Abstract
Objectives
To investigate the prevalence of metallo-β-lactamases (MBLs) and Tn6001 in carbapenem-non-susceptible Pseudomonas aeruginosa (CNSPA). The CNSPA included extensively drug-resistant P. aeruginosa (XDRPA) and non-XDRPA isolates in Taiwan.
Methods
A total of 308 CNSPA isolates collected at a medical centre from 2000 to 2005 and 26 XDRPA collected from six medical centres in different regions of Taiwan in 2003 were included. MBL genes and Tn6001 were detected by PCR. Clonal relatedness was determined by PFGE.
Results
Of the 308 CNSPA isolates, 30 (10%) were XDRPA, including 27 (9%) colistin-only-susceptible (COS) and 3 (1%) colistin-only-intermediate (COI) P. aeruginosa. blaVIM-3 was found in 16 (53%) isolates of the XDRPA (n = 30), whereas only 72 (26%) of the non-XDRPA (n = 278) carried the gene. In450 was higher in COS P. aeruginosa (12/27; 44%) than in non-XDRPA isolates (53/278; 19%). Tn6001 was highest in COS P. aeruginosa (11/27; 41%), followed by COI P. aeruginosa (1/3; 33%), and lowest in non-XDRPA (46/278; 17%). Of 26 XDRPA from six medical centres, higher prevalences of blaVIM-3 (16/26; 62%), In450 (16/26; 62%) and Tn6001 (12/26; 46%) were found. Genotyping by PFGE revealed 60 pulsotypes. Hybridization of a blaVIM-3-specific probe following PFGE suggested that the mobile element Tn6001 might have transferred horizontally.
Conclusions
Tn6001 and In450 play an important role in the dissemination of CNSPA and XDRPA. The prevalence of these genetic constituents was higher in XDRPA than in non-XDRPA isolates, suggesting that the mobile element Tn6001 might have transferred horizontally.
Keywords: carbapenem-non-susceptible Pseudomonas aeruginosa (CNSPA), extensively drug-resistant P. aeruginosa (XDRPA), metallo-β-lactamases
Introduction
Carbapenem resistance in Pseudomonas aeruginosa has become more common in many countries and is correlated with multidrug resistance (MDR). It is known that carbapenem resistance in P. aeruginosa is usually associated with decreased permeability, efflux pump up-regulation and/or the production of metallo-β-lactamases (MBLs).1–3 Different types of MBL have been identified, including IMP, VIM, GIM, SIM and SPM.2,3 The VIM-3 MBL was first reported in Taiwan in 2001 and was later found on gene cassettes of the class 1 integron.1,4,5,7 Recently, we reported a transposon, Tn6001, which contains a blaVIM-3-harbouring integron In450, in a clinical isolate of extensively drug-resistant (XDR) P. aeruginosa (XDRPA).6 However, the overall prevalence of In450 and Tn6001 in VIM-3-carrying carbapenem-non-susceptible P. aeruginosa (CNSPA) is unknown.
Materials and methods
Bacterial isolates
The 308 non-duplicate CNSPA isolates, defined as isolates resistant to imipenem or meropenem, were collected from 2000 to 2005 at the National Taiwan University Hospital (NTUH), a 2500 bed medical centre in northern Taiwan. If more than one isolate of CNSPA was recovered from the same patient, only the first isolate was included. One of the 308 isolates, NTUH-PA450, was previously reported to carry a transposon Tn6001.6 Colistin-only-susceptible (COS) P. aeruginosa was defined as resistance to all antibiotics except colistin (MICs of colistin, ≤2 mg/L) and colistin-only-intermediate (COI) P. aeruginosa was defined as resistance to all antibiotics with intermediate resistance to colistin (MIC of colistin, 4 mg/L). The 26 XDRPA isolates were collected in 2003 from six medical centres located in different regions of Taiwan.5
Antimicrobial susceptibility testing
MICs of nine antimicrobial agents were determined using an agar dilution method according to CLSI guidelines.8 The following antimicrobial agents were provided by their manufacturers for use in this study: piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, amikacin, ciprofloxacin, imipenem, meropenem and colistin. XDRPA was defined as P. aeruginosa isolates resistant to antimicrobial agents piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, amikacin, ciprofloxacin, imipenem and meropenem, and susceptible or intermediately resistant to colistin.
