Abstract
Context: FGFR1 mutations have been identified in about 10% of patients with Kallmann syndrome. Recently cases of idiopathic hypogonadotropic hypogonadism (IHH) with a normal sense of smell (nIHH) have been reported.
Aims: The objective of the study was to define the frequency of FGFR1 mutations in a large cohort of nIHH, delineate the spectrum of reproductive phenotypes, assess functionality of the FGFR1 mutant alleles in vitro, and investigate genotype-phenotype relationships.
Design: FGFR1 sequencing of 134 well-characterized nIHH patients (112 men and 22 women) and 270 healthy controls was performed. The impact of the identified mutations on FGFR1 function was assessed using structural prediction and in vitro studies.
Results: Nine nIHH subjects (five males and four females; 7%) harbor a heterozygous mutation in FGFR1 and exhibit a wide spectrum of pubertal development, ranging from absent puberty to reversal of IHH in both sexes. All mutations impair receptor function. The Y99C, Y228D, and I239T mutants impair the tertiary folding, resulting in incomplete glycosylation and reduced cell surface expression. The R250Q mutant reduces receptor affinity for FGF. The K618N, A671P, and Q680X mutants impair tyrosine kinase activity. However, the degree of functional impairment of the mutant receptors did not always correlate with the reproductive phenotype, and variable expressivity of the disease was noted within family members carrying the same FGFR1 mutation. These discrepancies were partially explained by additional mutations in known IHH loci.
Conclusions: Loss-of-function mutations in FGFR1 underlie 7% of nIHH with different degrees of impairment in vitro. These mutations act in concert with other gene defects in several cases, consistent with oligogenicity.
Loss-of-function mutations in fibroblast growth factor receptor 1 underlie normosmic idiopathic hypogonadotropic hypogonadism, either alone or in combination with other gene defects.
Idiopathic hypogonadotropic hypogonadism (IHH) is a rare genetic disorder defined by absent or incomplete sexual maturation in conjunction with low circulating gonadotropin and sex steroid levels and otherwise normal pituitary function/imaging (1,2). This disorder may occur with anosmia, termed Kallmann syndrome (KS), or with normal olfaction, termed normosmic IHH (nIHH). Associated nonreproductive phenotypes such as cleft palate and hearing loss occur with varying frequency (3,4). The underlying pathology is believed to be caused by defects in GnRH neuron fate specification or migration as well as anomalies in GnRH secretion or action (2,5,6). The multiple rare loci underlying IHH, acting alone or in combination include KAL1 (7,8), fibroblast growth factor receptor 1 (FGFR1) (9), fibroblast growth factor 8 (FGF8) (10), GnRH receptor (GNRHR) (11), nasal embryonic LHRH factor (NELF) (12), G protein-coupled receptor 54 (GPR54) (13,14), prokineticin 2 (PROK2) (15), prokineticin receptor 2 (PROKR2) (15), chromodomain helicase DNA-binding protein (CHD7) (16), neurokinin B (TAC3) (17), and neurokinin B receptor (TAC3R) (17).
FGFR1 mutations in KS were thought to result in a defect in GnRH neuron migration via abnormal olfactory bulb morphogenesis (9,18). FGFR1 is one of the four members of the conserved FGFR tyrosine kinase receptor subfamily. Upon binding fibroblast growth factor (FGF) and heparan sulfate, FGFRs undergo dimerization and trans-autophosphorylation to activate Ras-MAPK and phospholipase Cγ pathways (19). FGFR1 is an N-glycosylated protein. Glycosylation is thought to have a role in folding and quality control in the endoplasm reticulum (ER) (20) and may control subcellular localization and trafficking of proteins (21).
KS families harboring heterozygous FGFR1 mutations display variable olfactory phenotypes (9,22). In addition, a few cases of heterozygous FGFR1 mutations underlie IHH patients with a normal sense of smell (nIHH) (22,23,24,25). However, the frequency of FGFR1 mutations among nIHH patients is currently unknown. Therefore, this study aims to investigate the prevalence of FGFR1 mutations within a large cohort of nIHH patients, analyze the molecular mechanisms underlying receptor dysfunction through both structural and functional studies, and define genotype-phenotype relationships.
Subjects and Methods
Subjects
Normosmic IHH
Normosmic IHH was diagnosed according to the following criteria: 1) absent/incomplete puberty by age 18 yr; 2) serum testosterone 100 ng/dl or less in men or estradiol 20 pg/ml or less in women in association with low or normal levels of serum gonadotropins; 3) otherwise normal pituitary function; 4) normal serum ferritin concentrations; 5) normal magnetic resonance imaging (MRI) of the hypothalamic-pituitary region; and 6) normal olfaction as defined by history. When possible, olfaction was assessed by the University of Pennsylvania Smell Identification Test (UPSIT). A score of fifth percentile or greater adjusted for age and sex was considered to be a normal sense of smell (26).
One hundred thirty-four nIHH unrelated probands were included. The cohort comprised 112 male and 22 female probands. One third of the probands had a history of partial puberty, whereas the remainder had absent puberty. Additionally, 33% of the cases were familial. By definition, all 134 probands reported a normal sense of smell.
Controls
Two hundred seventy healthy Caucasian controls were studied from Massachusetts General Hospital. All controls had normal reproductive function and normal sense of smell by history. They provided blood for DNA analysis. The study was approved by the Partners Human Research Committee and all subjects provided written informed consent.
Clinical studies
Medical history and physical examination
Detailed medical histories and physical examination were performed as previously described (27). When available, family members were recruited for these clinical phenotyping studies.
Neuroendocrine evaluation
Probands agreeing to undergo detailed neuroendocrine studies were admitted to the Massachusetts General Hospital General Clinical Research Center for an overnight 12-h frequent sampling study to assess endogenous LH pulsatility (every 10 min) as previously described (28). Pulsatile hormone secretion was analyzed using a modification of the Santen and Bardin method (28), and study pools were measured for FSH, testosterone, and estradiol as previously reported (28). A renal ultrasound, dual-energy x-ray absorptiometry scan, and cranial three-dimensional MRI scan to assess the olfactory system were performed (22).
