core and expanded by the investigators for further screening.
PCR and Southern blot screening of ES cells
Nested PCR was used for the screening of ES cells that were electroporated with targeting vectors with long HAs. Purified BAC DNA containing a desired targeting construct was diluted with wild-type mouse genomic DNA and used as a positive control for the optimization of PCR conditions. PCR primers with the expected size of the PCR products are listed in Supplemental Table S4. The 25-μL PCR reaction included: 2.5 μL of 10× PCR buffer, 0.25 μL of KlenTaqLA (Sigma), 0.5 μL of 10 mM dNTP mixture, 1 μL each of 20 μM upstream and downstream primers, 8.25 μL of 4 M betaine, and 1 μL of DNA template of unknown concentration. The PCR cycle conditions were 5 min at 94°C, followed by 35 cycles of 94°C (1 min), 50°C ∼65°C (1 min, based on primer sequence), 68°C (2–5 min depending on the amplicon size at 1 min/kb), and finally 10 min at 68°C. The Southern blot was done as described (Sambrook and Maniatis 1989) with [α-32P32]dCTP labeled probes (GE Healthcare).
Pyrosequencing
For each measurement, we PCR-amplified the SNV-containing region with two target-specific primers flanking the desired SNV, one of which (a tailed primer) included a universal sequence complementary to a third, the universal biotinylated primer (UBP) (Fig. 2A; Fakhrai-Rad et al. 2002; Royo et al. 2007). All primers were designed using Pyrosequencing Assay Design Software (Biotage) (Supplemental Table S2). It is worth noting that the PCR amplification efficiency of fragments that differ by only a single SNV in the same PCR reaction mixture is the same unless the SNV creates or disrupts a stable secondary structure, a complication that can be avoided when the SNV is chosen by the investigator. Single-strand DNA PCR products are then purified with streptavidin-conjugated Sepharose beads and used for pyrosequencing analysis. The whole procedure is high-throughput-friendly, and 96 samples could be processed within 3–4 h (from the setup of PCR reactions to the obtaining of pyrosequencing results). This three-primer PCR reaction system (Fakhrai-Rad et al. 2002; Royo et al. 2007) was used for the amplification of the DNA fragments containing an SNV in a PCR reaction mixture containing 2 μL of DNA template, 12.5 μL of Bullseye Taq DNA Polymerase Mix (Midsci), 1 μL of untailed gene-specific primer, 0.2 μL of tailed gene-specific primer, 0.8 μL of UBP-labeled (all at a concentration of 5 μM), and 8 μL of distilled water (primer sequences are provided in Supplemental Table S2). PCR conditions were 8 min at 95°C followed by 40 cycles at 95°C (30 sec), 60°C (30 sec), and 72°C (30 sec).
Single-strand PCR products were purified as described (Bruinsma et al. 2008). Briefly, single-stranded biotinylated PCR products were immobilized on streptavidin-coated Sepharose beads in a vacuum prep tool (Biotage). For each reaction, 40 μL of Binding Buffer (10 mM Tris-HCl, 2 M NaCl, 1 mM EDTA, 0.1% Tween 20 at pH 7.6), 20 μL of double-distilled water, and 3 μL of Streptavidin Sepharose High Performance beads (GE Healthcare) were mixed together with 20 μL of PCR product for 10 min at room temperature using an Eppendorf Thermomixer. After immobilization, vacuum was applied to the prep tool and the template was captured on filter probes. The non-biotinylated template was then denatured by alkali treatment and removed by washing for 5 sec in 70% EtOH, 0.2 M NaOH denaturation solution (5 sec), and 10 mM tris-acetate washing buffer (pH 7.6) for 5 sec. The last washing step neutralized the pH. Next, the vacuum was released, and the beads were transferred into a Biotage PSQ 96 well plate containing the sequencing primer (5 pmol) and 40 μL of annealing buffer (20 mM tris-acetate, 2 mM MgAc2 at pH 7.6). The pyrosequencing reactions were performed using a PSQ96-MA machine (Biotage) according to the manufacturer's instructions using the Pyro Gold reagent kit (Biotage). Assays were performed using cyclic enzyme and nucleotide dispensation, followed by analysis using the SNV Software (Biotage).