Detection of MBL genes by PCR amplification and sequencing
PCR amplification of MBL genes, including blaIMP-1-9, blaVIM-1 and blaVIM-2 genes, was performed according to previously reported protocols [Table S1, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)].4,9 The types of MBL were confirmed by sequence.
Detection of Tn6001 and blaVIM-2-harbouring integron cassette by multiplex PCR
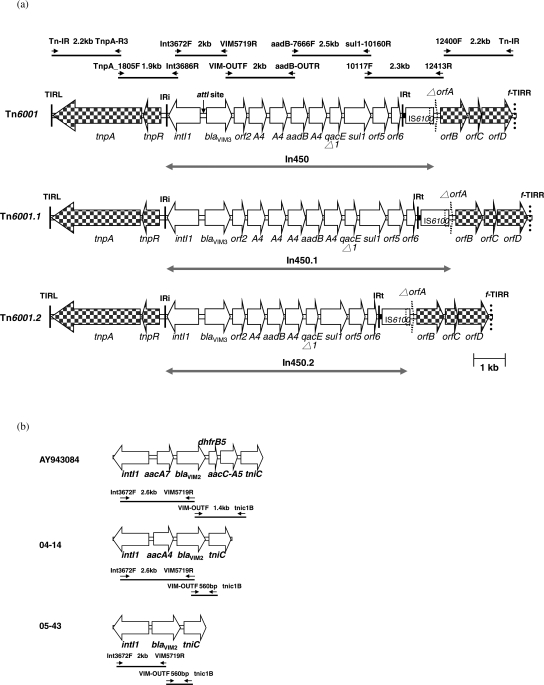
The dissemination of Tn6001 in clinical isolates was detected by multiplex PCR (the primers are listed in Table S1) targeting the junctions of the In450 and Tn6001 backbone structure (GenBank accession number EF138817). The amplicon sizes and locations are shown in Figure 1(a). The blaVIM-2-harbouring integron cassette was detected by two sets of PCR (Figure 1b) and confirmed by sequencing.
Figure 1.
(a) Genetic organization of transposons Tn6001, Tn6001.1 and Tn6001.2 in P. aeruginosa isolates. Genes are shown as arrows, with their orientation of transcription indicated by the arrowheads. A4, aacA4 gene. The sizes and location of the multiplex PCR amplicons are displayed in Tn6001. IRi, left-hand integron inverted repeat (25 bp); IRt, right-hand integron inverted repeat (25 bp); TIRL: left-hand transposon inverted repeat; f-TIRR: potential right-hand transposon inverted repeat. (b) Three different blaVIM-2-harbouring integron cassettes were amplified by two primer sets.
Genotyping by PFGE
Genotypes of 109 of the 308 CNSPA isolates (30 isolates of XDRPA, including 27 COS P. aeruginosa and 3 COI P. aeruginosa, and 79 blaVIM-2 and blaVIM-3-producing non-XDR CNSPA) from NTUH and 26 XDRPA from six medical centres were determined by PFGE. PFGE analysis was carried out as described previously.1
Southern blot hybridization
The SpeI-digested chromosome fragments of four clinical isolates were analysed by PFGE. The PFGE gel was then subjected to Southern blotting and hybridized with a DIG (digoxigenin)-labelled blaVIM-3-specific probe. The hybridization assay was performed as described previously.10
Results
Antimicrobial susceptibility of CNSPA isolates from NTUH
A high percentage of cross-resistance to ceftazidime (66%–89%) and cefepime (63%–90%) was found in the CNSPA isolates [Table S2, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)]. The rate of resistance to monobactam (aztreonam) was also high (52%–90%). It is noted that resistance to ciprofloxacin increased quickly from 2002 to 2004 (76%–93%) while resistance to amikacin ranged from 24% to 56% and there was no resistance to colistin noted in these isolates. Difficulty in treating COS and COI P. aeruginosa was found from 2003 to 2005, but the percentages (11%–19% and 0%–3%, respectively) were low. Neither a significant increase in the prevalence of COS or COI P. aeruginosa nor shifts of resistance to categories of medically useful antibiotics were noted.