Mutation analyses
Normosmic IHH probands were prospectively screened for mutations in FGFR1 within a 3-yr period. Sequencing of the coding regions and intron-exon boundaries of the FGFR1 gene (GenBank accession NM_023110) was performed (22). Nonsense changes as well as nucleotides changes that were nonsynonymous or led to splicing errors and absent in more than 340 ethnically matched alleles were defined as mutations. All genes and proteins are described by using standard nomenclature (29).
Normosmic IHH probands with a mutation in FGFR1 were subsequently screened for other gene defects if not previously performed [GNRHR (30), GPR54 (14), PROK2 (31), PROKR2 (32), and FGF8 (10)]. In addition, family members of probands with FGFR1 mutations underwent detailed phenotyping and mutational screening when possible.
Predictions of the functional impacts of FGFR1 mutations based on the available structures of FGFRs
The crystal structures of the FGFR1 extracellular ligand binding region in complex with FGF2 and heparin (Protein Data Bank entry 1FQ9) (33), the FGFR1 kinase domain (Protein Data Bank entry 1FGK) (34), and the solution structures of D1 of human and mouse FGFR1 (Protein Data Bank entries 2CR3 and 2CKN) were used to explore the effects of the mutations on the function-structure of FGFR1 (35). The impact of the K618N mutation on FGFR1 function was investigated using the crystal structure of A-loop phosphorylated active FGFR2 kinase domain (Protein Data Bank entry 2PVF) (36), which has 90% sequence identity to FGFR1 kinase domain. Structures were visualized by using the program O (37), and structural representations were made using Pymol (www.pymol.org, Delano Scientific, LLC).
Site-directed mutagenesis
N-terminal myc-tagged FGFR1c cDNA (38) in pcDNA3.1+ (Invitrogen, Carlsbad, CA) was used as a template for site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutant constructs were sequenced on both strands to verify nucleotide changes. The L83V GNRHR mutation was similarly introduced into an expression vector encoding the wild-type human GnRH receptor (hGnRHR) (39).
Receptor expression and maturation studies
Although antibodies directed against FGFR1 ectoderm can be used to detect cell surface expression of FGFR1, we chose not to use antibodies against FGFR1 ectodomain because some of the identified mutations map to this region and could potentially eliminate some epitopes. Instead we fused a myc-tag to the ectodomain of the receptor and used anti-myc antibodies to detect cell surface expression. Herein we performed endoglycosidase digestion, Western analysis, and cell surface expression (40). Details of the methods used are found in supplemental data S1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org.
FGF reporter gene assay
L6 myoblasts (American Type Culture Collection, Manassas, VA), which are largely devoid of endogenous FGFRs and FGFs (41,42), were maintained in DMEM containing penicillin (100 U/liter), streptomycin (100 μg/liter), and 10% fetal calf serum (Invitrogen). Transient transfections were performed at 10% cell confluency in 24-well plates with 5 ng of wild-type (WT) or mutant FGFR1 cDNA with 100 ng of the osteocalcin FGF response element-luciferase reporter (kind gift from Dr. David Ornitz, Washington University Medical School, St. Louis, MO), and 295 ng of empty vector DNA, using FuGene6 reagent (Roche Diagnostics, Indianapolis, IN). After 24 h of serum starvation, cells were treated for 16 h with increasing doses of FGF2 (from 0 to 2000 pm) in DMEM containing 0.1% BSA. The cells were lysed with passive lysis buffer (Promega, Madison, WI), and assayed for luciferase activity using Promega luciferase assay system. Results were plotted and fitted with four-parameter sigmoidal dose response curves using Prism4 software (GraphPad Software Inc., San Diego, CA). Experiments were performed in triplicate and repeated at least three times.
Kinase autophosphorylation assay
The tyrosine autophosphorylation activity of the wild-type FGFR2 kinase domain and K621N and A674P (analogous to K618N and A671P in FGFR1, respectively) mutant FGFR2 kinase domains were quantified using a continuous spectrophotometric assay, as previously described (43). In this assay, hydrolysis of ATP to ADP is coupled to the oxidation of nicotinamide adenine dinucleotide hydroxide (NADH) to oxidized of nicotinamide adenine dinucleotide and measured as a reduction in NADH absorbance at 340 nm. Reactions were carried out at 30 C in 50 μl of buffer containing 100 mm Tris-HCl (pH 7.5), 1.0 mm ATP, 5 mm MgCl2, 1.5 mm phosphoenolpyruvate, 1.2 mg/ml NADH, 89 U/ml pyruvate kinase, 124 U/ml lactate dehydrogenase, and 5 μm recombinant WT or mutant FGFR2 kinase for 1 h.
Inositol phosphate (IP) assay
WT or L83V mutant hGnRHR cDNA expression vectors were transiently transfected into COS7 cells. The response of the transiently transfected cells to a GnRH agonist (des-Gly10[d-Ala6] GnRH ethylamide; Sigma Chemicals, St. Louis, MO) was measured by IP accumulation as previously described (39). Assay points were performed in triplicate, and the experiment was repeated three times.
Statistical analyses
Frequency of FGFR1 mutations in IHH males or females were compared using Fisher’s exact test. Cell surface expression levels of WT and mutant receptors were compared using a Mann Whitney nonparametric t test on all 12 observations (three experiments, four repeats). In the transcriptional assay, the activity of each FGFR1 mutant was compared with WT using the WT EC50 dose of FGF2. P < 0.05 was considered significant for all analyses.
Results
Sequence analysis of the FGFR1 gene
Sequencing of the FGFR1 gene in 134 unrelated nIHH probands revealed the presence of nine heterozygous mutations (Fig. 1A): c.296 A>G in exon 3 (p.Y99C), c.350 A>G in exon 3 (p.N117S), c.682 T>G in exon 6 (p.Y228D), c.716 T>C in exon 6 (p.I239T), c.749 G>A in exon 7 (p.R250Q), c.1409 G>T in exon 10 (p.R470L), c.1854 G>T in exon 13 (p.K618N), c.2011 G>C in exon 15 (p.A671P), and c. 2038 C>T in exon 15 (p.Q680X), four of which have been previously published (10,12,22,44). Thus, the frequency of FGFR1 mutations was 7% (nine of 134), and higher among females (four of 22) compared with males (five of 112) (P < 0.05).
