Abstract
The Singapore Genome Variation Project (SGVP) provides a publicly available resource of 1.6 million single nucleotide polymorphisms (SNPs) genotyped in 268 individuals from the Chinese, Malay, and Indian population groups in Southeast Asia. This online database catalogs information and summaries on genotype and phased haplotype data, including allele frequencies, assessment of linkage disequilibrium (LD), and recombination rates in a format similar to the International HapMap Project. Here, we introduce this resource and describe the analysis of human genomic variation upon agglomerating data from the HapMap and the Human Genome Diversity Project, providing useful insights into the population structure of the three major population groups in Asia. In addition, this resource also surveyed across the genome for variation in regional patterns of LD between the HapMap and SGVP populations, and for signatures of positive natural selection using two well-established metrics: iHS and XP-EHH. The raw and processed genetic data, together with all population genetic summaries, are publicly available for download and browsing through a web browser modeled with the Generic Genome Browser.
The detailed survey of human genomic variation across four populations globally from the International HapMap Project (The International HapMap Consortium 2005, 2007) has yielded valuable insights into the design (de Bakker et al. 2005; Pe'er et al. 2006) and analysis (Marchini et al. 2007) of studies that examine the entire genomic landscape for correlation with the onset of diseases or traits. These genome-wide association studies (GWAS) typically detect indirect associations, where the identified genetic variants by themselves are not biologically functional but are in the neighborhood and thus are correlated or are in linkage disequilibrium (LD) with the causal polymorphisms. Commercial genotyping arrays for genome-wide studies utilize these informative markers for providing suitably dense genomic coverage, which with the appropriate use of sophisticated imputation methods can increase the effective genomic coverage of these arrays to that of the HapMap by statistically inferring the genotypes of the remaining unobserved markers in the HapMap (Marchini et al. 2007; Servins and Stephens 2007). The accuracy of genotype imputation, however, relies on having reference databases that are representative of the target populations to be imputed. While it has been shown that tagging SNPs identified from the HapMap are expected to be portable across other non-African populations (de Bakker et al. 2006; Conrad et al. 2006; Huang et al. 2009), imputation performance is expected to be optimized if local reference haplotypes are used (Huang et al. 2009; Jallow et al. 2009). The ability to reproduce an association finding in other populations through replication studies or meta-analyses is a prerequisite to validating the authenticity of the discovery (NCI-NHGRI Working Group on Replication in Association Studies 2007), and this fundamentally relies on having a similar LD structure between the identified variant and the functional polymorphism in these populations (Teo et al. 2009a). The success of imputation procedures, meta-analyses, and replication studies thus hinges critically on possessing sufficient knowledge on the extent of genomic variation between multiple populations. The Singapore Genome Variation Project (SGVP) is established with this aim of characterizing genomic variation and positive natural selection in three major population groups in Asia.
Singapore is a relatively young country with a migratory history predominantly consisting of immigrants with Chinese, Malay, and Indian genetic ancestries from neighboring countries such as China, India, Indonesia, and Malaysia (Saw 2007). The Chinese community consists mainly of descendents of Han Chinese settlers from the southern provinces of China, such as Fujian and Guangdong, and currently represents the dominant racial population in Singapore, accounting for 76.7% of the resident population from the Singapore Census conducted in 2000 (Saw 2007). While Han Chinese represents the largest ethnic group amongst the Chinese globally, there are a considerable number of sub-ethnicities within the Han classification with a diverse range of dialects and cultural diversity, with established genetic heterogeneity following a geographical north–south cline (Chu et al. 1998; Wen et al. 2004). The majority of the early Chinese immigrants to Singapore were mainly attributed to the dialect groups of Hokkien, Teochew, Cantonese, Hakka, and Hainanese (Saw 2007) that are predominantly found in Southern China. While Malays formed the dominant race in Singapore prior to the colonization by British settlers, the proportion of indigenous Malays has been surpassed by migrant Malays from Peninsula Malaysia, as well as Javanese and Boyanese people from Indonesia. Cultural and religious similarities have resulted in intermarriages between the immigrant and local Malays, whose descendents are now collectively known as Malays and account for 13.9% of the Singapore population (Saw 2007). The British colonization of Singapore also brought Indian migrants from the Indian subcontinent, with the majority consisting of Telugas and Tamils from southeastern India and a minority of Sikhs and Pathans from north India. The definition of Indians in Singapore comprises people with paternal ancestries tracing back to the Indian subcontinent, and, as a race, Indians represent 7.9% of the Singapore population. Cumulatively, the SGVP resource has the potential for representing the genetic diversity across multiple large populations in Asia while serving as a useful complement to the HapMap database.