Distribution of MBL genes, integron cassette and Tn6001
blaVIM-2 and blaVIM-3 genes were detected in 6 (1.9%) and 88 (28.6%) of the 308 CNSPA from NTUH. The prevalence of the blaVIM-3 gene ranged from 13% to 32% in non-XDRPA isolates collected during 2000–05 except in 2002. The prevalence of the blaVIM-3 gene was exceptionally high (52%) in 2002 (Table 1). COS and COI P. aeruginosa were first isolated in 2003 and persisted in 2004 and 2005. Furthermore, significant increases in VIM-3 (40% to 67%), In450 (40% to 56%) and Tn6001 (40% to 50%) from 2003 to 2005 were also noted among COS isolates. Six CNSPA carried a blaVIM-2-harbouring integron cassette (Figure 1b). Of these integrons, four were identical to AY943084, whereas two (04-14 and 05-43) contained different types of blaVIM-2-harbouring integron.
Table 1.
Prevalence of VIM-3, In450 and Tn6001 in 308 CNSPA isolates at NTUH from 2000 to 2005
| No. (%) of isolates |
||||
|---|---|---|---|---|
| Year (CNSPA isolates) | Isolates (no.) | VIM-3 | In450 | Tn6001 |
| 2000 (n = 60) | non-XDRa (n = 60) | 10 (17) | 8 (13) | 5 (8) |
| 2001 (n = 41) | non-XDRa (n = 41) | 13 (32) | 9 (23) | 9 (23) |
| 2002 (n = 21) | non-XDRa (n = 21) | 11 (52) | 5 (24) | 3 (14) |
| 2003 (n = 63) | non-XDRa (n = 50) | 16 (32) | 12 (24)d | 12 (24)d |
| XDR | ||||
| COSb (n = 12) | 4 (40) | 4 (40) | 4 (40) | |
| COIc (n = 1) | 1 (100) | 0 (0) | 0 (0) | |
| 2004 (n = 70) | non-XDRa (n = 59) | 16 (27) | 14 (24) | 13 (22) |
| XDR | ||||
| COSb (n = 9) | 5 (56) | 5 (56) | 4 (44) | |
| COIc (n = 2) | 2 (100) | 1 (50) | 1 (50) | |
| 2005 (n = 53) | non-XDRa (n = 47) | 6 (13) | 5 (11)e | 4 (9)f |
| XDR | ||||
| COSb (n = 6) | 4 (67) | 3 (50) | 3 (50) | |
| Total (n = 308) | 88 (29) | 66 (21) | 58 (19) | |
aNon-XDR, non-COS and non-COI.
bCOS, colistin-only-susceptible, resistant to all antibiotics except colistin.
cCOI, colistin-only-intermediate, resistant to all antibiotics except intermediate to colistin.
dIn450.1 (n = 1) and Tn6001.1 (n = 1) were found in the same clinical isolate.
eIn450.1 (n = 1) and In450.2 (n = 1) were found in two clinical isolates.
fTn6001.1 (n = 1) and Tn6001.2 (n = 1) were found in two clinical isolates.
blaVIM-2 and blaVIM-3 genes were detected in 4 (15.4%) and 16 (61.5%) of the 26 XDRPA isolates collected from six medical centres in Taiwan. Twenty-five (96%) were COS and only one (4%) was COI [Table S3, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)]. Neither In450 nor Tn6001 was found in the COI isolate. High percentages of In450 and Tn6001 [100% (11/11) and 64% (7/11), respectively] were detected in COS isolates from hospital N1. Similar high percentages (4/10, 40%) were also noted in hospital N2. These findings suggest that the high prevalences of In450 and Tn6001 are associated with MDR P. aeruginosa in Taiwan.