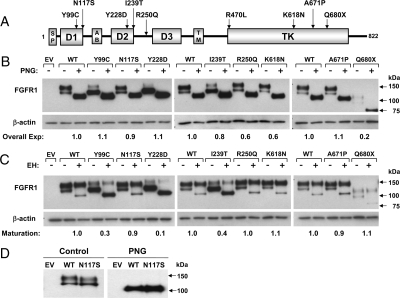
Figure 1.
A, Schematic of the FGFR1 mutations. The nine FGFR1 mutations span several functional domains of the receptor. AB, Acid box. B–D, Endoglycosidase analysis of FGFR1 mutants. COS-7 cells were transiently transfected with 5 ng of Myc-tagged WT or mutated FGFR1 cDNA. Cell lysates were subjected to PNGase (PNG, B) or EndoH (EH, C) digestion and then processed for FGFR1 immunoblotting using anti-myc antibodies. Overall expression levels were determined by normalizing PNGase-treated bands to their respective β-actin densities (B). Receptor maturation levels were estimated by calculating the fraction of the upper band (mature) out of the total FGFR1 immunoreactivity of EndoH-treated samples (C). In both analyses, the calculated mutant values are expressed as a ratio of WT. Untreated and PNGase-treated WT and N117S mutant are subjected to longer electrophoretic migration (D).
Mapping the FGFR1 mutations onto the crystal structure
Unlike the craniosynostosis mutations, which map to certain hotspot regions of FGFRs, the IHH mutations are scattered over several domains of the receptor (Fig. 1A). The p.Y99C and p.N117S mutations localize to the immunoglobulin 1 (D1) domain of FGFR1 (Fig. 2). Mapping these mutations onto the solution structure of FGFR1 D1 predicts both to be loss of function (Fig. 2). The side chain of Y99, located on αF strand, points into the hydrophobic interior of D1 and plays a critical role in D1 folding (Fig. 2C). Substitution of this tyrosine for a smaller cysteine should destabilize D1 folding, leading to impaired glycosylation and thus retention of the misfolded mutant receptor in the ER. Consistent with its structural role, Y99 is fully conserved in all four human FGFRs. N117, situated on αG strand, is solvent exposed and a putative glycosylation site. Substitution of asparagine for a serine should interfere with FGFR1 glycosylation and lead to reduced cell surface expression of the mutant receptor.
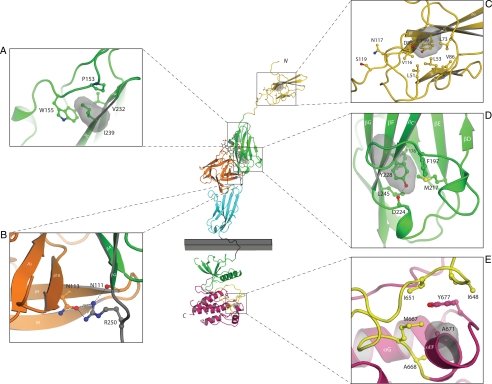
Figure 2.
Mapping of the nIHH mutations onto the known FGFR crystal structures suggest that they impair the activity of FGFR1c. The various mutants are mapped onto the ribbon diagram of D1 solution structure (Protein Data Bank entry 2CR3, 2CKN) or FGF2-FGFR1c-heparin complex (Protein Data Bank entry 1FQ9) and FGFR1 kinase domain (Protein Data Bank entry 1FGK) crystal structures. D1 is colored in gold. FGF is colored in orange and the extracellular ligand binding region of FGFR is colored as follows: D2, green; D3, cyan; D2–D3 linker, gray. The coloring of the intracellular tyrosine kinase domain is as follows: the N-terminal lobe of kinase in green; the C-terminal lobe, purple; the activation loop, yellow; and the kinase hinge region, gray. Note that ATP (not shown) binds in the cleft between the N-lobe and C-lobe of the kinase domain. Only relevant β-strands and α-helices of FGFR1c are labeled. A–E, Close-up of the microenvironment of Y99C, N117S, I239, Y228, R250, and A671 subject to mutation in probands with nIHH. In each panel, in addition to the mutated residue, other relevant receptor residues are shown as ball and sticks. A, Side chain of I239 pointing into the hydrophobic core of D2 and interacting with hydrophobic residues P153, W155, and V232, thus contributing to tertiary fold of D2. The surface of I239 is shown as red mesh and P153, W155, and V232 are shown as sticks. B, Network of hydrogen bonds between R250 of FGFR1 and FGF2 is shown. C, Side chain of Y99 pointing into the hydrophobic core of D1 and thus contributing to tertiary fold of D1. The surface of Y99 is shown as red mesh and nearby interacting hydrophobic residues L51, L53, L73, V86, and V116 are shown as sticks. N117 is a potential glycosylation site and is rendered in sticks. D, Side chain of Y228 engages in intramolecular hydrophobic interactions with F176, F197, M217, and L245 in the core of D2. The surface of Y228 is shown as red mesh and F176, F197, M217, and L245 are shown as sticks. E, Surface of A671 is shown in red mesh and the surfaces of hydrophobic residues in the vicinity of A671 are shown in blue mesh. Atom coloring is as follows: nitrogen in blue; oxygen, red; sulfur, yellow. Hydrogen bonds are shown as dashed lines. Letters N and C, N and C termini of FGFR1c, respectively. The membrane bilayer is represented as a gray rectangle.