This paper aims to describe the SGVP resource, which genotyped in excess of 2 million polymorphisms across 99 Chinese, 98 Malay, and 95 Indian individuals. The genotype data, phased haplotypes, and other data summaries for this resource have been modeled after the format of the International HapMap Project and are publicly available online. In addition, this paper details the extent of population differences between the SGVP, the HapMap, and the populations from the Human Genome Diversity Project (HGDP) (Rosenberg et al. 2002; Jakobsson et al. 2008; Li et al. 2008). We also compared the diversity of SNPs and haplotypes between the populations in the HapMap and SGVP, with a particular focus on the extent of LD variations between these populations. A genome-wide survey for candidate signatures of recent positive natural selection was also performed in the SGVP populations, replicating a number of previous findings from HapMap while identifying novel candidates, particularly in the Malay and Indian population groups.
Results
Sample and SNP quality control
A total of 292 individuals comprising of 99 Chinese, 98 Malays, and 95 Indians were genotyped across 2,007,788 SNPs on the Affymetrix SNP6.0 and Illumina 1M arrays, of which 268,667 SNPs overlap between the two platforms. The fidelity and accuracy of the genotype data are of paramount importance in establishing reference haplotype maps. We implemented a hierarchical quality control (QC) procedure that begins with an initial round of SNP QC to identify a set of “pseudo-cleaned” SNPs for detecting problematic samples. Samples with high levels of missingness, potential relatedness, and discordance between self-reported and genetically inferred population membership were identified and excluded from further analyses (Supplemental Table S1). A final round of SNP QC was performed within each population separately on the basis of missingness, departures from Hardy–Weinberg equilibrium (HWE), excessive discordance in the genotypes for the duplicated samples, and annotation failures. A total of 96 Chinese, 89 Malays, and 83 Indians remained after merging the SNP data from both arrays. Here, we further excluded SNPs that were common on both arrays but with <95% concordant genotypes, and SNPs that mapped to different alleles on the forward strand according to the SNP manifests from Affymetrix and Illumina. This yielded a final post-QC set with 1,584,040 autosomal SNPs for Singapore Chinese (CHS); 1,580,905 SNPs for Singapore Malays (MAS); and 1,583,454 SNPs for Singapore Indians (INS) (Supplemental Table S1), with an average inter-SNP distance of 2 kb across most of the genome (Supplemental Figs. S1, S2). The overall concordance in the genotype calls for the sample duplicates was 99.899%, at an overall call rate of 99.285%. Details of the QC process can be found in the Methods and Supplemental material.
Population structure
Principal components analysis (Price et al. 2006) and Wright's FST statistic (Wright 1951) were used to explore the extent of population differentiation between the SGVP, HapMap, and HGDP populations (Supplemental Table S2).
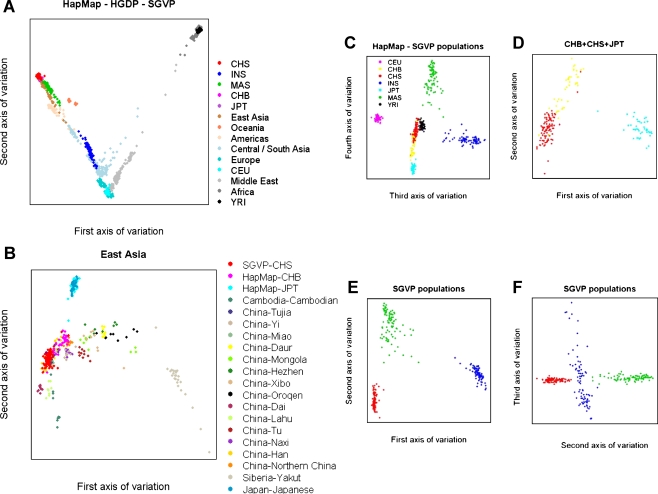
In the context of global genetic diversity, Singapore Chinese, the HapMap Han Chinese in Beijing, China (CHB), and HapMap Japanese in Tokyo, Japan (JPT) were virtually indistinguishable, while Singapore Malays were observed to be highly similar to the East Asian populations in general (Fig. 1A). Singapore Indians were comparable to samples from Central and South Asia, and genetically closer to the samples with European ancestries than to the East Asian samples from HGDP. As with the non-African populations in HapMap and HGDP, all three Singapore groups were considerably distinct from the HapMap Yoruba samples from the Ibadan region of Nigeria (YRI) and the African samples in the HGDP. The first axis of variation at this global level effectively distinguished samples from the Far East from Africans, while the second axis of variation addressed the difference between European and African ancestries. Comparing between the East Asian populations, the first axis separated the Yakut people of Siberia from Chinese sub-ethnic groups mainly located in Southern China (Dai, Lahu) and Southeast Asia (Cambodian, CHS) (Fig. 1B). When we consider only the HapMap and SGVP populations, the third axis of variation separated INS from the HapMap Utah samples with ancestry from Northern and Western Europe (CEU), while MAS was differentiated from the Far East Asian cluster (comprising CHB, CHS, and JPT) by the fourth axis of variation (Fig. 1C).