PFGE analysis
PFGE analysis of the 135 P. aeruginosa isolates revealed 60 different pulsotypes. There were 51 different pulsotypes found in 109 CNSPA isolates from NTUH and 16 from 26 XDRPA from six medical centres (seven pulsotypes were distributed among isolates from both NTUH and six medical centres). Four of 15 pulsotypes from the 30 XDRPA isolates and 18 of 49 pulsotypes from 79 non-XDR CNSPA from NTUH and 5 of 16 pulsotypes of the 26 XDRPA from six medical centres had multiple isolates (≥2 isolates).
Structures of two variants of In450
Two variant structures of In450 were found in three non-XDRPA isolates. One integron cassette, designated In450.1, contained the following seven genes: blaVIM-3, orf2, three copies of aacA4, aadB and a copy of aacA4. The other integron cassette, designated In450.2, contained the following five genes: blaVIM-3, orf2, aacA4, aadB and a copy of aacA4 (Figure 1a). Both In450.1 and In450.2 were harboured in the Tn6001 backbone structure, designated Tn6001.1 and Tn6001.2, respectively (Figure 1a).
Mobile DNA Tn6001
Although it was difficult to demonstrate that gene transfer had occurred in the MDR isolate, the results from two closely related clinical isolates from patient A could support the transfer of Tn6001. In patient A, an imipenem-susceptible isolate (MIC, 0.12 mg/L) recovered 3 months before the isolation of the carbapenem-resistant isolate (MIC, >32 mg/L) was used for comparison. PFGE/hybridization analysis revealed only a one band difference between these two isolates [Figure S1a, lanes 1 and 2, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)]. This DNA fragment contained the blaVIM-3 gene, which was associated with imipenem resistance (Figure S1b, lanes 1 and 2). Furthermore, this imipenem-resistant isolate showed the presence of Tn6001 by multiplex PCR. These findings indicate that the second clinical isolate developed its imipenem resistance phenotype through DNA acquisition and was probably transferred by transposon.
Discussion
Consistent with previous studies, the blaVIM-3 gene was the most common MBL gene in MBL-producing P. aeruginosa isolates in Taiwan and was usually harboured by class 1 integrons.1,4,5 The prevalence of blaVIM-3 in COI or COS P. aeruginosa was higher than that in non-XDRPA isolates. Higher prevalences of In450 and Tn6001 were also detected in COI and COS isolates, both from NTUH and from the six hospitals located in various areas in Taiwan (Tables 1 and S3). These findings indicate that In450 and Tn6001 may account for the transfer of drug resistance genes. However, other mechanisms may also play a role. Since as high as 52% (11/21) of CNSPA contained blaVIM-3 in 2002, but only a disproportionately low percentage (24%, 5/21) of them carried In450 and 14% (3/21) carried Tn6001. These results suggest that unknown mechanisms other than In450 and Tn6001 may be responsible for the spread of blaVIM-3.
Tn6001 was found in different pulsotypes of P. aeruginosa isolates. Although it is difficult to demonstrate the gene transfer property of Tn6001 in the laboratory, results from this study still provide two pieces of evidence supporting the possibility that Tn6001 is a mobile DNA element and thus disseminated among different isolates. First, the finding that there was only one band difference identified by PFGE and hybridization with a blaVIM-3-specific probe may suggest that Tn6001 could be acquired by a susceptible isolate and thus become resistant (Figure S1). Second, the heterogeneity of pulsotypes for Tn6001-containing P. aeruginosa isolates revealed polyclonal relatedness, suggesting that horizontal transfer may have occurred. However, several isolates recovered from NTUH (2000–05) and six medical centres (2003) exhibited identical pulsotypes and contained Tn6001 suggesting the presence of intra-hospital and inter-hospital clonal dissemination.
