Abstract
Context
Recent evidence from linkage analyses and follow-up candidate gene studies supports the involvement of SLC1A1, which encodes the neuronal glutamate transporter, in the development of obsessive-compulsive disorder (OCD).
Objectives
To determine the role of genetic variation of SLC1A1 in OCD in a large case-control study and to better understand how SLC1A1 variation affects functionality.
Design
A case-control study.
Setting
Publicly accessible SLC1A1 expression and genotype data.
Patients
Three hundred twenty-five OCD probands and 662 ethnically and sex-matched controls.
Interventions
Probands were assessed with the Structured Clinical Interview for DSM-IV, the Yale-Brown Obsessive Compulsive Scale, and the Saving Inventory–Revised. Six single-nucleotide polymorphisms (SNPs) were genotyped. Multiple testing corrections for single-marker and haplotype analyses were performed by permutation.
Results
Gene expression of SLC1A1 is heritable in lymphoblastoid cell lines. We identified 3 SNPs in or near SLC1A1 that correlated with gene expression levels, 1 of which had previously been associated with OCD. Two of these SNPs also predicted expression levels in human brain tissue, and 1 SNP was further functional in reporter gene studies. Two haplotypes at 3 SNPs, rs3087879, rs301430, and rs7858819, were significantly associated with OCD after multiple-testing correction and contained 2 SNPs associated with expression levels. In addition, another SNP correlating with SLC1A1 gene expression, rs3933331, was associated with an OCD-hoarding subphenotype as assessed by 2 independent, validated scales.
Conclusions
Our case-control data corroborate previous smaller family-based studies that indicated that SLC1A1 is a susceptibility locus for OCD. The expression and genotype database–mining approach we used provides a potentially useful complementary approach to strengthen future candidate gene studies in neuropsychiatric and other disorders.
Dysfunctional glutamatergic neurotransmission in the pathogenesis of obsessive-compulsive disorder (OCD) was postulated on the basis of magnetic resonance imaging studies.1,2 This hypothesis was subsequently supported by animal models based on abnormal glutamate neurotransmission that contributed to OCD spectrum–like phenotypes,3,4 case reports of beneficial therapeutic effects of riluzole (an antiglutamatergic agent) on treatment-resistant OCD,5 and elevated glutamate levels in the cerebrospinal fluid of OCD probands.6 From a genetic perspective, investigation of glutamatergic genes in OCD therefore represents a plausible biological candidate approach given this disorder's high heritability and risk to relatives.7,8 Indeed, studies have shown positive associations of glutamate receptor genes with OCD.9,10
More recently, a glutamate transporter gene, SLC1A1 (GenBank NM_004170), has been proposed to be involved with OCD in particular, as this gene is also a chromosome 9p24 positional candidate from linkage analyses.11,12 This transporter (also known as EAAC1 and EAAT3) is widely expressed in neurons and also involved in cysteine transport.13,14 While an initial mutation screen of SLC1A1 and family association study did not detect biased transmission in OCD,15 3 subsequent studies with larger numbers of tested polymorphisms and somewhat larger proband samples yielded positive associations in both single-marker and haplotypic tests, though neither specific single-nucleotide polymorphisms (SNPs) nor haplotypes closely corresponded.16-18 These 3 family-based studies motivated us to analyze several SLC1A1 polymorphisms in our large and phenotypically well-characterized collection of OCD probands using a case-control study design. Case-control studies, while susceptible to population stratification, provide greater statistical power than family-based designs. We focused on SLC1A1 SNPs that were significant in at least 1 previous study and we included additional SNPs that predicted gene expression levels as determined by analyzing deposited expression and genotype data from nonpatient sources. We present evidence for a susceptibility haplotype spanning large portions of SLC1A1 and also for a SNP located 100 kilobases (kb) upstream of SLC1A1 that was associated with an OCD subphenotype, compulsive hoarding, which has some genetic linkage and clinical differences from the more common OCD phenotype.19-21
METHODS
SLC1A1 GENE EXPRESSION ANALYSES
For heritability calculations, we extracted lymphoblastoid cell line SLC1A1 gene expression values (probe No. 213664_at) from the Gene Expression Omnibus data set GSE1485.22 All individuals had a “present” call for this probe; technical replicates were averaged and then log2-transformed. Family structures for the 14 three-generational families represented in this data set were obtained from the Centre d'Etude du Polymorphisme Humain database (http://www.cephb.fr/en/cephdb/browser.php). Pedigree and expression data were then analyzed with the Sequential Oligogenic Linkage Analysis Routines software package by performing a standard quantitative genetic analysis.23 The residual kurtosis of 0.125 was within the normal range.