Figure 1.
Principal component analysis plots of genetic diversity across HapMap, HGDP, and SGVP populations. Each figure represents the genetic diversity seen across the populations considered, with each sample mapped onto a spectrum of genetic variation represented by two axes of variations corresponding to two eigenvectors of the PCA. (A) Individuals from each population in the HapMap and SGVP are represented by a unique color, while samples from HGDP are broadly grouped by geography in which a unique color is assigned to each geographical location. (B) Comparison between CHS and samples from Far East Asia found in the HapMap and HGDP. (C) A plot of the third and fourth axes of variation for the seven populations from HapMap and SGVP. (D) A plot of the first two axes of variation when the PCA is run on only the three Far East Asian populations comprising the Singapore Chinese, HapMap Han Chinese in Beijing, China, and Japanese in Tokyo, Japan. (E) A plot of the first two principal components in a separate analysis within the three SGVP populations. (F) A plot of the second and third principal components within the SGVP populations. The same color scheme has been used in C–F; the legend for the color assignment can be found in C.
Comparing within the three Far East Asian populations from HapMap and SGVP, the JPT samples were clearly more different from the two Chinese cohorts (FST = 0.3% with CHB; 0.4% with CHS) than between the two Chinese cohorts themselves, although substantial dissimilarities exist to distinguish between the two Chinese cohorts (FST = 0.2%; Fig. 1D). In the latter analysis, a few CHB samples were clustered together with most of the CHS samples and vice versa (see also Supplemental Fig. S3). The separation seen between samples from CHB and CHS may be indicative of a north–south genetic cline, as Singapore Chinese are predominantly descendents of immigrants from southern provinces in China, while we expect the HapMap Han Chinese in Beijing samples to mainly reflect the genetic ancestry from northern China. It is possible that the HapMap Han Chinese samples from Beijing have included individuals with genetic ancestries more commonly seen in Southern China, and likewise with the Singapore Chinese samples, as it is evident from the Chinese samples in HGDP (Fig. 1B) that the designation of Han Chinese encompasses people from genetically distinguishable sub-groups or sub-ethnicities. Within the SGVP populations, the INS was more differentiated compared with CHS (FST = 3.9%) and MAS (FST = 2.7%), than between the Chinese and the Malay samples (FST = 0.6%, Fig. 1E). Interestingly, the third axis of variation indicated there was substantial genetic variability within the Indian samples (Fig. 1F), which may be attributed to the numerous ethnicities that comprise the Indian population.
SNP and haplotype diversity
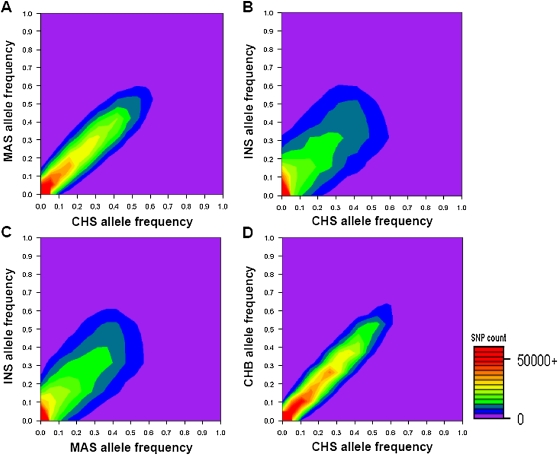
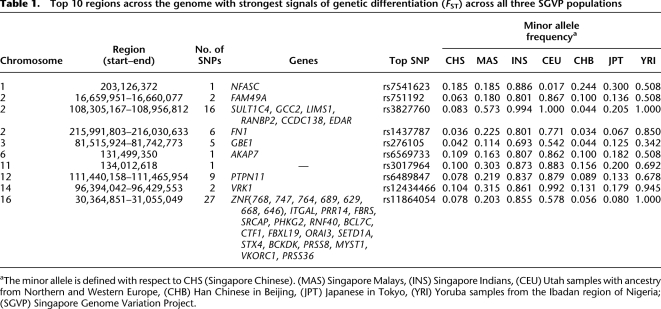
The availability of accurate genome-wide data allows the assessment of genetic diversity across the SGVP populations. At the SNP level, there was considerably less variance in the allelic spectrum between CHS and MAS, relative to comparisons between either population and INS (Fig. 2A–C), while, expectedly, CHS was most similar to CHB (Fig. 2D; Supplemental Figs. S4, S5). In a genome-wide survey for regions that are highly differentiated in the SGVP populations, the top 10 regions were attributed mainly to allele frequency variations between INS and the two other populations and encapsulated well-documented regions of genomic differentiation between East Asian and other global populations, including EDAR (Sabeti et al. 2007) and VKORC1 (Lal et al. 2006; Lee et al. 2006) (Table 1).
Figure 2.