The impacts of the remaining mutations were investigated in the context of the crystal structures of FGFR1-FGF2-heparin complex (1FQ9), the unphosphorylated FGFR1 kinase domain (Protein Data Bank entry 1FGK), and the A-loop phosphorylated active FGFR2 kinase domain (Protein Data Bank entry 2PVF) (36). Both Y228 and I239 map to D2, and the crystal structure shows that these two residues are essential for structural integrity of D2 domain as their side chains contribute to the hydrophobic core of D2 (Fig. 2, A and D). Therefore, similar to the p.Y99C D1 mutation, the p.Y228D and p.I239T mutations should impair the D2 fold and thus manifest in glycosylation defects and retention of the misfolded mutant receptors in the ER. In contrast, the p.R250Q mutation should lead to loss of function by reducing the binding affinity of FGFR1 toward all FGFs because R250 of FGFR and the homologous arginines in other FGFRs engage in FGFR-invariant hydrogen bonds with ligand (Fig. 2B). The mechanisms by which the p.R470L and p.Q680X kinase domain mutations lead to loss of function have been previously described (12,22). The newly identified p.A671P and p.K618N kinase domain mutations should also lead to loss of function. The p.A671P mutation maps to αEF helix in the C-lobe of TK domain and should compromise the formation of helix EF, which is a critical structural element involved in substrate recognition (Fig. 2E). The p.K618N mutation, also located in the C-terminal lobe of the TK domain, is predicted to lead to loss of function. Crystal structure of the A-loop phosphorylated active FGFR2 kinase domain shows that K621, the corresponding residue in FGFR2 kinase domain, supports the active A-loop conformation by making hydrogen bonds with residues in the A-loop (data not shown).
Biological assays
To test our structural predictions, we first compared cell surface expression of WT and mutant receptors transiently expressed in COS-7 cells.
Western blotting analysis shows two immunoreactive-specific bands for WT FGFR1c at 140 and 120 kDa (Fig. 1B). peptide N-glycosidase digestion, which removes all types of N-linked carbohydrate chains from a glycoprotein, reduced the two bands into a single one of about 100 kDa, confirming that the two bands are differently N-glycosylated receptor. The overall expression levels of the receptors were calculated from the PNGase-treated samples. The overall expression of the R250Q and K618N mutants were slightly decreased compared with WT, whereas the expression of the Q680X mutant was dramatically decreased (Fig. 1B).
In contrast, treatment with endoglycosidase H (EndoH), which removes only high mannose N-linked sugars, causes only the minor 120-kDa band of the WT FGFR1c receptor to change its mobility (Fig. 1C), indicating that this band represents the high mannose partially processed receptor, whereas the 140-kDa EndoH-resistant band represents the fully glycosylated mature form of FGFR1c. Densitometric analysis revealed that 80% of the WT FGFR1 was expressed as a mature form (Fig. 1C). Consistent with our structural analysis predicting that Y99C, Y228D, and I239T mutations destabilize the tertiary structure of FGFR1 ectodomain, these mutant receptors were expressed mostly as the high mannose 120-kDa immature form (0.3, 0.1, 0.4 relative to WT, respectively; Fig 1C). In contrast, the R250Q, K618N, and A671P mutants had similar expression levels of mature form as WT receptor. Importantly, these data are also harmonious with our structural analysis predicting that R250Q, K618N, and A671P mutations do not have any adverse effects on the tertiary fold of neither FGFR1 ectodomain nor kinase domain. Lastly, mature N117S migrates slightly faster than the WT, consistent with the loss of the N-glycosylation site at position 117. Notably, after PNGase digestion, both bands run to the same level (Fig. 1D).
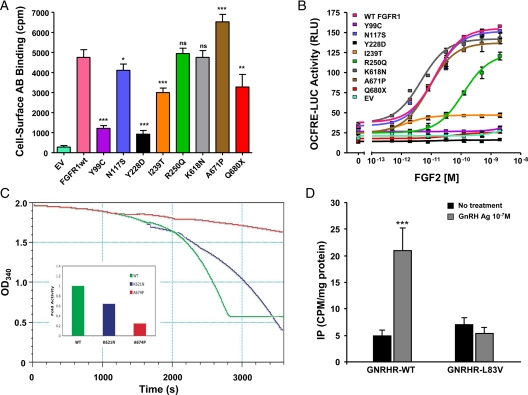
Consistent with the results of deglycosylation experiments, the mutants Y99C, Y228D, and I239T exhibited a major reduction in cell surface expression levels (0.2, 0.14, 0.61 relative to WT, respectively; P < 0.05) (Fig. 3A). The N117S mutant, which eliminates a glycosylation site, showed a small but significant reduction in cell surface expression of the receptor (0.85 relative to WT; P < 0.05) (Fig. 3A). The R250Q and K618N mutants had similar cell surface expression levels as WT. The A671P mutant was detected at significantly higher levels (1.4 relative to WT; P < 0.05; Fig. 3A), suggesting a potential defect in receptor turnover rate. A large decrease in cell surface expression was also observed for Q680X (0.65 relative to WT, P < 0.05), consistent with its low overall expression levels. The mutation Q680X is located 13 bp upstream of the end of exon 17. Therefore, this mutated allele is not predicted to undergo an mRNA nonsense-mediated decay (45).
Figure 3.
A, Cell surface expression of FGFR1 mutants. COS-7 cells were transiently transfected with 5 ng of Myc-tagged WT or mutated FGFR1 cDNA. Cell surface receptor levels were assessed using anti-Myc monoclonal antibodies and [125I]rabbit antimouse IgG. Data shown are means ± sem of three experiments, each performed in quadruplicates. Statistical difference between expression of mutants vs. WT receptors were analyzed using repeated-measures ANOVA followed by Dunnett’s multiple comparison test. ns, Not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001. B, Transcription reporter activity of WT and mutants FGFR1. WT and FGFR1c mutants were transiently transfected into L6 myoblasts with an FGFR1-responsive osteocalcin promoter luciferase construct. FGF2 treatment of WT FGFR1c induced a 5-fold increase in LUC reporter gene expression, whereas Y99C, Y228D, I239T, and Q680X remained silent. The mutant R250Q had a right-shifted curve and did not reach maximum activity of WT. In contrast, N117S, K618N, and A671P activities do not differ from WT. C, Kinase activity of WT and mutants FGFR1. The tyrosine autophosphorylation activity of wild-type (green) and the K621N (blue) and A674P (red) mutant FGFR2 kinases (corresponding to K618N and A671P in FGFR1, respectively) were quantified by using a continuous spectrophotometric assay as described by Barker et al. (43). In this assay, hydrolysis of ATP to ADP is measured as a reduction in NADH absorbance at 340 nm. Relative to wild-type kinase (normalized as 1), the activity of mutant kinases are determined as 0.64 for K621N and 0.25 for A674P consistent with loss of function. D, The L83V hGnRHR mutation blocks GnRH-induced IP accumulation. IP accumulation was measured in COS-7 cells transiently transfected with cDNA encoding wild-type or L83V hGnRHR and stimulated with 10−7 m GnRH agonist. Data points represent the mean ± sem of triplicate samples. The figure is a representative graph from three individual experiments.