For expression quantitative trait locus (eQTL) analyses, we retrieved lymphoblastoid cell line SLC1A1 expression values for the 60 HapMap Centre d'Etude du Polymorphisme Humain founders with northern and western European ancestry from 2 independent platforms: the Gene Expression Omnibus data set GSE5859 (Affymetrix platform, probe No. 213664_at; replicates were averaged and transformed as above)24 and from the Gene Expression Variation Web site hosted by the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/humgen/genevar; Illumina platform, probe No. GI_31543625-S).25 Genotypes for all 90 individuals from the Centre d'Etude du Polymorphisme Humain HapMap data were downloaded from the HapMap Web site (http://www.hapmap.org, release 22) for the entire SLC1A1 region plus 1 megabase (Mb) flanking both sides (corresponding to position 3 480 444 to 5 577 469 on chromosome 9 in the University of California–Santa Cruz Genome Browser Human, March 2006 assembly). Markers inconsistent with mendelian inheritance patterns, with minor allele frequencies of less than 1%, or with more than 10% missing genotypes were removed, resulting in a total of 2467 polymorphisms that were analyzed. These SNPs were then tested for quantitative association with SLC1A1 expression using PLINK,26 Microsoft Excel (Microsoft, Redmond, Washington), and Prism 4 (GraphPad Software, San Diego, California).
REAL-TIME REVERSE TRANSCRIPTASE–POLYMERASE CHAIN REACTION QUANTITATION OF SLC1A1 IN POSTMORTEM BRAIN TISSUE
We obtained RNA and genomic DNA from 105 postmortem dorsolateral prefrontal cortex samples through the Stanley Medical Research Institute, Chevy Chase, Maryland. This collection is composed of 3 diagnostic groups (35 normal, 35 bipolar, and 35 schizophrenic donors). With the exception of 2, all samples were from white donors. Detailed demographics of this sample are available at http://www.stanleyresearch.org. Six of these 105 samples did not provide sufficient RNA for quantitation, and the 2 non-white samples were quantitated but not analyzed, resulting in a total of 97 samples, which are presented herein. All samples were successfully genotyped for rs3933331, rs7858819, and rs301430.
