Abstract
Context: Autoimmunity associated with Addison’s disease (AD) can be detected by measuring 21-hydroxylase (21OH) autoantibodies. Subjects with type 1 diabetes (T1D) are at increased risk for AD. Genetic factors including HLA-DRB1*0404 and MICA have been associated with AD in populations with and without T1D.
Objective: The objective of the study was to examine the effect of the MICA5.1 allele in subjects with 21OH autoantibodies on progression to AD.
Design: Two components were used: 1) a cross-sectional study with subjects with AD identified and enrolled from September 1993 to November 2008 and 2) a cohort study prospectively following up patients with T1D who screened positive for 21OH autoantibodies.
Setting: Subjects were identified from the Barbara Davis Center and through the National Adrenal Diseases Foundation.
Patients: Sixty-three subjects with AD were referred through the National Adrenal Diseases Foundation (AD referrals). Sixty-three subjects with positive 21OH antibodies from the Barbara Davis Center were followed up for progression to AD, and 11 were diagnosed with AD (progressors).
Results: Seventy-three percent of progressors (eight of 11) and 57% of AD referrals (36 of 63) were MICA5.1 homozygous (P = ns). Overall, 59% of patients with AD (44 of 74) were MICA5.1/5.1 compared with 17% of nonprogressors (nine of 52) (P < 0.0001) and 19% of normal DR3/4-DQB1*0302 controls (64 of 336) (P < 0.0001).
Conclusions: Identifying extreme risk should facilitate monitoring of progression from 21OH antibody positivity to overt AD. The HLA-DR3/0404 genotype defines high-risk subjects for adrenal autoimmunity. MICA5.1/5.1 may define those at highest risk for progression to overt AD, a feature unique to AD and distinct from T1D.
MICA5.1 homozygosity is associated with an increased risk for Addison’s disease in subjects positive for 21-hydroxylase autoantibodies.
The majority of Addison’s disease (AD) in the developed world is caused by adrenal autoimmunity. The autoimmunity associated with AD can be identified by the presence of antibodies against 21-hydroxylase (21OH) (1). Subjects with 21OH antibodies may develop AD over many years, and long-term follow-up of subjects is warranted (2). AD may present alone or with other organ-specific autoimmune diseases in the autoimmune polyendocrine syndromes (APS)-1 and -2 (3). APS-1 is caused by mutations in the autoimmune regulator gene (4); inherited in an autosomal recessive fashion; and is characterized by the triad of AD, hypoparathyroidism, and mucocutaneous candidiasis (5). In contrast, APS-2 is characterized by the association of AD, type 1 diabetes, and autoimmune thyroid disease and is inherited in a polygenic fashion (6). Genes within the immune system have been implicated in the risk for AD (6,7,8).
The major histocompatibility complex (MHC), located on chromosome 6, exhibits significant long-range linkage disequilibrium and contains more than 100 genes, many of which influence immune function. Genes within the MHC include the human leukocyte antigens (HLA) (class I and class II) and MHC class I (MIC)-related genes (MICA, MICB, MICC, MICD, and MICE). Class II HLA genes and MICA have been associated with autoimmune diseases. Specifically, the HLA genotype DRB1*0301-DQA1*0501-DQB1*0201/DRB1*04-DQA1*0301-DQB1*0302 (DR3/4-DQB1*0302) has been associated with type 1 diabetes and AD. We and others have shown that DRB1*0404 vs. other DRB1*04 alleles is found in a large proportion of AD subjects with DR3/4-DQB1*0302 both in the United States (29%) (7) and Europe (31%) (8), compared with 0.7% in the general population (9).
Polymorphisms of the MICA gene have also been associated with autoimmune diseases, including Graves’ disease, type 1 diabetes (10,11), celiac disease, and AD (12,13). MICA alleles are designated by the number of GCT repeats (4, 5, 6, and 9). The allele designated 5.1 has an additional bp and results in the introduction of a premature stop codon (14), which results in the truncation of the protein within the transmembrane region. MICA proteins are ligands for NKG2D receptors that are expressed on the surface of CD8 and NK cells. Normally, MICA is expressed in the intestinal and thymic epithelium (15,16), and its expression is induced by cellular stress. It is also expressed in increased levels in tumor cells (17). MICA can be released as a soluble molecule. Increased levels of soluble MICA have been identified in the serum of subjects with celiac disease (17) and are also correlated with malignancy (18).