Allele frequency comparison between pairs of populations. The axes in each figure represent the allele frequencies for each of the two represented populations. For each SNP, we define the minor allele after agglomerating the genotype data from all three SGVP populations and subsequently calculate the frequency of this allele in each population. Twenty allele frequency bins each spanning 0.05 units are constructed for each population, and we tabulate the number of SNPs found in each bin. The intensity of the contour represents the number of SNPs that displayed the corresponding allele frequencies in the two populations, from a low number of SNPs (purple) to a higher number of SNPs (red). The figure panels compare the allelic spectrum among CHS-MAS (A), CHS-INS (B), MAS-INS (C), and CHS-CHB (D).
Table 1.
Top 10 regions across the genome with strongest signals of genetic differentiation (FST) across all three SGVP populations
aThe minor allele is defined with respect to CHS (Singapore Chinese). (MAS) Singapore Malays, (INS) Singapore Indians, (CEU) Utah samples with ancestry from Northern and Western Europe, (CHB) Han Chinese in Beijing, (JPT) Japanese in Tokyo, (YRI) Yoruba samples from the Ibadan region of Nigeria; (SGVP) Singapore Genome Variation Project.
To investigate the extent of haplotype diversity across the seven SGVP and HapMap populations, we calculated the percentage of the chromosomes within each population that can be accounted for by a specified number of distinct haplotypes across 22 regions of 500 kb. We observed that there was considerably higher haplotype diversity in YRI compared with the rest, while the populations with Far East Asian ancestries (CHB, CHS, and JPT) have the lowest haplotype diversity (Supplemental Fig. S6). For example, 12 haplotypes accounted for only 43% of the YRI chromosomes, and between 73% (for JPT) and 79% (for CHS) for the three populations with Far East Asian ancestries. Among the SGVP populations, INS has the greatest haplotype diversity, with 12 haplotypes accounting for 57% of the INS chromosomes. This is followed by MAS, with 68% of the chromosomes accounted for by 12 haplotypes. The pattern of haplotype sharing between these populations was very similar across the 22 regions, and we illustrate this with chromosome 1 (Supplemental Fig. S6). A high degree of haplotypes was shared between CHB, CHS, and JPT, and it was evident that there were different haplotypes present in CEU, INS, and YRI that were either absent or at low frequencies in the rest of the populations. These analyses concurred with the observations from the analysis of population structure that CHB, CHS, and JPT are more genetically similar compared with the rest of the populations, with INS being the most genetically diverse among the SGVP populations.
Linkage disequilibrium, tagging efficiency, and LD variation
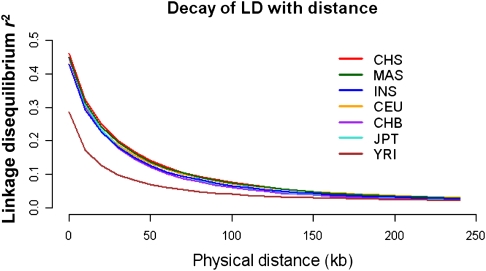
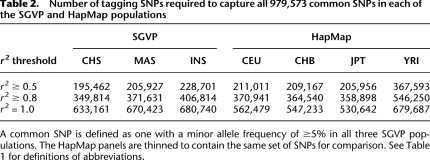
One important utility of the SGVP resource is the comparison of the extent of LD between the SGVP and HapMap populations, as this reflects the tagging efficiency for genotyping arrays that were designed using the patterns of LD that were observed in the HapMap populations. Overall, the SGVP populations exhibited similar rates of LD decay with increasing distance as compared with the HapMap non-African populations, with CHS and INS having the greatest and least conservation of LD, respectively, with distance amongst the three SGVP populations (Fig. 3). This is similarly reflected in the number of tagging SNPs that are required to capture all the common SNPs in the SGVP panels at a pairwise r2 threshold of 0.8, where between 349,800 and 406,900 SNPs are required for CHS and INS, respectively (Table 2). For comparison, the corresponding range for the HapMap populations is between 358,800 and 546,300 for JPT and YRI, respectively. Intriguingly, we observed the number of tagging SNPs required at a pairwise r2 threshold of 1 for each SGVP population is almost comparable to the number required for YRI, although this is likely to be a consequence of designing commercial genotyping microarrays utilizing LD patterns observed in the HapMap populations.
Figure 3.
Decay of LD with distance. Decay of LD as measured by the r2 statistic with increasing distance up to 250 kb for each of the HapMap and SGVP populations, where 90 chromosomes were chosen from each population to perform the LD calculation. Only SNPs with minor allele frequencies ≥5% in each population were considered in this analysis.
Table 2.
Number of tagging SNPs required to capture all 979,573 common SNPs in each of the SGVP and HapMap populations
A common SNP is defined as one with a minor allele frequency of ≥5% in all three SGVP populations. The HapMap panels are thinned to contain the same set of SNPs for comparison. See Table 1 for definitions of abbreviations.