We reverse-transcribed 1 μg of total RNA with oligo-dT primers using the Transcriptor First Strand cDNA Synthesis kit according to the manufacturer's instructions (Roche Applied Science, Indianapolis, Indiana). Next, we amplified a total of 5 ng of reverse-transcribed RNA in triplicate reactions with 1 assay for SLC1A1 and 1 endogenous control assay for PGK1. The SLC1A1 assay was designed with the Universal Probe Library Assay Design Center, version 2.4 (Roche Applied Science) using primers GCT GTC GAC TGG CTC CTG and GGA GAG CTT TTC CAC AAT GC (Operon Biotechnologies, Huntsville, Alabama) and Universal Probe Library probe No. 9 (Roche Applied Science). The PGK1 assay was predesigned and purchased directly from the vendor (Universal Probe Library Human PGK1 Gene Assay, Roche Applied Science). Both assays span introns and do not contain known SNPs in primers or probes. Amplification was carried out in a 10-μL total reaction volume with 500 nM of each primer and 100 nM of fluorescent probe using PerfeCta qPCR FastMix (Quanta BioSciences, Gaithersburg, Maryland). Thermocycling consisted of 2 minutes at 45°C for uracil-N-glycosylase incubation, 30 seconds at 95°C for hot-start activation, and 40 amplification cycles of 2 seconds at 95°C and 20 seconds at 60°C followed by fluorescence acquisition in a LightCycler 480 instrument (Roche Applied Science). Crossing-point determination by second derivative maximum was carried out with LightCycler 480 software, release 1.3. Relative abundances of SLC1A1 messenger RNA were then calculated using the efficiency-corrected comparative threshold method and log2-transformed.27 Efficiency calculations were performed with 2-fold serially diluted pooled complementary DNA (range, 30-0.47 ng per reaction). Amplicon size and assay specificity were verified by agarose electrophoresis. No-template controls did not display detectable amplification (data not shown). All experiments were done after the specimen code was broken and they were thus unblinded. The entire data set has been uploaded to the Stanley Medical Research Institute data bank (http://www.stanleyresearch.org/brain).
REPORTER GENE EXPERIMENTS
We performed luciferase assay experiments for 3 SNPs within or near SLC1A1 as recently described.28 Briefly, genomic DNA was amplified with primers flanking rs3933331 (TTC TGC CCA AGA CAA TCA CA and TCT GGG TTT AGA CTG CCA CA), rs7858819 (TGC AAA TGG TTA TGG GAC AA and TCA AAT GGT CCT TGG CTA CC), and rs301430 (CAT GTC TTC AGG CAG GGA CT and CAC TGC GAC ATT CTT GGT GT; all primers were obtained through Operon Biotechnologies). Amplicons were then TA-cloned into pCR2.1 (Invitrogen, Carlsbad, California) and subcloned into the pGL3 reporter vector (Promega, Madison, Wisconsin). Equal orientation and sequence specificity, including absence of additional SNPs, were confirmed through sequencing of pGL constructs at the National Institute of Neurological Disorders and Stroke intramural DNA sequencing core facility. Allele-specific luminescence was determined with the Dual Luciferase Reporter Assay System (Promega). For each of the 6 reporter constructs, 5 independent transfections were performed in both human neuroblastoma (SH-SY5Y) and rat pheochromocytoma (PC12) cell lines grown under standard conditions. For each SNP, data were normalized to the geometric mean of 1 allele and tested against the other allele with a 2-sided, unpaired t test using Prism 4 (GraphPad Software).
SUBJECTS
Adult OCD probands (325 white subjects) were recruited through an ongoing outpatient OCD program at the National Institute of Mental Health, Bethesda, Maryland. All probands were aged at least 18 years at the time of inclusion (mean age at time of interview, 38 years; standard deviation [SD], 16 years) and gave written informed consent. The diagnosis was made on the basis of the Structured Clinical Interview for DSM-IV. Exclusion criteria were active psychosis or schizophrenia, severe mental retardation that would prohibit an evaluation of OCD, and OCD symptoms that occurred exclusively in the context of major depression.
The sample consisted of 186 women (57%) and 139 men (43%), with a mean age at OCD onset of 14 years (SD, 9.6 years), which was similar to that in the SLC1A1 OCD study by Arnold et al (14.4 years)16 and higher than those in the studies by Dickel et al (8 years)17 and Stewart et al (7.9 years).18 As reported previously in our sample,29 mood (81%) and anxiety (53%) disorders were most frequently represented among comorbid disorders, with alcohol abuse disorder (24%), substance abuse disorders (14%), and tics (6%) less common. Hoarding, as we will define, was present in 27% of this patient sample. Total mean Yale-Brown Obsessive Compulsive Severity Scale (Y-BOCS) lifetime severity scale score was 29.6 (SD, 15.4), which is approximately equally contributed to by obsession and compulsion scores.