In this report we seek to describe the HLA and MICA genotypes in a relatively large cohort of subjects with AD and to determine the influence of MICA polymorphisms on the prediction of AD in subjects with 21OH antibodies. We examined HLA genotypes and MICA alleles within two cohorts: subjects with AD (AD referrals) and subjects with positive 21OH antibodies followed for AD (progressors and nonprogressors).
Subjects and Methods
Enrollment of subjects
Subjects were recruited through the National Adrenal Diseases Foundation (www.nadf.us) and the clinic at the Barbara Davis Center for Childhood Diabetes. All subjects were enrolled with informed consent, and the study was approved by the institutional review board at the University of Colorado Denver. For subjects identified through the National Adrenal Diseases Foundation, diagnosis of AD was made by self-report. To exclude those with autoimmune regulator gene mutations, interferon (IFN)-α antibodies were measured in all subjects (23). To detect autoimmunity associated with AD, 21OH antibodies were measured and subjects were studied prospectively if positive. For details of affected subjects, see Supplemental Table, published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org.
Subjects with type 1 diabetes were screened for 21OH antibodies at onset of diabetes and every 1–2 yr after. In addition, a subset of relatives of subjects with type 1 diabetes was screened. If positive autoantibodies were detected, results were confirmed on a second sample. With confirmed positive 21OH antibodies, ACTH stimulation testing was performed every 1–2 yr or more often based on symptoms. Subjects and parents were advised of signs and symptoms of AD and instructed to call the endocrinologist and/or study coordinator if the signs and symptoms developed.
For genetic comparisons, we compared those referred with AD (AD referrals), 21OH antibody-positive subjects followed up to progression to AD (progressors), and 21OH antibody-positive subjects followed for AD but not yet developing disease (nonprogressors). General population estimate data for HLA genotypes were obtained from general population children screened in Denver through the Diabetes Autoimmunity Study in the Young (DAISY) (9). HLA genotype data for type 1 diabetes comparison subjects was obtained from Noble et al. (19). Subjects with DR3/4-DQB1*0302 and type 1 diabetes participating in the DAISY study were used for DRB1*0404 and MICA comparisons.
Endocrine testing
Endocrine testing was performed as previously described (7). Individuals were admitted to the Clinical Translational Research Center and placed in a supine position, and an iv catheter was inserted 30 min before baseline laboratory work. At baseline, blood was drawn for 21OH antibodies, plasma renin activity, ACTH, cortisol, and HLA genotyping. At time 0, 1 μg Cortrosyn (low dose Cortrosyn stimulation) was given iv; 30 min later, serum was obtained for cortisol measurement. Then a dose of 250 μg Cortrosyn (high dose Cortrosyn stimulation) was given iv, and cortisol was measured 30 and 60 min later. The normal range for ACTH is 7–48 pg/ml (1.54–10.56 pmol/liter), and the plasma renin activity normal range is 0–3 ng/ml · h. Diagnosis of AD was made with a peak cortisol level after 250 μg Cortrosyn of less than 18 μg/dl (496.6 nmol/liter) or a fasting cortisol of 3 μg/dl (82.8 nmol/liter) or less (20). The highest cortisol level after stimulation was considered the peak level. Subjects were evaluated at baseline and then every 1–2 yr. Subjects are advised on the signs and symptoms of AD and advised to seek medical attention for vomiting illnesses and advised to inform the medical staff that they are at risk for AD and ask the staff to contact an endocrinologist from our group for advise regarding evaluation and management.
21OH assay
Autoantibodies against 21OH were measured by a method previously described (21). In brief, 21OH cDNA was transcribed and translated in vitro with a commercially available kit (Promega Corp., Madison, WI). [35S]methionine was incorporated, and 20,000 cpm of the labeled product were incubated with 2 μl serum overnight. Protein A-Sepharose (Amersham Biosciences, Piscataway, NJ) was used to capture autoantibody-bound radioactivity in 96-well filtration plates (Millipore Corp., Bedford, MA). Scintillation fluid was added directly to the 96-well plate for counting with a Top-Counter (Packard, Downers Grove, IL). Autoantibodies are reported as an index relative to a positive control sample. The cutoff (>0.150) was established at the 100th percentile of 241 normal controls. The inter- and intraassay coefficient of variation was 11 and 7.6%, respectively.
Competitive europium-IFN-α assay
IFN-α antibodies were measured as previously described (22). Samples were run with and without competition and performed in duplicate. The intra- and interassay coefficient of variation was 10.5 (n = 10) and 8.7% (n = 11), respectively.