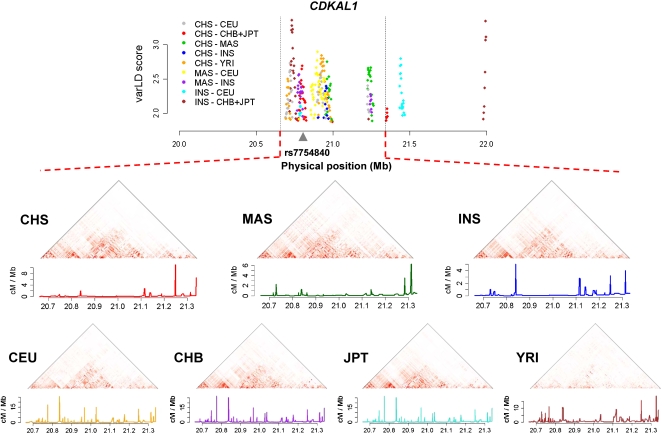
One of the factors that affects the reproducibility of the association results from GWAS is the degree of similarity in the correlation structure between the causal variants and the reported SNPs in these populations (Teo et al. 2009a). By comparing the extent of LD differences in a sliding-window approach between any two populations, we identified the regions that are found in the top 5% of the distribution of LD differences as candidate regions of LD variation, where consecutive signals in the top 5% within 25 kb are binned as a single region (see Methods). As a significant proportion of GWAS has been performed in populations of European descent, Supplemental Table S3 shows the top 10 candidate regions of LD variation between each SGVP population and CEU, while a complete listing of the identified regions in the top 0.1% of the distribution between pairs of populations from SGVP and HapMap can be found in Supplemental Table S4. Perhaps unsurprisingly, one of these regions observed between INS and CEU spans the SLC24A5 gene, which has been established to be functionally involved with skin pigmentation (Lamason et al. 2005). A region that shows considerable signals of LD variations from multiple pairs of populations and that coincided with reported association signals from GWAS spans the CDKAL1 gene, which has been implicated with Type 2 diabetes in populations with European ancestry (Saxena et al. 2007; Scott et al. 2007; Steinthorsdottir et al. 2007; Zeggini et al. 2007) and also in Asian populations such as the Chinese (Liu et al. 2008; Wu et al. 2008), Koreans (Ng et al. 2008), and Japanese (Tabara et al. 2009). Our analysis indicates that the implicated variant rs7754840 is found in a region with extensive LD differences between multiple groups (Fig. 4). The population-specific recombination profiles differed between the SGVP and HapMap populations as the higher SNP density from the HapMap data allowed inference of the recombination rates at a finer scale compared with the SGVP (Myers et al. 2005).
Figure 4.
LD variation and population-specific recombination rates at CDKAL1. The extent of LD variation between pairs of SGVP and HapMap populations at the CDKAL1 gene, with separate LD heatmaps and recombination rates estimated from genotype data at each population. Population-specific recombination rates are shown except for CHB and JPT, where the same HapMap estimated recombination rates for JPT+CHB are used.
Comparing the genome-wide LD patterns between the two Chinese populations (CHB and CHS), the top 10 regions identified contain an olfactory cluster on chromosome 1 as well as two HLA gene clusters in the major histocompatibility complex (MHC) region on chromosome 6 (Supplemental Table S5), suggesting that these regions are highly polymorphic even between two relatively homogeneous populations. Intriguingly, we observed that three regions outside the top 10 were in the vicinity of candidate genes for common metabolic disorders (FABP2, PCSK1, CLOCK) that have been implicated for climate adaptations (Hancock et al. 2008). The frequencies of the derived allele associated with greater tolerance to cold climate at the A54T (rs1799883) polymorphism in FABP2 were significantly lower in CHS (22.4%) and MAS (15.7%) when compared with CHB (31.4%) and JPT (30.0%), consistent with reported findings of a significant correlation with latitude (Hancock et al. 2008). For comparison, the frequencies for CEU, INS, and YRI were 37.3%, 30.7%, and 20.8%, respectively.
Signatures of positive natural selection
Genome-wide data on the three SGVP populations also permit the survey of signatures of recent positive natural selection through the detection of uncharacteristically long haplotypes in the genome. Using the single-SNP integrated haplotype score (iHS) and the XP-EHH score (see Methods and Supplemental material), we observed that most of the signals detected by iHS in the SGVP populations concur with those established in the HapMap populations, particularly for signals that span multiple SNPs (Supplemental Table S6). Novel candidates for positive selection were identified in each of the three SGVP populations, with the largest number observed in INS. Supplemental Table S7 lists the top 10 candidate regions for recent positive selection in each SGVP population. Across the genome, selection signals that corroborated with earlier findings from the HapMap in genes with well-documented functions include the alcohol dehydrogenase (ADH) gene cluster in CHS and INS, genes involved in skin pigmentation (SLC24A5 in INS, OCA2 in CHS and MAS, TYRP1 in CHS and INS, MYO5A in all three populations), sucrose metabolism (SI in CHS and MAS), brain development and function (CENPJ in CHS and INS, MCPH1 in MAS and INS, CDK5RAP2 in all three), regulation of energy and appetite (LEPR in CHS and MAS), and low-density lipoprotein cholesterol (LDLR in CHS and INS, APOB in all three) (Supplemental Table S8). The concurrence of positive selection across multiple populations is reassuring, although we advocate caution in drawing immediate relevance to the biological interpretations.