Our control samples (662 white subjects [46% male and 54% female]) originated from 3 independent sources: (1) undergraduate students (270 self-declared healthy white students) from a large Southeastern university who participated in a separate study of genes and personality in return for partial course credit; (2) Coriell Cell Repository, Camden, New Jersey (200 self-declared healthy white Americans); and (3) European Collection of Cell Cultures (Sigma-Aldrich, St Louis, Missouri; 192 apparently healthy white blood donors in the United Kingdom). The male to female ratio did not significantly differ between cases and controls (; P>.05). We cannot rule out the occurrence of OCD within the second and third control groups, as self-report and standard scales were used for evaluation of only the first group. Genotypic and allelic frequencies did not significantly differ between these 3 control groups (data not shown). All studies were conducted under protocols approved by the institutional review board at the National Institute of Mental Health Division of Intramural Research Programs and by the Human Subjects Committee at Florida State University. Additional details on proband and control samples have been described previously.29,30
GENOTYPING AND STATISTICAL GENETICS
Genomic DNA was extracted from whole blood obtained through venipuncture or from saliva samples (Oragene discs; DNA Genotek, Ottawa, Ontario, Canada). All SNPs were genotyped with 5′ exonuclease assays (Applied Biosystems Taq-Man predesigned or custom assays; Applied Biosystems, Foster City, California) (assay identification numbers and primer/probe sequences are available from the corresponding author) under standard conditions described previously.31 The overall genotyping completion rate exceeded 98% for each assay; the per-individual genotyping completion rate was at least 5 of 6 SNPs. None of the SNPs deviated from Hardy-Weinberg equilibrium as determined by contingency-table statistics (nominal P>.05; data not shown). Duplicate samples (at least 10% of all samples, randomly chosen, for each of the 6 SNPs) and no-template controls consistently yielded expected results.
Genotype data from probands and controls were analyzed for single-locus and haplotypic associations with the software packages PLINK26 and WHAP.32 Conditional haplotype analyses were carried out as recently described after detection of a significant omnibus association.28 Calculation of intermarker linkage disequilibrium and correction for multiple testing with 100 000 permutations for the 6 individual markers as well as for the 3-locus haplotype was conducted using HaploView, version 4.0.33
Using the Genetic Power Calculator,34 we determined that our sample had 78% power at P=.05 with unscreened controls (risk allele frequency=0.3; disease prevalence=0.025; genotypic relative risk Aa=1.4 and AA=1.8; marker allele frequency=0.3 at D′=0.9 with risk variant). When we selected screened controls and used otherwise identical parameters, power rose only slightly, to 80%. We conclude from these calculations that our partially unscreened control sample did not significantly affect our study results.
SUBPHENOTYPE ANALYSES
We examined 2 subphenotypes in this article: (1) we measured lifetime OCD symptom severity using Y-BOCS35; and (2) we further considered hoarding as a second subphenotype based on 2 separate, validated measures of the hoarding construct. We focused on hoarding as a subphenotype, since its familiality is stronger than other OCD subphenotypes,36 which is further reflected by 2 recent significant linkage analyses.19,20 The Y-BOCS symptom checklist was used to derive factor scores for the hoarding symptom dimension using principal component analysis and the Varimax procedure, as previously reported.37 The factor analysis generated a factor score for each subject representing the correlation of their symptom profile with the hoarding factor, factor IV, as reported previously.37 Because the hoarding factor was essentially independent (factor loading=0.85) from the other 3 factors identified in our sample (factor loading on the hoarding factor by other factors, 0.01-0.16) and likewise by others,36-38 the hoarding factor scores were standardized, with a mean of 0 and an SD of 1. Subjects were dichotomized on the basis of scores greater than 0 or less than 0 on the hoarding factor. Details of this procedure have been presented elsewhere.37,38 We also assessed the severity of hoarding symptoms with the Saving Inventory–Revised (SIR).39 This measure is a validated tool for examining the magnitude of hoarding symptoms and includes facets of this phenomenon not considered in the Y-BOCS symptom checklist.40 Both of these measures of hoarding have been previously characterized and evaluated in our clinical OCD sample.37,40
Statistical differences in these subphenotypes according to genotype were ascertained using linear regression and SPSS, version 14.0 (SPSS Inc, Chicago, Illinois), for Microsoft Windows XP (Microsoft). All tests were performed with 2-sided P<.05. Given missing data, the populations for each of the analyses varied slightly (SIR, n=198; hoarding factor, n=216).