Signaling activity of the FGFR1 mutants was assayed using the FGF-responsive reporter osteocalcin FGF response element-luciferase in L6 myoblasts, which acts downstream of the MAPK pathway (Fig. 3B) (46). Cells expressing WT receptor showed a typical sigmoid dose-response curve with about 5-fold maximum induction and EC50 around 15 pm after treatment with increasing doses of FGF2. As was expected from their low cell surface expression levels, mutants Y99C, Y228D, I239T, and Q680X did not respond to FGF stimulation, even at the highest FGF2 dose tested (2 nm). The N117S, K618N, and A671P mutants responded similarly to the WT, whereas R250Q generated a dose-response curve with an approximately 10-fold increase in the EC50, consistent with decreased binding affinity of the receptor to the ligand, thus confirming our structural prediction.
To sort out the discrepancy between the structural analysis and transcriptional assay for the K618N and A671P mutations, we studied the effect of these mutations in an in vitro kinase assay. The catalytic kinase domains of FGFR1 and FGFR2 have greater than 90% sequence similarity and are both regulated by a FGFR-invariant molecular brake at the kinase hinge/interlobe region (36). Because FGFR2 kinase domain can be produced in Escherichia coli in larger quantities and better purity than FGFR1 kinase domain, we chose the FGFR2 kinase domain as the template. The kinase activity of mutant FGFR2 kinase domains harboring K621N or A674P mutations, analogous to K618N and A671P mutations in FGFR1, was compared with WT FGFR2 kinase using a continuous spectrophotometric assay (Fig. 3C). In agreement with our structural prediction, the activities of K621N or A674P mutant kinases were 0.64 and 0.25, respectively, relative to WT FGFR2 kinase (normalized as 1).
Genotype phenotype correlation
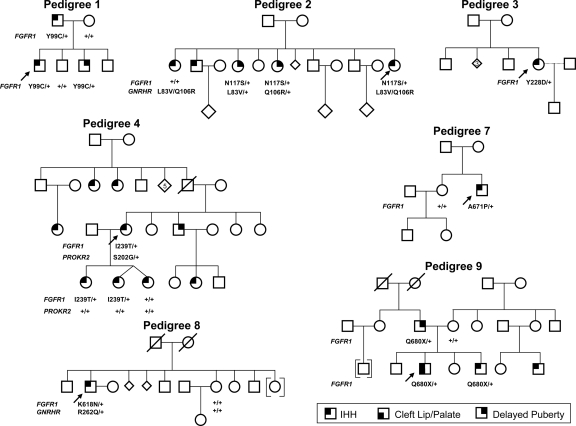
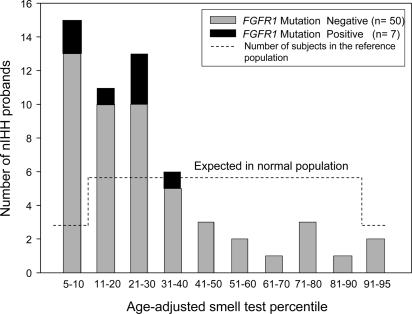
The clinical data of the nine nIHH probands with an FGFR1 mutation are summarized in Table 1 and Fig. 4, and each pedigree is described in detail in supplementary data S2. Of note, seven probands (73%) had absent puberty, whereas two had partial puberty. Seven of nine cases (73%) were familial. This is greater than the familial cases noted within the entire cohort of nIHH (35%). A subset of 57 probands performed a UPSIT 40-item smell test. All IHH subjects had score greater than fifth percentile adjusted for age and sex. Thus, the UPSIT results agree with the history of normosmia. However, the UPSIT tests results were lower in the nIHH (with or without FGFR1 mutation) compared with the normal reference population (Fig. 5). This FGFR1 nIHH cohort includes two cases of HH reversal, one male (no. 1) and one female (no. 4) and a variety of skeletal phenotypes (Table 1). Furthermore, variable expressivity was observed within family members carrying the FGFR1 mutation (Fig. , supplemental data S2). In vitro studies of the FGFR1 mutants did not correlate with the severity of GnRH deficiency in all probands (Table 2). Notably, additional gene defects were identified in five of nine nIHH probands carrying an FGFR1 mutation (Table 1). Subjects 2, 6, and 8 harbor additional GNRHR mutations, [p.L83V + p.Q106R] (Fig. 4D), [p.Q106R + p.R262Q] (12), and heterozygous p.R262Q, respectively, whereas subject 5 carries an additional heterozygous loss-of-function FGF8 mutation previously reported (p.K100E) (10). In addition, subject 4 harbors an additional heterozygous PROKR2 mutation (p.S202G), the functionality of which has not been assessed in vitro.
Table 1.
Clinical presentation of nIHH subjects carrying a FGFR1 mutation
| Proband | Sex | Puberty | Inheritance | LH secretion pattern | MRI of OB | Other phenotypes | Additional gene defects |
|---|---|---|---|---|---|---|---|
| 1a | M | Partial | Familial | Pulsatile | OB and sulci normal | HH reversal | |
| 2 | F | Absent | Familial | Undetectable | NA | None | GnRHR |
| 3 | F | Absent | Sporadic | Undetectable | NA | Osteoporosis | |
| 4 | F | Absent | Familial | Undetectable | NA | HH reversal | PROKR2 |
| 5a | M | Absent | Familial | Undetectable | NA | None | FGF8 |
| 6a | F | Absent | Familial | Undetectable | OB and sulci normal | None | GNRHR |
| 7 | M | Absent | Sporadic | Undetectable | OB and sulci normal | Frontal bossing | |
| 8 | M | Partial | Familial | Apulsatile | OB and sulci normal | Clinodactily, osteopenia | GNRHR |
| 9a | M | Absent | Familial | Undetectable | OB and sulci normal | Cleft lip/palate |
Figure 4.