Discussion
The aim of the Singapore Genome Variation Project is primarily to provide a genomic resource for studying the three major population groups in Southeast Asia that can be used in the design and analysis of studies for discovering the genetic basis of complex traits and common diseases. This is achieved by making a database of 1.6 million polymorphisms assayed in 268 individuals publicly available for researchers, with the data intentionally formatted in a similar structure to complement the International HapMap Project.
It has been historically documented that Chinese migrants into early Singapore predominantly consisted of people from the southern provinces of China. Our analysis of population structure in East Asia where CHS clustered together with Chinese sub-ethnicities from Southern China and Southeast Asia supported this claim, together with the observation at FABP2 that Singapore Chinese are less likely to carry the genetic variant that confers greater tolerance to cold climates compared with the Han Chinese in Beijing from Northern China. As this variant is similarly found at low frequency in the Malays with equatorial habitats, this suggests that the difference between Singapore Chinese and the Han Chinese from Beijing reflects the genetic diversity found between the northern and southern parts of China.
One of the main motivations in establishing this genomic resource is to explore the possibility of localizing functional polymorphisms through combining association signals across populations with diverse genetic backgrounds. Preliminary findings from targeted sequencing and sequence-resolution imputation studies have suggested that the presence of long LD in populations of European and East Asian descent is a hindrance to this process of fine-mapping, as what emerges from these sequencing studies are sets of SNPs in perfect or almost perfect LD that are virtually impossible to distinguish between for isolating the causal variants. However, as the patterns of LD between the causal variants and the neighboring SNPs can vary across populations, pooling GWAS results with dense population-specific reference haplotypes across multiple populations can be expected to minimize the number of SNPs that are potential candidates to be functional. While the full merit of such transethnic fine-mapping approaches will only be realized with sequence-level haplotypes in the relevant populations, we expect the availability of dense genome-wide data for more populations will at least serve a few purposes: (1) to serve as reference panels to impute against for the purpose of extending the coverage of current genome-wide experiments in Southeast Asia to at least 1.6 million SNPs; (2) to prioritize SNPs that emerged from genome-wide scans for replication in Southeast Asia; and (3) to perform genome-wide comparisons of LD between populations, which will be valuable in identifying regions where transethnic fine-mapping holds the greatest promise.
To date, most genetic research and genomic databases (other than the HapMap) have either focused on populations of European descent or have surveyed comparatively few samples in each Asian population (e.g., the Human Genome Diversity Project). The SGVP provides a timely complement to these databases by providing a publicly available resource of 1.6 million polymorphisms genotyped in 268 samples from three major population groups in Asia. To facilitate the access, analysis, and display of the SGVP data, we have designed a genome browser that is publicly available at http://www.nus-cme.org.sg/SGVP/ (Supplemental Fig. S7). We expect this resource will be valuable for advancing genetic and genomics science in Asia.
Methods
Samples
Subjects enrolled in the SGVP were originally recruited for an interpopulation study on the genetic variability to drug response, where 100 individuals from each of the Chinese, Malay, and Indian population groups were anonymously and randomly chosen from the manifest to partake in SGVP, with only gender and population information. Of these 300 samples, genomic DNA samples for 99 Chinese, 98 Malay, and 95 Indians were chosen for genotyping. Population membership was ascertained on the basis that all four grandparents belong to the same population group. Ethical consent for the original study on drug response and further ethical approval for the extension to genome-wide genotyping were granted by two independent Institutional Review Boards at the National University Hospital (Singapore) and the National University of Singapore, respectively.
SNP genotyping
Genomic DNA for all 292 individuals was assayed on the Affymetrix SNP6.0 Genotyping Chip and the Illumina 1M-single DNA Analysis BeadChip. Preliminary genotypes for 3022 control probes on the Affymetrix array were called using the DM algorithm (Di et al. 2005) for sample QC. The set of genotype data from the Affymetrix array used in downstream analyses was called using the BirdSeed algorithm (Korn et al. 2008). Genotypes for the Illumina array were assigned using the proprietary calling algorithm GenCall in the BeadStudio Suite (Oliphant et al. 2002; Fan et al. 2004). We implemented a threshold of 0.15 on the GC score during the calling process: a valid genotype was assigned if the GC score was ≥0.15; otherwise, a missing genotype was assigned.