ES cells from individual clones were trypsinized and harvested. Genomic DNA was purified as described (Laird et al. 1991) and dissolved in 50 μL of TE (pH 8.0). For pyroscreening, 2 μL of ES cell genomic DNA (unknown concentration) was used for PCR reaction, followed by single-strand DNA purification and pyrosequencing. Clones with an SNV ratio between 40% and 60% were considered as positive candidates for further screening.
RQPS
For RQPS, the concentration of both genomic DNA and the linearized RQPS probe was measured. At least two mixtures of genomic DNA/RQPS probe were then prepared, in which the molar ratio between the two was between 9:1 and 1:9 (we used 1:1 and 1:2 when two mixtures were done). When 30 ng of mouse or human genomic DNA is used, for an RQPS probe with the size of 3–4 kb, 0.03–0.04 pg is required to get a roughly 1:1 molar ratio. For the detailed calculation method, please see http://www.med.umich.edu/tamc/spike.html. A 2-μL sample from each mixture was used as the template for the PCR reactions with reference or query specific primers, followed by single-strand DNA purification and pyrosequencing.
Mathematical derivation of gene copy numbers with RQPS
To calculate the copy number of a query gene in a genome (t), we created a mixture containing (a) genomes and (b) molecules of RQPS probe. Since each RQPS probe contains one reference sequence, there are (b) R-SNV nucleotides in the mixture. In contrast, each diploid genome contains two alleles of the autosomal reference gene and (t) copies of the query gene. Thus, the number of Rs-SNV nucleotide is (2a) and the number of Qs-SNV is (ta). Therefore, the fraction of the R-SNV in the mixture (m) is equal to the molar ratio R-SNV/(R-SNV + Rs-SNV) or:
Likewise, the fraction of the Q-SNV in the mixture (n) is
Equation 1 could be solved for b:
and, similarly, Equation 2 could be solved for b:
Thus,
and can be further solved to:
and
When the value [m/(1 − m)] is plotted against [n/(1 − n)], a line passing the origin will have the slope k that equals half of the transgene copy number in the genome (k = 0.5t). If a single-copy gene is used as reference, k = t.
For the detection of the loss of one allele in ES cells, contamination from feeder cells needs to be considered. If MEF cells are present in cases in which only nonhomologous recombination occurs, the copy number of targeted alleles per genome will remain 2. If the ES cell undergoes homologous recombination, there is only one targeted allele in each genome of the ES cell, whereas two copies of the targeted allele are present in the MEF genome. This will result in higher t values than the expected 0.5, resulting in a false-negative.
Data process
All pyrosequencing data were processed with Microsoft Excel. m/(1 − m) was plotted against n/(1 − n), and linear data trend lines were forced to have a zero intercept.
Generation of animals with defined copy number of Notch1 exon 30
To generate animals with one, two, three, and four copies of exon 30 but with a diploid number of the Notch1 extracellular domain (exon 23), we used the N1∷Cre line (Vooijs et al. 2007) in which Cre replaced exon 30 in one allele, and the Gt(ROSA)26SorNotch (Murtaugh et al. 2003) line in which one copy of the Gt(ROSA)26Sor locus is replaced with a cDNA encoding N1ICD to generate a mouse with three copies of exon 30. Wild-type animals have two alleles of Notch1, and homozygous N1+/+; Gt(ROSA)26SorNotch/Notch has four copies of exon 30.
Array-CGH
Array-CGH was carried out as described (Schwarzbraun et al. 2009).
Acknowledgments
We thank Daniel Schneider and Kerry Kornfeld for their great help with pyrosequencing and Mary Fullbright for critically reading the manuscript. RO1 DK066408 to R.K. supported this research. R.K., Z.L., and Washington University filed a patent application No. 61/110,073 describing this technology.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.093856.109.