Pedigrees of probands with an FGFR1 mutation. Pedigrees 2 and 4 show additional gene defects (GNRHR and PROKR2, respectively). Circles, Females; squares, males; diamonds, additional siblings; arrows, proband; and +, WT for the gene.
Figure 5.
Histogram of age-adjusted smell test percentile in nIHH. UPSIT results from seven probands carrying an FGFR1 mutation are contrasted with 50 nIHH probands negative for FGFR1 mutations. The dashed line reflects the number of subjects in the reference population.
Table 2.
Summary of functional studies in nIHH subjects with a FGFR1 mutation
| Proband | Mutation | Domain | Structural prediction | Overall cell exp. | Protein maturation | Cell surface exp. | Reporter-luciferase assay | Kinase activity |
|---|---|---|---|---|---|---|---|---|
| 1 | p.Y99C | D1 | Disrupts D1 folding | = | ⇓ | ⇓ | ⇓ | |
| 2 | p.N117S | D1 | Elimates a glycosylation site | = | = | ↓ | = | |
| 3 | p.Y228D | D2 | Disrupts D2 ligand-binding site | = | ⇓ | ⇓ | ⇓ | |
| 4 | p.I239T | D2 | Destabilize D2 | = | ⇓ | ⇓ | ⇓ | |
| 5 | p.R250Q | D1–D2 linker | Disrupts ligand binding | ↓ | = | = | ↓ | |
| 7 | p.K618N | TK | Disrupts kinase A-loop | ↓ | = | = | = | ↓ |
| 8 | p.A671P | TK | Disrupts kinase A-loop | = | = | ↑ | = | ⇓ |
| 9 | p.Q680X | TK | Eliminates portion of catalytic domain | ⇓ | = | ⇓ | ⇓ |
D1, Domain 1; D2, domain 2; TK, tyrosine kinase; Exp, expression; =, equal to WT; ↓, moderate decrease; ⇓, severe decrease; and ↑, moderate increase.
Discussion
The frequency of FGFR1 mutations in this large cohort of probands with nIHH was 7% (nine of 134), compared with 3% in the previous Brazilian report (one of 34) (24), and approximately 10% in prior reports of KS cohorts (9,47). In light of these findings, FGFR1 appears to be a key locus for both KS and nIHH and should be included with GNRHR (11), GPR54 (13), PROK2 (31), PROKR2 (32), FGF8 (10), CHD7 (16), TAC3 (17), and TAC3R (17) in the genetic screening panel for nIHH patients. Our results also support the notion that KS and nIHH are not two entirely distinct clinical entities but can share a common genetic basis (22). Of note, the frequency of FGFR1 mutations was higher among nIHH females compared with males. This gender discrepancy could be in part explained by the effect of putative X-chromosomal modifier genes on the IHH phenotype.
Probands carrying an FGFR1 mutation displayed a wide degree of pubertal development ranging from absent puberty to reversal of IHH in two probands (44), one of whom is a female with severe nIHH. To our knowledge, there is only one previous case report describing a spontaneous pregnancy in a woman with partial nIHH after sex steroid exposure, suggesting a reversal of her IHH (48). Interestingly, that woman harbors the identical homozygous GNRHR mutation described in a male nIHH reversal subject (49).
The broad range of reproductive phenotypes in our nIHH cohort indicates varying degrees of endogenous GnRH secretion. Structural and functional consequences of the FGFR1 mutations revealed loss of function through different mechanisms including: 1) decreased ligand binding affinity (R250Q); 2) destabilization of the receptor ectodomain leading to impaired glycosylation and hence decreased cell surface expression of the receptor (Y99C, Y228D, and I239T) (21); 3) altered glycosylation, resulting in decreased cell surface expression and likely abnormal receptor trafficking (N117S); 4) decreased tyrosine kinase activity (A671P and K618N); and 5) the truncated Q680X mutant displays substantially decreased overall expression, suggesting folding defects. Of note, the FGFR1 gene encodes two major isoforms, FGFR1b and FGFR1c, generated through alternative splicing in D3. All the nIHH FGFR1 mutations identified so far map to the nonspliced regions of the receptor and therefore are predicted to affect both FGFR1c and FGFR1b isoforms. This contrasts with KS in which the FGFR1c isoform has been implicated (47,50,51). Thus, we decided to select FGFR1c for our biochemical investigations.
The gradations in loss of function did not accurately predict the severity of the reproductive phenotypes (i.e. the degree of pubertal development and GnRH deficiency) in all cases (Tables 1 and 2). Furthermore, several family members carrying an identical FGFR1 mutation exhibited variable expressivity of reproductive phenotypes. This finding could be partially explained by multiple gene defects in five pedigrees, three of which are novel, which support the emerging model of oligogenicity in IHH (10,12,15). Alternatively, the inconsistent correlations between in vitro activity of the FGFR1 mutants and the corresponding phenotypes may be explained by differences in endogenous hormonal effects (i.e. in utero sex steroid exposure) and/or environmental cues. Indeed, there are examples of identical twins (both males and females) discordant for reproductive phenotypes (52,53). Finally, our description of IHH reversal in probands carrying a FGFR1 mutation further supports a critical role for gene-environment interaction in this disorder.
In conclusion, inactivating FGFR1 mutations, either alone or in combination with other gene defects underlying IHH, occur at a high frequency in nIHH probands.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grants R01 HD015788-21, R01 HD19938, U54HD028138-16, and R01 DE13686-09; Foundation for Pediatric Research, Finland; and the Academy of Finland.
Disclosure Summary: The authors have nothing to declare.