Quality assessment
The quality of the genotypes for data from both arrays was assessed independently, in the following four phases in sequential order: (1) preliminary SNP QC on the autosomal chromosomes to identify a set of “pseudo-cleaned” SNPs for sample QC; (2) sample QC to remove sample duplicates, related samples, or samples with high rates of missing data; (3) identification of samples with inconsistent population membership or inconsistent gender when comparing between the self-reported and genetically inferred data; (4) another round of SNP QC after excluding samples identified by (2) and (3) to yield the set of SNPs for inclusion in the SGVP database. Post-QC data for both arrays were available for 96 Chinese, 89 Malay, and 83 Indian samples. For SNPs that are common to both Affymetrix and Illumina, only those with ≥95% concordant genotypes between the two arrays were retained.
Assessing population structure
Population structure between the HapMap and SGVP populations was assessed by principal components analysis (PCA) with EIGENSTAT (Price et al. 2006). We thinned the available SNPs by using every tenth SNP out of the 1,423,464 SNPs that were common between HapMap and SGVP, consisting of 142,347 SNPs, to reduce the extent of LD between the SNPs used in the PCA. The FST calculation uses the same formula as that used by the International HapMap Project (The International HapMap Consortium 2005), which accounts for the different number of samples in each population (see Supplemental material).
Haplotype phasing and LD calculation
The software fastPhase (Scheet and Stephens 2006) was used to perform the phasing of the genotype data within each population separately. The parameters used in the analysis were optimized to yield minimal error rates within realistic running time of the analysis. The LD between a focal SNP and any SNP found within 250 kb upstream and downstream of the focal SNP was calculated using the software Haploview (Barrett et al. 2005). LD was measured by the square of the genetic correlation coefficient r2, D′, and the LOD score, and was calculated off the phased haplotype data. Comparisons of LD across populations utilized 45 samples from each population to avoid the effects of different sample sizes.
Comparing allele frequency spectrum
We considered the same set of SNPs that passed QC across all the SGVP panels. For each SNP, the minor allele was identified after agglomerating the genotypes from all three SGVP populations. The frequencies of the minor alleles were subsequently calculated within each SGVP populations and categorized in 20 bins of size 0.05 spanning 0 to 1.
Quantifying haplotype diversity
For each chromosome, we randomly selected a 500-kb region, avoiding centromeres and genomic regions with low SNP density. For an unbiased comparison across all seven population panels from HapMap and SGVP, we considered only the SNPs that were common to all seven panels. In each of the 500-kb regions, we identified the number of distinct haplotype forms. We then quantified haplotype diversity by the proportion of chromosomes from each population that had been accounted for by a specific number of haplotypes. This procedure is similar to that established for quantifying haplotype diversity across multiple populations (Bonnen et al. 2006). In order to investigate the extent of haplotype sharing, chromosomes from the region in chromosome 11 were clustered and visualized with the use of haplosim and hapvisual from the R package haplosuite (Teo and Small 2009). Briefly, haplosim identifies the canonical haplotypes in each region across all seven populations, where each canonical haplotype is defined as a specific haplotype configuration to which a substantial proportion of the individuals are highly similar. Each chromosome is subsequently mapped either uniquely to one of these canonical haplotypes, or as a mosaic of these haplotypes. We explicitly chose to implement an upper limit of seven possible canonical haplotypes in our analysis of the HapMap and SGVP populations. The outcome of the haplotype clustering was subsequently fed into hapvisual, which produced a visualization of the haplotype clustering for each population, where each canonical haplotype is assigned a unique color that remains consistent across the populations.
Analysis of LD variation
Comparison of regional LD between two populations was performed with the varLD algorithm (Teo et al. 2009b). Briefly, we considered windows of 50 consecutive SNPs found in both populations and calculated the signed r2, defined as the r2 with the sign of the D′ metric, between all possible pairs of these SNPs. Consequently, we constructed a 50 × 50 symmetric matrix for each population where the (i, j)th element represents the signed r2 metric between the ith and jth SNPs calculated. We compared the equality between the two matrices by comparing the extent of departures between the eigenvalues, given by the sum of the absolute difference between the ranked eigenvalues for the two matrices that yields a score for each window of 50 SNPs. The extent of LD differences in each window was assessed by comparing the relative rank of the score obtained against the distribution of scores in the genome, and we identified regions that constituted the top 5% of the distribution of the scores. For visualizing the signals from comparisons across multiple population pairs, we standardized the scores to have a mean of zero and a standard deviation of one. Signals in the top 5% of the distribution were binned into regions if two consecutive signals were found within 25 kb.