MICA
Genotypes were determined using a fluorescent-based method as reported previously (10,12). Briefly, PCR fragments were generated using primers (MICA forward, 5′-/6-FAM/CCTTTTTTTCAGGGAAAGTGC-3′; MICA reverse, 5′-CCTTACCATCTCCAGAAACTGC-3′) that flank the microsatellite repeat polymorphism in the transmembrane region of the MICA gene (exon 5). Reactions (25 μl) were assembled using FailSafe PCR PreMix J, 2.5 U MasterAmp Taq polymerase (Epicentre, Madison, WI), 10 nmol each primer, and 100 ng of genomic template. The PCR product was amplified via 35 PCR cycles of 94 C for 30 sec, 57 C for 35 sec, 72 C for 1 min, and a final extension of 72 C for 45 min. Products were diluted 1:50 and separated by capillary electrophoresis on an ABI3100 Avant genetic analyzer (Applied Biosystems, Foster City, CA). Alleles were identified using GeneMapper version 3.5 (Applied Biosystems). The approximate peak sizes corresponding to each allele are 180 for allele 4, 182.7 for allele 5, 184.1 for allele 5.1, 185.7 for allele 6, and 194.5 for allele 9. The nomenclature used to define the HLA-DR, DQ, and MICA alleles was that of the Official Nomenclature for Factors of the HLA System, 1991 (23), and that of the MICA sequences, 2000 (24).
HLA-DQB1
HLA-DQB1 typing was performed using linear array kits from Roche Molecular Systems (Alameda, CA). Amplified DNA using primers (one biotinylated) of the second exon of DQB1 was hybridized to an array of specific oligonucleotide probes. After a series of incubations and washing, at different temperatures, the strips were developed for color with horseradish peroxidase conjugate and a chromogen. A control band is included on all strips. StripScan (Roche Molecular Systems, Pleasanton, CA) was used to call the alleles.
HLA-DRB1
HLA-DRB1 typing was performed using a kit from Abbott Molecular Diagnostics (Abbott Park, IL). The amplified second exon of DRB1 was cleaned of primers and sequenced on ABI 3130xl or 3100Avant genetic analyzers. The alleles were called with Assign software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego CA; www.graphpad.com) and SAS Software version 9.1.3 (Cary, NC). For normally distributed variables, descriptive statistics are reported as the mean and sd. Comparisons for continuous variables were made using a t test. Proportions were compared using a χ2 test unless 20% of the cells had an expected value less than 5 in which case Fisher’s exact test was used. Survival analysis for progression of AD was performed for 21OH antibody-positive DR3/4-DQB1*0302 individuals stratified by MICA typing using life table analysis with the log rank test. Cox proportional hazards modeling was performed to assess the influence of MICA 5.1 on the progression to AD after controlling for age, gender, and presence of diabetes. Results were considered statistically significant with an α < 0.05.
Results
Baseline
21OH antibody-positive individuals (n = 63) were followed up for progression to AD with ACTH stimulation testing. These 63 subjects were identified out of a total of 5368 screened and represent 1.1% of the group. Subjects with positive antibodies were similar in age, gender distribution, and presence of diabetes compared with those with negative antibodies (data not shown).
Of the 63 subjects with 21-hydroxylase autoantibodies, 11 developed AD (progressors). Overall follow-up was 2.78 yr for progressors and 3.84 for nonprogressors. Progressors and nonprogressors were compared for baseline characteristics. The gender distribution (54.5 vs. 42.3% male); race (100 vs. 90% non-Hispanic white); age at 21OH antibody detection (17.4 vs. 21.4 yr)]; first positive 21OH antibody level (1.24 vs. 0.957) (P = ns); percent with type 1 diabetes (90.9 vs. 86.5%), and duration of follow-up (2.78 vs. 3.84 yr) were not statistically significantly different between progressors and nonprogressors for each characteristic.
Endocrine testing
The median ACTH at diagnosis for progressors was 218 pg/ml (48 pmol/liter) with a range of 92–1013 pg/ml (20.2–222.9 pmol/liter). The median most recent ACTH for nonprogressors was 21 pg/ml (4.6 pmol/liter) with a range of 6–53 pg/ml (1.3–11.7 pmol/liter) (P < 0.0001). One subject was diagnosed with AD 6 months after having a normal ACTH stimulation test after presenting to the emergency department with a vomiting illness.