RESULTS
SLC1A1 GENE EXPRESSION ANALYSES IN LYMPHOBLASTOID CELL LINES
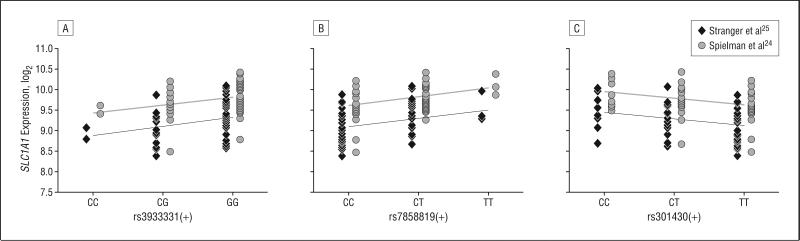
Using publicly available gene expression data, we observed that SLC1A1 is robustly expressed in lymphoblastoid cell lines. This prompted us to investigate whether SLC1A1 expression constitutes a heritable trait. By analyzing 194 lymphoblastoid cell lines from 14 three-generational families,22 we determined an additive (narrow-sense or h2) heritability of 0.23 (P<.001) for SLC1A1 gene expression, with no significant sex effects when sex was included as a covariate. We then sought to identify eQTLs for SLC1A1 acting in cis within 1 Mb of the SLC1A1 genomic locus by performing a linear regression analysis of expression data on genotypes recoded as 0, 1, or 2 on the basis of minor allele occurrences. Of 2467 markers tested, 3 SNPs that were not in complete linkage disequilibrium scored a nominal P≤.02 and had the same direction of effect in both data sets (ie, positive or negative regression coefficient; r2 value, 9%-14%) (Figure 1). One of these SNPs, rs3933331, is located about 100 kb upstream of the SLC1A1 transcription initiation site, while the 2 other eQTL SNPs, rs7858819 and rs301430, are in the 3′ end of SLC1A1. Remarkably, rs301430 has recently been associated with early-onset OCD in a collection of 71 trios both individually and as part of a haplotype,17 and the haplotypic association has been replicated subsequently.18
Figure 1.
Regression of SLC1A1 expression on single-nucleotide polymorphisms using publicly accessible lymphoblastoid cell line data. Depicted here are 3 cis-acting expression quantitative trait loci that were nominally significant and had the same direction of effect in 2 independent data sets.24,25 The single-nucleotide polymorphism rs301430 has previously been shown to be associated with obsessive-compulsive disorder both as a single marker and as part of a haplotype.17,18 A, Stranger et al25 and Spielman et al24 data, P=.02. B, Stranger et al25 data, P=.02; Spielman et al24 data, P=.005. C, Stranger et al25 data, P=.02; Spielman et al24 data, P=.01.