Detecting signatures of positive selection
We used the single-SNP integrated haplotype score (iHS) statistic introduced by Voight et al. (2006) to identify signals of positive selection within each of the HapMap and SGVP populations. This analysis followed the set-up described in Sabeti et al. (2007). To compare signals of positive natural selection that differ between populations, we used the XP-EHH test with the same set-up as introduced and described by Sabeti and colleagues (Sabeti et al. 2007).
A full description of the methods with additional figures and tables for the methodologies can be found in the Supplemental material.
Acknowledgments
We thank three anonymous reviewers and E.S. Tai for their insightful comments that helped improve the manuscript. We thank all the subjects in this study for their participation. This project also acknowledges the support of the Yong Loo Lin School of Medicine, the National University Health System, the Life Science Institute and Office of Deputy President (Research and Technology) from the National University of Singapore. We also acknowledge the support of the Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore.
Author contributions: Y.Y.T., M.S., and K.S.C. jointly conceived and designed the experiment; Y.Y.T., X.S., and R.T.H.O. wrote the paper; Y.Y.T., X.S., R.T.H.O., A.K.S.T., C.S.K., E.T., K.S.S., and J.C. analyzed the data; R.T.H.O. and X.S. designed the website; E.J.D.L. contributed samples; C.S.K. and M.S. coordinated the genotyping; M.S. and K.S.C. jointly directed the project.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.095000.109.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bonnen PE, Pe'er I, Plenge RM, Salit J, Lowe JK, Shapero MH, Lifton RP, Breslow JL, Daly M, Reich DE, et al. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat Genet. 2006;38:214–217. doi: 10.1038/ng1712. [DOI] [PubMed] [Google Scholar]
- Chu JY, Huang W, Kuang SQ, Wang JM, Xu JJ, Chu ZT, Yang ZQ, Lin KQ, Li P, Wu M, et al. Genetic relationship of populations in China. Proc Natl Acad Sci. 1998;95:11763–11768. doi: 10.1073/pnas.95.20.11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Prichard JK. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–1260. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1213. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–1303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- Di X, Matsuzaki H, Webster TA, Hubbell E, Liu G, Dong S, Bartell D, Huang J, Chiles R, Yang G, et al. Dynamic model based algorithms for screening and genotyping over 100K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21:1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen MS, Steemers F, Butler SL, Deloukas P, et al. High parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2004;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard J, Coop G, Di Reinzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;427:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M, et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S, Jada SR, Xiang X, Lim WT, Lee EJ, Chowbay B. Pharmacogenetics of target genes across the warfarin pharmacological pathway. Clin Pharmacokinet. 2006;45:1189–1200. doi: 10.2165/00003088-200645120-00004. [DOI] [PubMed] [Google Scholar]
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Lee SC, Ng SS, Oldenburg J, Chong PY, Rost S, Guo YJ, Yap HL, Rankin SC, Khor HB, Yeo TC, et al. Inter-ethnic variability in warfarin requirement is explained by VKORC1 genotype in an Asian population. Clin Pharmacol Ther. 2006;79:197–205. doi: 10.1016/j.clpt.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yu L, Zhang D, Chen Z, Zhou DZ, Zhao T, Li S, Wang T, Hu X, Feng GY, et al. Positive association between variations in CDKAL1 and type 2 diabetes in Han Chinese individuals. Diabetologia. 2008;51:2134–2137. doi: 10.1007/s00125-008-1141-6. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- NCI-NHGRI Working Group on Replication in Association Studies. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, Lam VK, Ma RC, So WY, Cho YS, et al. Implications of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2 and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57:2226–2233. doi: 10.2337/db07-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;32:S56–S61. [PubMed] [Google Scholar]
- Pe'er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw SH. The population of Singapore. 2nd edition. Institute of South East Asian Studies; Singapore: 2007. [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: Candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- Tabara Y, Osawa H, Kawamoto R, Onuma H, Shimizu I, Miki T, Kohara K, Makino H. Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes. 2009;58:493–498. doi: 10.2337/db07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Small KS. A novel method for haplotype clustering and visualization. Genet Epidemiol. 2009 doi: 10.1002/gepi.20432. [DOI] [PubMed] [Google Scholar]
- Teo YY, Small KS, Fry AE, Wu Y, Kwiatkowski DP, Clark TG. Power consequences of linkage disequilibrium variation between populations. Genet Epidemiol. 2009a;33:128–135. doi: 10.1002/gepi.20366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, Fry AE, Bhattacharya K, Small KS, Kwiatkowski DP, Clark TG. Genome-wide comparisons of variation in linkage disequilibrium. Genome Res. 2009b;19:1849–1860. doi: 10.1101/gr.092189.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, et al. Genetic evidence supports demic diffusion of Han culture. Nature. 2004;431:302–305. doi: 10.1038/nature02878. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li H, Loos RJ, Yu Z, Ye X, Chen L, Pan A, Hu FB, Lin X. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired casting glucose in a Chinese Han population. Diabetes. 2008;57:2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]