The median baseline cortisol at diagnosis for progressors was 6.9 μg/dl (189.1 nmol/liter) with a range of 0.99–10.0 pg/ml (27.3–276 nmol/liter). The most recent baseline cortisol median for nonprogressors was 10.0 pg/ml (276 nmol/liter) with a range of 3–30 pg/ml (82.8–828 nmol/liter) (P = 0.0263). Four nonprogressors with baseline cortisol less than 5 pg/ml (138 nmol/liter) continued to not fulfill stimulated cortisol diagnosis of AD on the remainder of their stimulation tests in the presence of normal ACTH.
HLA
The high-risk HLA genotype, DR3/4-DQB1*0302, was present at an increased frequency in AD referrals (29 of 63; 46%), progressors (seven of 11; 63.6%), and nonprogressors (22 of 52; 42.3%) compared with the general population of Denver newborns report [74 of 3063; 2.4% (9)] (P < 0.0001, Table 1). However, the frequency of DR3/4-DQB1*0302 was not significantly different in AD referrals, progressors, and nonprogressors compared with subjects with type 1 diabetes (39.4%) (19). The highest-risk genotype DR3/0404-DQB1*0302 was present in six of seven (85.7%) DR3/4-DQB1*0302 progressors and 14 of 22 (63.6%) DR3/4-DQB1*0302 nonprogressors (P = ns) (Table 2). Within DR3/4-DQB1*0302 progressors and nonprogressors, 86% (six of seven) and 64% (14 of 22), respectively, had the DRB1*0404 allele vs. 30% (97 of 326) of DR3/4-DQB1*0302 control individuals (P = 0.004 progressors; P = 0.002 nonprogressors).
Table 1.
HLA genotypes by population
| Genotype | AD referral (n = 63) | 21OH + progressors (n = 11) | 21OH + non-progressors (n = 52) | Non-Hispanic white general populationb | Type 1 diabetesc |
|---|---|---|---|---|---|
| DR3/4 | 29 (46%)a | 7 (63.6%) | 22 (42.3%) | 2.4% | 39.4% |
| DR3/3 | 5 (7.9%) | 2 (18.2%) | 1 (1.9%) | 1.7% | 5.6% |
| DR4/4 | 1 (1.6%) | 0 | 5 (9.6%) | 3.6% | 6.7% |
| DR3/X | 13 (20.6%) | 2 (18.2%) | 6 (11.5%) | 18.7% | 9.4% |
| DR4/X | 6 (9.5%) | 0 | 13 (25%) | 14.9% | 13.3% |
| All other genotypes | 9 (14.3%) | 0 | 5 (9.6%) | 58.7% | 25.6%d |
DR3/4 frequencies are significantly different between AD referrals, progressors, and nonprogressors compared with general population subjects (P < 0.0001). DR4, DRB1*04-DQB1*0302.
Data from DAISY general population newborns in Denver (9).
Data from type 1 diabetes patients (19).
Includes all other genotypes in addition to any genotype present at less than 0.83% in the cohort.
Table 2.
HLA/MICA genotypes by population
| DR3/4 | DRB1*0404 of DR3/4 | MICA5.1/5.1 of DR3/0404 | Overall estimate DR3/0404-5.1/5.1 | |
|---|---|---|---|---|
| General population estimatea | 2.4% | 30% (97/326) | 40% (38/95) | 0.96% |
| Type 1 diabetes (n = 31)b | 39.4%c | 10% (3/31) | 33% (1/3) | 1.3% |
| AD referral (n = 63) | 46% (29/63) | 72% (21/29)d | 71% (15/21) | 24% (15/63) |
| 21OH + progressor (n = 11) | 64% (7/11) | 86% (6/7)d | 67% (4/6) | 36% (4/11) |
| 21OH + nonprogressor (n = 52) | 42% (22/52) | 64% (14/22)d | 29% (4/14) | 7.7% (4/52) |
MICA
Nonprogressors (47 of 104; 45.2%) had a lower MICA5.1 allele frequency compared with both patients with known AD (AD referrals 95 of 126; 75%, P < 0.0001) and progressors (19 of 22; 86.4%, P = 0.0004) (Table 3). Of the patients with DR3/4-DQB1*0302 and AD (AD referrals and progressors) 97% (35 of 36) had at least one MICA5.1 allele, whereas 77% (259 of 336) of control DR3/4-DQB1*0302 subjects had at least one 5.1 allele (P = 0.002) and 84% (26 of 31) of DR3/4-DQB1*0302 type 1 diabetes subjects had at least one MICA 5.1 allele (P = ns).
Table 3.