First Published Online October 9, 2009
Abbreviations: EndoH, Endoglycosidase H; ER, endoplasm reticulum; FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; hGnRHR, human GnRH receptor; IHH, idiopathic hypogonadotropic hypogonadism; IP, inositol phosphate; KS, Kallmann syndrome; MRI, magnetic resonance imaging; nIHH, normosmic IHH; UPSIT, University of Pennsylvania Smell Identification Test; WT, wild type.
References
- Santoro N, Filicori M, Crowley Jr WF 1986 Hypogonadotropic disorders in men and women: diagnosis and therapy with pulsatile gonadotropin-releasing hormone. Endocr Rev 7:11–23 [DOI] [PubMed] [Google Scholar]
- Seminara SB, Hayes FJ, Crowley Jr WF 1998 Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann’s syndrome): pathophysiological and genetic considerations. Endocr Rev 19:521–539 [DOI] [PubMed] [Google Scholar]
- Quinton R, Duke VM, Robertson A, Kirk JM, Matfin G, de Zoysa PA, Azcona C, MacColl GS, Jacobs HS, Conway GS, Besser M, Stanhope RG, Bouloux PM 2001 Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin Endocrinol (Oxf) 55:163–174 [DOI] [PubMed] [Google Scholar]
- Sato N, Katsumata N, Kagami M, Hasegawa T, Hori N, Kawakita S, Minowada S, Shimotsuka A, Shishiba Y, Yokozawa M, Yasuda T, Nagasaki K, Hasegawa D, Hasegawa Y, Tachibana K, Naiki Y, Horikawa R, Tanaka T, Ogata T 2004 Clinical assessment and mutation analysis of Kallmann syndrome 1 (KAL1) and fibroblast growth factor receptor 1 (FGFR1, or KAL2) in five families and 18 sporadic patients. J Clin Endocrinol Metab 89:1079–1088 [DOI] [PubMed] [Google Scholar]
- Kim SH, Hu Y, Cadman S, Bouloux P 2008 Diversity in fibroblast growth factor receptor 1 regulation: learning from the investigation of Kallmann syndrome. J Neuroendocrinol 20:141–163 [DOI] [PubMed] [Google Scholar]
- Chung WC, Moyle SS, Tsai PS 2008 Fibroblast growth factor 8 signaling through fibroblast growth factor receptor 1 is required for the emergence of gonadotropin-releasing hormone neurons. Endocrinology 149:4997–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, Bougueleret L, Delemarre-van de Waal HA, Lutfalla G, Weissenbach J, Petit C 1991 The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell 67:423–435 [DOI] [PubMed] [Google Scholar]
- Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, Brown CJ, Willard HF, Lawrence C, Persico MG, Camerino G, Ballabio A 1991 A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature 353:529–536 [DOI] [PubMed] [Google Scholar]
- Dodé C, Levilliers J, Dupont JM, De Paepe A, Le Dû N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pêcheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP 2003 Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet 33:463–465 [DOI] [PubMed] [Google Scholar]
- Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, Tsai P, Pitteloud N 2008 Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest 118:2822–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E 1997 A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med 337:1597–1602 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W 2007 Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest 117:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E 2003 Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA 100:10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno Jr JS, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley Jr WF, Aparicio SA, Colledge WH 2003 The GPR54 gene as a regulator of puberty. N Engl J Med 349:1614–1627 [DOI] [PubMed] [Google Scholar]
- Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP 2006 Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet 2:e175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC 2008 Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet 83:511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O'Rahilly S, Semple RK 2009 TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet 41:354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, Lin M, Partanen J, Rossant J, McConnell SK 2003 FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development 130:1101–1111 [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Olsen SK, Ibrahimi OA 2005 Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev 16:107–137 [DOI] [PubMed] [Google Scholar]
- Parodi AJ 2000 Protein glucosylation and its role in protein folding. Annu Rev Biochem 69:69–93 [DOI] [PubMed] [Google Scholar]
- Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW 2004 Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science 306:120–124 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Acierno Jr JS, Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, Metzger DL, Hayes FJ, Dwyer AA, Hughes VA, Yialamas M, Hall JE, Grant E, Mohammadi M, Crowley Jr WF 2006 Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 103:6281–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Herrick SR, Lemyre E, Kishikawa S, Salisz JA, Seminara S, MacDonald ME, Bruns GA, Morton CC, Quade BJ, Gusella JF 2005 Hypogonadotropic hypogonadism and cleft lip and palate caused by a balanced translocation producing haploinsufficiency for FGFR1. J Med Genet 42:666–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trarbach EB, Costa EM, Versiani B, de Castro M, Baptista MT, Garmes HM, de Mendonca BB, Latronico AC 2006 Novel fibroblast growth factor receptor 1 mutations in patients with congenital hypogonadotropic hypogonadism with and without anosmia. J Clin Endocrinol Metab 91:4006–4012 [DOI] [PubMed] [Google Scholar]
- Xu N, Qin Y, Reindollar RH, Tho SP, McDonough PG, Layman LC 2007 A mutation in the fibroblast growth factor receptor 1 gene causes fully penetrant normosmic isolated hypogonadotropic hypogonadism. J Clin Endocrinol Metab 92:1155–1158 [DOI] [PubMed] [Google Scholar]
- Doty RL, Applebaum S, Zusho H, Settle RG 1985 Sex differences in odor identification ability: a cross-cultural analysis. Neuropsychologia 23:667–672 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Hayes FJ, Boepple PA, DeCruz S, Seminara SB, MacLaughlin DT, Crowley Jr WF 2002 The role of prior pubertal development, biochemical markers of testicular maturation, and genetics in elucidating the phenotypic heterogeneity of idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab 87:152–160 [DOI] [PubMed] [Google Scholar]