References
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Armour JA, Palla R, Zeeuwen PL, den Heijer M, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten E, White SJ, Kalf ME, Vossen RH, Thygesen HH, Ruivenkamp CA, Kriek M, Breuning MH, den Dunnen JT. Methods to detect CNVs in the human genome. Cytogenet Genome Res. 2008;123:313–321. doi: 10.1159/000184723. [DOI] [PubMed] [Google Scholar]
- Bruinsma JJ, Schneider DL, Davis DE, Kornfeld K. Identification of mutations in Caenorhabditis elegans that cause resistance to high levels of dietary zinc and analysis using a genomewide map of single nucleotide polymorphisms scored by pyrosequencing. Genetics. 2008;179:811–828. doi: 10.1534/genetics.107.084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier F, Raux G, Wang Q, Drouot N, Cordier F, Limacher JM, Saurin JC, Puisieux A, Olschwang S, Frebourg T. Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res. 2000;60:2760–2763. [PubMed] [Google Scholar]
- Clarke SC. Pyrosequencing: Nucleotide sequencing technology with bacterial genotyping applications. Expert Rev Mol Diagn. 2005;5:947–953. doi: 10.1586/14737159.5.6.947. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Cotta-de-Almeida V, Schonhoff S, Shibata T, Leiter A, Snapper SB. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: An accurate detection platform for single nucleotide polymorphisms. Hum Mutat. 2002;19:479–485. doi: 10.1002/humu.10078. [DOI] [PubMed] [Google Scholar]
- Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, Heward JM, Gough SC, de Smith A, Blakemore AI, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rodriguez J, Washington V, Cheng J, Dutra A, Pak E, Liu P, McVicar DW, Schwartzberg PL. Advantages of q-PCR as a method of screening for gene targeting in mammalian cells using conventional and whole BAC-based constructs. Nucleic Acids Res. 2008;36:e117. doi: 10.1093/nar/gkn523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, van de Kerkhof PC, Traupe H, de Jongh G, den Heijer M, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langaee T, Ronaghi M. Genetic variation analyses by Pyrosequencing. Mutat Res. 2005;573:96–102. doi: 10.1016/j.mrfmmm.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Langerak P, Nygren AO, Schouten JP, Jacobs H. Rapid and quantitative detection of homologous and non-homologous recombination events using three oligonucleotide MLPA. Nucleic Acids Res. 2005;33:e188. doi: 10.1093/nar/gni187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Jeon JT. Methods to detect and analyze copy number variations at the genome-wide and locus-specific levels. Cytogenet Genome Res. 2008;123:333–342. doi: 10.1159/000184725. [DOI] [PubMed] [Google Scholar]
- Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–164. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110:1341–1348. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]
- Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielberg G, Day AE, Plastow GS, Andersson L. A sensitive method for detecting variation in copy numbers of duplicated genes. Genome Res. 2003;13:2171–2177. doi: 10.1101/gr.1188203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Shokralla S, Gharizadeh B. Pyrosequencing for discovery and analysis of DNA sequence variations. Pharmacogenomics. 2007;8:1437–1441. doi: 10.2217/14622416.8.10.1437. [DOI] [PubMed] [Google Scholar]
- Royo JL, Hidalgo M, Ruiz A. Pyrosequencing protocol using a universal biotinylated primer for mutation detection and SNP genotyping. Nat Protoc. 2007;2:1734–1739. doi: 10.1038/nprot.2007.244. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Maniatis EF. Molecular Cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbraun T, Obenauf AC, Langmann A, Gruber-Sedlmayr U, Wagner K, Speicher MR, Kroisel PM. Predictive diagnosis of the cancer prone Li-Fraumeni syndrome by accident: New challenges through whole genome array testing. J Med Genet. 2009;46:341–344. doi: 10.1136/jmg.2008.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderbäck E, Zackrisson AL, Lindblom B, Alderborn A. Determination of CYP2D6 gene copy number by pyrosequencing. Clin Chem. 2005;51:522–531. doi: 10.1373/clinchem.2004.043182. [DOI] [PubMed] [Google Scholar]
- Testa G, Zhang Y, Vintersten K, Benes V, Pijnappel WW, Chambers I, Smith AJ, Smith AG, Stewart AF. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat Biotechnol. 2003;21:443–447. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, van den Born M, Stappenbeck T, Wu Y, Clevers H, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Elbein SC. Detection of allelic imbalance in gene expression using pyrosequencing. Methods Mol Biol. 2007;373:157–176. doi: 10.1385/1-59745-377-3:157. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson J, Skolnick G, Love-Gregory L, Permutt MA. Assessing allele frequencies of single nucleotide polymorphisms in DNA pools by pyrosequencing technology. Biotechniques. 2002;32:1144–1146. doi: 10.2144/02325dd04. 1148, 1150. [DOI] [PubMed] [Google Scholar]
- Yang Y, Seed B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat Biotechnol. 2003;21:447–451. doi: 10.1038/nbt803. [DOI] [PubMed] [Google Scholar]