MICA alleles and genotypes by population
| AD referral | 21OH + progressors | 21OH + nonprogressors | Type 1 diabetes DR3/4 | General population DR3/4 controls | |
|---|---|---|---|---|---|
| Allele | (n = 126) | (n = 22) | (n = 104) | (n = 62) | (n = 672) |
| MICA4 | 10 (7.9%) | 0 | 10 (9.6%) | 3 (4.8%) | 63 (9.4%) |
| MICA5 | 3 (2.4%) | 0 | 21 (20.2) | 10 (16.1%) | 115 (17.1%) |
| MICA6 | 8 (6.3%) | 2 (9.1%) | 12 (11.5%) | 7 (11.3%) | 86 (12.8%) |
| MICA5.1 | 95 (75.4%) | 19 (86.4%) | 47 (45.2%)a | 35 (56.5%) | 323 (48.1%) |
| MICA9 | 10 (7.9%) | 1 (4.5%) | 14 (13.5%) | 7 (11.3%) | 85 (12.6%) |
| Genotypeb | (n = 63) | (n = 11) | (n = 52) | (n = 31) | (n = 336) |
| MICA5.1/5.1 | 36 (57.1%) | 8 (72.7%) | 9 (17.3%)a | 9 (29%) | 64 (19%) |
| MICA5.1/X | 23 (36.5%) | 3 (27.3%) | 29 (55.8%) | 17 (55%) | 195 (58%) |
| MICAX/X | 4 (6.3%) | 0 | 14 (26.9%) | 5 (16%) | 77 (22.9%) |
DR4, DRB1*04-DQB1*0302.
P < 0.001 compared with progressor and AD referrals.
X = not MICA5.1.
The frequency of the MICA5.1/5.1 genotype was significantly higher in AD referrals (36 of 63; 57%) compared with nonprogressors (nine of 52; 17.3%) (P < 0.0001). Additionally, the frequency of the MICA5.1/5.1 genotype was higher in progressors (eight of 11; 72.7%) vs. nonprogressors (P = 0.0006).
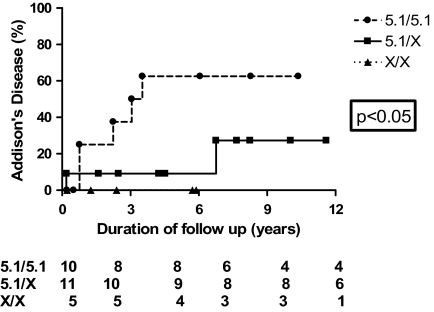
By life table analysis, with a mean of 4.18 yr of follow-up, 50% of MICA5.1/5.1, DR3/4-DQB1*0302, 21OH antibody-positive patients progress to overt AD vs. 12.5% (two of 16) of DR3/4-DQB1*0302 non-MICA5.1/5.1 patients (P = 0.04) at 3 yr follow-up (Fig. 1). To assess the influence of DR3/0404 on progression to AD in 21-hydroxylase-positive individuals, life table analysis was performed. There was no difference in AD-free survival in subjects with DR3/0404 vs. all other genotypes (P = ns). Using Cox proportional hazards modeling, after controlling for HLA, gender, and presence of diabetes, the influence of MICA 5.1 on the development of AD remained strong with a hazard ratio of 8.628 (95% confidence interval 2.029–36.696).
Figure 1.
Proportion of DR3/4-DQB1*0302 subjects who develop AD over time from first positive 21OH antibody measurement by MICA genotypes. MICA 5.1/5.1 vs. MICA5.1/not 5.1 vs. MICA not 5.1/not 5.1 (P = 0.04). Twenty-six DR3/4-DQB1*0302 subjects were typed and followed up.
Discussion
In conclusion, we have demonstrated that homozygosity of MICA5.1, or other unknown polymorphisms in linkage disequilibrium with MICA, is associated with progression of high-risk DR3/4-DQB1*0302, 21OH antibody-positive individuals to AD. Screening for adrenal autoimmunity with the 21OH antibody assay identifies individuals who are at risk for the development of AD before clinical presentation. The natural history of adrenal autoimmunity and factors contributing to progression to AD continue to be defined. Others have identified characteristics such as 21OH antibody levels (25), the high risk HLA haplotype DR3/4-DQB1*0302 genotype (7,8), and younger age as risk factors for the development of clinical AD in 21OH antibody-positive subjects. We previously demonstrated that DRB1*0404 is a high-risk allele for 21OH autoimmunity (7). In this study, DR3/4-DQB1*0302 nonprogressors and progressors with type 1 diabetes have an increased frequency of the DRB1*0404 allele compared with DR3/4-DQB1*0302 type 1 diabetes subjects, which supports the finding that DRB1*0404 allele is associated with 21OH antibody positivity. Additionally, our data continue to support previous work that demonstrated homozygosity of MICA5.1 increased progression to overt AD among 21OH antibody-positive individuals and further emphasizes its effect independent of HLA DR3/4-DQB1*0302 because the effect was still observed when analysis was limited to subjects with DR3/4-DQB1*0302 (26). MICA5.1 allele was found in 22 of 24 Estonian patients with AD, but results from Finnish and Russian patients did not support its independent role in disease susceptibility (27).