- Hayes FJ, McNicholl DJ, Schoenfeld D, Marsh EE, Hall JE 1999 Free α-subunit is superior to luteinizing hormone as a marker of gonadotropin-releasing hormone despite desensitization at fast pulse frequencies. J Clin Endocrinal Metab 1999:1028–1036 [DOI] [PubMed] [Google Scholar]
- Antonarakis SE 1998 Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat 11:1–3 [DOI] [PubMed] [Google Scholar]
- Beranova M, Oliveira LM, Bédécarrats GY, Schipani E, Vallejo M, Ammini AC, Quintos JB, Hall JE, Martin KA, Hayes FJ, Pitteloud N, Kaiser UB, Crowley Jr WF, Seminara SB 2001 Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab 86:1580–1588 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Zhang C, Pignatelli D, Li JD, Raivio T, Cole LW, Plummer L, Jacobson-Dickman EE, Mellon PL, Zhou QY, Crowley Jr WF 2007 From the cover: loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 104:17447–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, Pignatelli D, Hughes VA, Dwyer AA, Raivio T, Hayes FJ, Seminara SB, Huot C, Alos N, Speiser P, Takeshita A, Van Vliet G, Pearce S, Crowley Jr WF, Zhou QY, Pitteloud N 2008 Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab 93:3551–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M 2000 Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell 6:743–750 [DOI] [PubMed] [Google Scholar]
- Mohammadi M, Schlessinger J, Hubbard SR 1996 Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell 86:577–587 [DOI] [PubMed] [Google Scholar]
- Kiselyov VV, Bock E, Berezin V, Poulsen FM 2006 NMR structure of the first Ig module of mouse FGFR1. Protein Sci 15:1512–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M 2007 A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol Cell 27:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL 2002 The PyMOL user’s manual. San Carlos, CA: DeLano Scientific [Google Scholar]
- Mohammadi M, Dionne CA, Li W, Li N, Spivak T, Honegger AM, Jaye M, Schlessinger J 1992 Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature 358:681–684 [DOI] [PubMed] [Google Scholar]
- Meysing AU, Kanasaki H, Bedecarrats GY, Acierno Jr JS, Conn PM, Martin KA, Seminara SB, Hall JE, Crowley Jr WF, Kaiser UB 2004 GNRHR mutations in a woman with idiopathic hypogonadotropic hypogonadism highlight the differential sensitivity of luteinizing hormone and follicle-stimulating hormone to gonadotropin-releasing hormone. J Clin Endocrinol Metab 89:3189–3198 [DOI] [PubMed] [Google Scholar]
- Lee C, Gardella TJ, Abou-Samra AB, Nussbaum SR, Segre GV, Potts Jr JT, Kronenberg HM, Jüppner H 1994 Role of the extracellular regions of the parathyroid hormone (PTH)/PTH-related peptide receptor in hormone binding. Endocrinology 135:1488–1495 [DOI] [PubMed] [Google Scholar]
- Olwin BB, Hauschka SD 1989 Cell type and tissue distribution of the fibroblast growth factor receptor. J Cell Biochem 39:443–454 [DOI] [PubMed] [Google Scholar]
- Roghani M, Mohammadi M, Schlessinger J, Moscatelli D 1996 Induction of urokinase-type plasminogen activator by fibroblast growth factor (FGF)-2 is dependent on expression of FGF receptors and does not require activation of phospholipase Cgamma1. J Biol Chem 271:31154–31159 [DOI] [PubMed] [Google Scholar]
- Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB 1995 Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry 34:14843–14851 [DOI] [PubMed] [Google Scholar]
- Raivio T, Falardeau J, Dwyer A, Quinton R, Hayes FJ, Hughes VA, Cole LW, Pearce SH, Lee H, Boepple P, Crowley Jr WF, Pitteloud N 2007 Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med 357:863–873 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG 2001 Quality control of mRNA function. Cell 104:173–176 [DOI] [PubMed] [Google Scholar]
- Newberry EP, Boudreaux JM, Towler DA 1996 The rat osteocalcin fibroblast growth factor (FGF)-responsive element: an okadaic acid-sensitive, FGF-selective transcriptional response motif. Mol Endocrinol 10:1029–1040 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Meysing A, Quinton R, Acierno Jr JS, Dwyer AA, Plummer L, Fliers E, Boepple P, Hayes F, Seminara S, Hughes VA, Ma J, Bouloux P, Mohammadi M, Crowley Jr WF 2006 Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol 254–255:60–69 [DOI] [PubMed] [Google Scholar]
- Dewailly D, Boucher A, Decanter C, Lagarde JP, Counis R, Kottler ML 2002 Spontaneous pregnancy in a patient who was homozygous for the Q106R mutation in the gonadotropin-releasing hormone receptor gene. Fertil Steril 77:1288–1291 [DOI] [PubMed] [Google Scholar]
- Pitteloud N, Boepple PA, DeCruz S, Valkenburgh SB, Crowley Jr WF, Hayes FJ 2001 The fertile eunuch variant of idiopathic hypogonadotropic hypogonadism: spontaneous reversal associated with a homozygous mutation in the gonadotropin-releasing hormone receptor. J Clin Endocrinol Metab 86:2470–2475 [DOI] [PubMed] [Google Scholar]
- González-Martinez D, Hu Y, Bouloux PM 2004 Ontogeny of GnRH and olfactory neuronal systems in man: novel insights from the investigation of inherited forms of Kallmann’s syndrome. Front Neuroendocrinol 25:108–130 [DOI] [PubMed] [Google Scholar]
- Dode C, Fouveaut C, Mortier G, Janssens S, Bertherat J, Mahoudeau J, Kottler ML, Chabrolle C, Gancel A, Francois I, Devriendt K, Wolczynski S, Pugeat M, Pineiro-Garcia A, Murat A, Bouchard P, Young J, Delpech M, Hardelin JP 2007 Novel FGFR1 sequence variants in Kallmann syndrome, and genetic evidence that the FGFR1c isoform is required in olfactory bulb and palate morphogenesis. Hum Mutat 28:97–98 [DOI] [PubMed] [Google Scholar]
- Hermanussen M, Sippell WG 1985 Heterogeneity of Kallmann’s syndrome. Clin Genet 28:106–111 [DOI] [PubMed] [Google Scholar]
- Hipkin LJ, Casson IF, Davis JC 1990 Identical twins discordant for Kallmann’s syndrome. J Med Genet 27:198–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.