In addition, Yan et al.5 described that after digestion with EcoRI in MDR P. aeruginosa isolates, there were three different DNA fragments (6.2, 6.6 and 8.8 kb) found with a blaVIM-3-specific probe. These isolates all contained Tn6001 and one EcoRI site (position 5942 of GenBank accession number EF138817), which was located at orf2 downstream of blaVIM-3. This finding suggested that the three different patterns of EcoRI-digested DNA fragments found by Yan et al.5 were due to different locations of Tn6001 integration. These inferences suggest that the distribution of Tn6001 might be associated with horizontal transfer.
XDRPA formation may involve antibiotic resistance mechanisms other than Tn6001 dissemination. A high percentage of aztreonam resistance could be correlated with blaOXA-10 and blaOXA-17 spreading,5 whereas fluoroquinolone resistance has been linked to amino acid changes in topoisomerase types II and IV. We also found that mutations in GyrA (Thr-83→Ile) and ParC (Ser-87→Leu) in the quinolone resistance-determining region may be associated with quinolone resistance (data not shown). Thus, further characterization of XDRPA isolates is a priority and an important issue in Taiwan.
Funding
This study was supported by internal funding.
Transparency declarations
None to declare.
Supplementary data
Supplementary Material
Acknowledgements
We would like to thank the following individuals for providing XDRPA isolates: Jainn-Ming Shyr at Taichung Veterans General Hospital, Taichung, Taiwan; Yin-Ching Chuang and Yi-Chueh Yang at Chi-Mei Medical Center, Tainan, Taiwan; Li-Shin Wang at Buddhist Tzu-Chi General Hospital, Hualien, Taiwan; and Chien-Fang Peng at Kaohsiung Medical University, Kaohsiung, Taiwan.
References
- 1.Hsueh PR, Tseng SP, Teng LJ, et al. Pan-drug-resistant Pseudomonas aeruginosa causing nosocomial infection at a university hospital in Taiwan. Clin Microbiol Infect. 2005;11:670–3. doi: 10.1111/j.1469-0691.2005.01196.x. [DOI] [PubMed] [Google Scholar]
- 2.Walsh TR, Toleman MA, Poirel L, et al. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–25. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan JJ, Hsueh PR, Ko WC, et al. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother. 2001;45:2224–8. doi: 10.1128/AAC.45.8.2224-2228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan JJ, Hsueh PR, Lu JJ, et al. Characterization of acquired β-lactamases and their genetic support in multidrug-resistant Pseudomonas aeruginosa isolates in Taiwan: the prevalence of unusual integrons. J Antimicrob Chemother. 2006;58:530–6. doi: 10.1093/jac/dkl266. [DOI] [PubMed] [Google Scholar]
- 6.Tseng SP, Hsueh PR, Tsai JC, et al. Tn6001, a transposon-like element containing the blaVIM-3-harbouring integron In450. Antimicrob Agents Chemother. 2007;51:4187–90. doi: 10.1128/AAC.00542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MF, Peng CF, Hsu HJ, et al. Molecular characterization of the metallo-β-lactamase genes in imipenem-resistant Gram-negative bacteria from a university hospital in southern Taiwan. Int J Antimicrob Agents. 2008;32:475–80. doi: 10.1016/j.ijantimicag.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Eighteenth Informational Supplement M100-S18. Wayne, PA, USA: CLSI; 2008. [Google Scholar]
- 9.Toleman MA, Biedenbach D, Bennett D, et al. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J Antimicrob Chemother. 2003;52:583–90. doi: 10.1093/jac/dkg410. [DOI] [PubMed] [Google Scholar]
- 10.Tsai JC, Hsueh PR, Chen HJ, et al. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob Agents Chemother. 2005;49:4347–50. doi: 10.1128/AAC.49.10.4347-4350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.