One quarter of control DR3/4-DQB1*0302 individuals had no MICA5.1 alleles compared with 2.8% overall of those with AD. As expected, both those that progressed to AD and those with 21OH antibodies had an increased percentage of the high-risk DR3/4-DQB1*0302 genotype as well as an excess of DRB1*0404.
Distribution of MICA alleles demonstrate the similarities of AD referrals to those that progress from adrenal autoimmunity to AD and the significant differences between 21OH antibody-positive nonprogressors and both AD referrals and 21OH antibody-positive progressors. Life table analysis of 21OH antibody-positive DR3/4-DQB1*0302 individuals demonstrates that half of the MICA5.1 homozygous individuals progress to overt AD, whereas significantly fewer patients heterozygous for the MICA5.1 allele develop AD. This may suggest that MICA5.1/5.1 affects progression to AD.
We plan on continuing to follow up this cohort over time. We anticipate that some proportion of the subjects with 21OH antibodies who at this point have not yet developed AD will go on to develop AD. It is entirely possible that these genetic markers influence the pace at which AD develops and subjects without the high risk markers will develop AD later on in life.
The MICA gene encodes a protein that is a ligand for the NKG2D T cell receptor. MICA can activate this receptor on NK cells, and this interaction may be important for thymic T cell maturation (16). The MICA5.1 allele has an additional base pair, which results in a stop codon and results in the truncation of the protein within the transmembrane region. The loss of membrane bound MICA, especially for homozygous individuals, may underlie the marked association with AD. Alternatively, given that soluble MICA is found in increased levels in the serum of those with celiac disease (17), further investigations should examine soluble MICA in the serum of those with AD.
Genes outside the MHC including the gene encoding lymphoid tyrosine phosphatase have been associated with AD (28) and NACHT leucine-rich-repeat protein 1 (29), and analysis of these and other genes in this cohort may provide more predictive information.
Identifying extreme risk should facilitate monitoring of progression from 21OH antibody positivity to overt AD. As previously described, DR3/DRB1*0404 defines a high-risk population of subjects for adrenal autoimmunity (7). Homozygosity for MICA5.1 defines those at higher risk for progression to overt AD, a feature unique to AD. In conclusion, whereas HLA-DRB1*0404 may contribute to the development of 21OH autoimmunity, homozygosity of the MICA5.1 allele may contribute to progression of overt AD.
Supplementary Material
Acknowledgments
We acknowledge the Clinical and Translational Research Centers for their contribution in performing the endocrine testing and the core laboratories at the University of Colorado Hospital and the Children’s Hospital of Denver for performing the endocrine assays. We also acknowledge the patients and families for their participation and the National Adrenal Diseases Foundation.
Footnotes
The Diabetes Autoimmunity Study in the Young research was supported by National Institute of Diabetes and Digestive and Kidney Disease Grant DK32493, Diabetes Endocrine Research Center, Clinical Investigations and Bioinformatics Core Grant P30 DK 57516, and the Children’s Diabetes Foundation. Research was supported by National Institutes of Health Grants DK 32083 and A146374. Clinical and Translational Research Center was supported by Grants RR-00051 and RR-00069. J.M.B. is supported by the Juvenile Diabetes Research Foundation Grant 11-2005-15.
Disclosure Summary: The authors have nothing to disclose.
First Published Online October 9, 2009
Abbreviations: AD, Addison’s disease; APS, autoimmune polyendocrine syndrome; DAISY, Diabetes Autoimmunity Study in the Young; HLA, human leukocyte antigens; MHC, major histocompatibility complex; MIC, MHC class I; 21OH, 21-hydroxylase.