SLC1A1 GENE EXPRESSION ANALYSES IN HUMAN POSTMORTEM BRAIN TISSUE
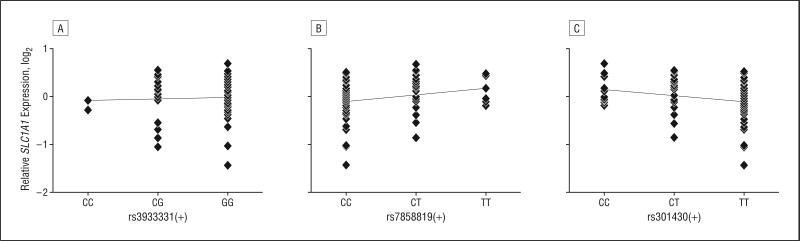
As a next step, we analyzed whether the 3 eQTLs we identified were also associated with SLC1A1 gene expression in neuronal tissue. To this end, we determined genotypes and relative SLC1A1 messenger RNA levels by quantitative polymerase chain reaction in 97 postmortem brain samples from normal, bipolar, and schizophrenic donors, all derived from the dorsolateral prefrontal cortex. There were no significant differences in SLC1A1 expression levels among the 3 diagnosis groups as calculated by analysis of variance (data not shown). We then analyzed all 3 groups jointly for association with eQTLs to have increased power. As shown in Figure 2, rs7858819 and rs301430 were nominally significantly associated with SLC1A1 messenger RNA levels in human brain tissue (P=.02 each). Moreover, the direction of effect (increased expression with increasing number of T alleles at rs7858819 and decreased expression at rs301430) was identical to the data from lymphoblastoid cell line data (Figure 1 and Figure 2).
Figure 2.
Regression of SLC1A1 expression on single-nucleotide polymorphisms in brain tissue. A total of 97 samples of human postmortem dorsolateral prefrontal cortex were genotyped and quantitated for SLC1A1 messenger RNA levels by real-time reverse-transcriptase–polymerase chain reaction. Two of the 3 expression quantitative trait loci single-nucleotide polymorphisms that were analyzed were nominally significant and had the same direction of effect in brain tissue as in lymphoblastoid cell lines (Figure 1). A, nonsignificant. B, P=.02. C, P=.02.
IN VITRO ANALYSES OF SLC1A1 EXPRESSION–ASSOCIATED SNPs WITH REPORTER GENE CONSTRUCTS
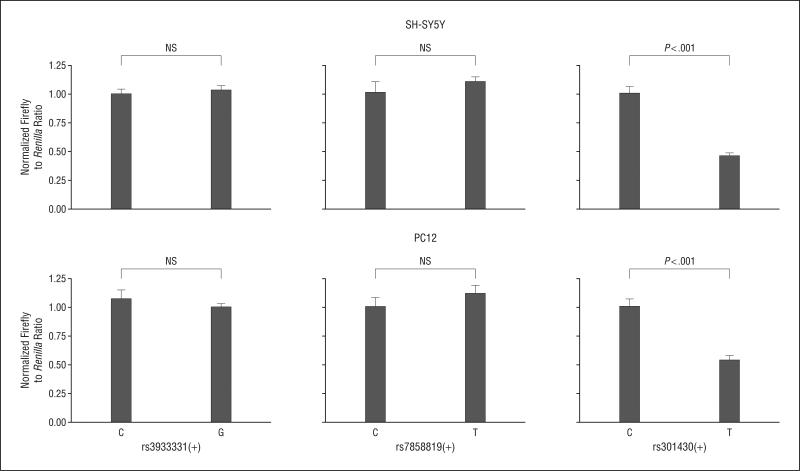
Following the replication of 2 of 3 eQTLs in neuronal tissue, we sought to directly assess functionality of these SNPs in vitro through the use of luciferase reporter gene assays. We generated constructs with both alleles at each of the 3 SNPs, rs3933331, rs7858819, and rs301430, and used human neuroblastoma (SH-SY5Y) and rat pheochromocytoma (PC12) cell lines for transfection. As depicted in Figure 3, the 2 alleles at rs301430 had a highly significant differential effect on reporter gene expression in both cell lines (P<.001 for both). In line with the T allele–associated decrease in SLC1A1 expression in lymphoblastoid cell lines and neuronal tissue, the rs301430 construct with the T allele showed about 50% decreased expression compared with the C allele (Figure 3).
Figure 3.