References
- Winqvist O, Karlsson FA, Kämpe O 1992 21-Hydroxylase, a major autoantigen in idiopathic Addison’s disease. Lancet 339: 1559–1562 [DOI] [PubMed] [Google Scholar]
- Betterle C, Coco G, Zanchetta R 2005 Adrenal cortex autoantibodies in subjects with normal adrenal function. Best Pract Res Clin Endocrinol Metab 19:85–99 [DOI] [PubMed] [Google Scholar]
- Neufeld M, Maclaren NK, Blizzard RM 1981 Two types of autoimmune Addison’s disease associated with different polyglandular autoimmune (PGA) syndromes. Medicine (Baltimore) 60:355–362 [DOI] [PubMed] [Google Scholar]
- Aaltonen J, Björses P, Perheentupa J, Horelli-Kuitunen N, Palotie A, Peltonen L, Lee YS, Francis F, Hennig S, Thiel C, Lehrach H, Yaspo M-L 1997 An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet 17:399–403 [DOI] [PubMed] [Google Scholar]
- Perheentupa J2002 APS-I/APECED: the clinical disease and therapy. Endocrinol Metab Clin North Am 31:295–320, vi [DOI] [PubMed] [Google Scholar]
- Eisenbarth GS, Gottlieb PA 2004 Autoimmune polyendocrine syndromes. N Engl J Med 350:2068–2079 [DOI] [PubMed] [Google Scholar]
- Yu L, Brewer KW, Gates S, Wu A, Wang T, Babu SR, Gottlieb PA, Freed BM, Noble J, Erlich HA, Rewers MJ, Eisenbarth GS 1999 DRB1*04 and DQ alleles: expression of 21-hydroxylase autoantibodies and risk of progression to Addison’s disease. J Clin Endocrinol Metab 84:328–335 [DOI] [PubMed] [Google Scholar]
- Myhre AG, Undlien DE, Løvås K, Uhlving S, Nedrebø BG, Fougner KJ, Trovik T, Sørheim JI, Husebye ES 2002 Autoimmune adrenocortical failure in Norway: autoantibodies and HLA class II associations related to clinical features. J Clin Endocrinol Metab 87:618–623 [DOI] [PubMed] [Google Scholar]
- Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, McDuffie Jr RS, Hamman RF, Klingensmith G, Eisenbarth GS, Erlich HA 1996 Newborn screening for HLA markers associated with IDDM: Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 39:807–812 [DOI] [PubMed] [Google Scholar]
- Zake LN, Ghaderi M, Park YS, Babu S, Eisenbarth G, Sanjeevi CB 2002 MHC class I chain-related gene alleles 5 and 5.1 are transmitted more frequently to type 1 diabetes offspring in HBDI families. Ann NY Acad Sci 958:309–311 [DOI] [PubMed] [Google Scholar]
- Alizadeh BZ, Eerligh P, van der Slik AR, Shastry A, Zhernakova A, Valdigem G, Bruining JG, Sanjeevi CB, Wijmenga C, Roep BO, Koeleman BP 2007 MICA marks additional risk factors for type 1 diabetes on extended HLA haplotypes: an association and meta-analysis. Mol Immunol 44:2806–2812 [DOI] [PubMed] [Google Scholar]
- Park YS, Sanjeevi CB, Robles D, Yu L, Rewers M, Gottlieb PA, Fain P, Eisenbarth GS 2002 Additional association of intra-MHC genes, MICA and D6S273, with Addison’s disease. Tissue Antigens 60:155–163 [DOI] [PubMed] [Google Scholar]
- Gambelunghe G, Falorni A, Ghaderi M, Laureti S, Tortoioli C, Santeusanio F, Brunetti P, Sanjeevi CB 1999 Microsatellite polymorphism of the MHC class I chain-related (MIC-A and MIC-B) genes marks the risk for autoimmune Addison’s disease. J Clin Endocrinol Metab 84:3701–3707 [DOI] [PubMed] [Google Scholar]
- Ota M, Katsuyama Y, Mizuki N, Ando H, Furihata K, Ono S, Pivetti-Pezzi P, Tabbara KF, Palimeris GD, Nikbin B, Davatchi F, Chams H, Geng Z, Bahram S, Inoko H 1997 Trinucleotide repeat polymorphism within exon 5 of the MICA gene (MHC class I chain-related gene A): allele frequency data in the nine population groups Japanese, Northern Han, Hui, Uygur, Kazakhstan, Iranian, Saudi Arabian, Greek and Italian. Tissue Antigens 49:448–454 [DOI] [PubMed] [Google Scholar]
- Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T 1996 Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci USA 93:12445–12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüe S, Monteiro RC, Berrih-Aknin S, Caillat-Zucman S 2003 Potential role of NKG2D/MHC class I-related chain A interaction in intrathymic maturation of single-positive CD8 T cells. J Immunol 171:1909–1917 [DOI] [PubMed] [Google Scholar]
- Skinningsrud B, Husebye ES, Gervin K, Lovas K, Blomhoff A, Wolff AB, Kemp EH, Egeland T, Undlien DE2008 Mutation screening of PTPN22: association of the 1858T-allele with Addison’s disease. Eur J Hum Genet 16:977–982 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Kawakami M, Yamanaka Y, Shimomura H, Imai Y, Ishida J, Yamamoto K, Ishitani A, Hatake K, Kirita T 2009 Relationship between soluble MICA and the MICA A5.1 homozygous genotype in patients with oral squamous cell carcinoma. Clin Immunol 130:331–337 [DOI] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Bugawan TL, Apple RJ, Thomson G, Erlich HA 2002 The HLA class I A locus affects susceptibility to type 1 diabetes. Hum Immunol 63:657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinspoon SK, Biller BM 1994 Clinical review 62: laboratory assessment of adrenal insufficiency. J Clin Endocrinol Metab 79:923–931 [DOI] [PubMed] [Google Scholar]
- Falorni A, Nikoshkov A, Laureti S, Grenbäck E, Hulting AL, Casucci G, Santeusanio F, Brunetti P, Luthman H, Lernmark Å1995 High diagnostic accuracy for idiopathic Addison’s disease with a sensitive radiobinding assay for autoantibodies against recombinant human 21-hydroxylase. J Clin Endocrinol Metab 80:2752–2755 [DOI] [PubMed] [Google Scholar]
- Zhang L, Barker JM, Babu S, Su M, Stenerson M, Cheng M, Shum A, Zamir E, Badolato R, Law A, Eisenbarth GS, Anderson MS2007 A robust immunoassay for anti-interferon autoantibodies that is highly specific for patients with autoimmune polyglandular syndrome type 1. Clin Immunol 125:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer JG, Marsh SGE, Albert ED, Bodmer WF, Dupont B, Erlich HA, Mach B, Mayr WR, Parham P, Sasazuki T, Schreuder GMT, Strominger JL, Svejgaard A, Terasaki PI1992 Nomenclature for factors of the HLA system, 1991. Eur J Immunogenet 19:327–344 [DOI] [PubMed] [Google Scholar]
- Robinson J, Pérez-Rodriguez M, Waller MJ, Cuillerier B, Bahram S, Yao Z, Albert ED, Madrigal JA, Marsh SG 2001 MICA sequences 2000. Immunogenetics 53:150–169 [DOI] [PubMed] [Google Scholar]
- Laureti S, De Bellis A, Muccitelli VI, Calcinaro F, Bizzarro A, Rossi R, Bellastella A, Santeusanio F, Falorni A 1998 Levels of adrenocortical autoantibodies correlate with the degree of adrenal dysfunction in subjects with preclinical Addison’s disease. J Clin Endocrinol Metab 83:3507–3511 [DOI] [PubMed] [Google Scholar]
- Barker JM, Ide A, Hostetler C, Yu L, Miao D, Fain PR, Eisenbarth GS, Gottlieb PA 2005 Endocrine and immunogenetic testing in individuals with type 1 diabetes and 21-hydroxylase autoantibodies: Addison’s disease in a high-risk population. J Clin Endocrinol Metab 90:128–134 [DOI] [PubMed] [Google Scholar]
- Gombos Z, Hermann R, Kiviniemi M, Nejentsev S, Reimand K, Fadeyev V, Peterson P, Uibo R, Ilonen J 2007 Analysis of extended human leukocyte antigen haplotype association with Addison’s disease in three populations. Eur J Endocrinol 157:757–761 [DOI] [PubMed] [Google Scholar]
- Hüe S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, Verkarre V, Fodil N, Bahram S, Cerf-Bensussan N, Caillat-Zucman S 2004 A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 21:367–377 [DOI] [PubMed] [Google Scholar]
- Magitta NF, Boe Wolff AS, Johansson S, Skinningsrud B, Lie BA, Myhr KM, Undlien DE, Joner G, Njolstad PR, Kvien TK, Forre O, Knappskog PM, Husebye ES 2009 A coding polymorphism in NALP1 confers risk for autoimmune Addison’s disease and type 1 diabetes. Genes Immun 10:120–124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.