Reporter gene analyses of putative expression quantitative trait loci. The 3 single-nucleotide polymorphisms identified in this study to be associated with SLC1A1 messenger RNA levels were tested in luciferase constructs. Six constructs were transfected 5 times in both human neuroblastoma (SH-SY5Y) and rat pheochromocytoma (PC12) cell lines. Error bars represent standard error of mean. One single-nucleotide polymorphism, rs301430, showed allele-specific differential luciferase expression that was highly significant in neuroblastoma (t8=8.6, P<.001) and in PC12 cells (t8=6.1, P<.001). Note that the T allele–mediated reduction in luciferase expression for rs301430 is in line with the T allele–associated decrease in SLC1A1 messenger RNA levels in lymphoblastoid cell lines and brain tissue (Figure 1 and Figure 2). NS indicates nonsignificant.
ASSOCIATION ANALYSES OF SLC1A1 AND OCD
We then genotyped the 3 eQTL SNPs we described and 3 additional SNPs within SLC1A1 that have been individually associated with OCD in prior family-based studies16,17 in our collection of 325 probands and 662 controls matched for ethnicity and sex. Table 1 details genomic location and linkage disequilibrium of the 6 analyzed markers. As presented in Table 1, none of these 6 SNPs were individually associated with OCD after permutation-based multiple-testing correction. We also performed these analyses separately for males and females and did not observe significant results (data not shown).
Table 1.
Polymorphisms, Linkage Disequilibrium Pattern, and Single-Locus Association Tests in 325 OCD Probands and 662 Controls
|
Location on Chromosome 9a |
Intermarker Linkage Disequilibrium, D′ |
Minor Allele |
Minor Allele Frequency |
Allelic Text, |
P Value |
Corrected P Valueb |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | rs7858819 | rs3780412 | rs301430 | rs301434 | rs3087879 | OCD | Control | ||||||
| rs3933331(+) | 4 379 941 | 100 kb upstream of SLC1A1 transcription initiation site | 18 | 4 | 1 | 1 | 1 | C | 0.183 | 0.166 | 0.89 | .34 | .87 |
| rs7858819(+) | 4 549 892 | Intron 2 of SLC1A1 | 94 | 91 | 17 | 88 | T | 0.219 | 0.252 | 2.46 | .12 | .46 | |
| rs3780412(+) | 4 562 480 | Intron 7 of SLC1A1 | 57 | 45 | 66 | C | 0.444 | 0.441 | 0.01 | .92 | >.99 | ||
| rs301430(+) | 4 566 680 | Exon 10 of SLC1A1 (coding synonymous) | 9 | 88 | C | 0.300 | 0.2995 | 0.06 | .81 | >.99 | |||
| rs301434(+) | 4 572 082 | Intron 10 of SLC1A1 | 97 | T | 0.439 | 0.480 | 2.96 | .09 | .36 | ||||
| rs3087879(+) | 4 576 808 | 3′ UTR of SLC1A1 | C | 0.306 | 0.357 | 5.09 | .02 | .12 | |||||
Abbreviations: kb, kilobase; OCD, obsessive-compulsive disorder; UTR, untranslated region.
The University of California–Santa Cruz Genome Browser Human (March 2006 assembly) was used.
Family-wise–corrected empirical P values as determined by permutation.
Haplotype analyses, in contrast, revealed a highly significant omnibus association of 3 of these markers, rs3087879, rs301430, and rs7858819, with OCD (Table 2). In haplotype-specific tests (ie, when tested individually against all other haplotypes, with df=1), 3 of the 4 common haplotypes were nominally significant at P<.05. Two of these haplotypes remained significant after multiple-testing correction by permutation (Table 2). One haplotype, H4 in Table 2, was highly significant and almost twice as common in OCD than in controls (haplotype frequency, 9.4% vs 5.1%, respectively; odds ratio, 1.89; P=.001, family-wise permuted). We obtained similar—though not always as highly significant—results when we conducted these haplotype analyses stratified by sex (Table 2).
Table 2.
Haplotypic Association Testing of SLC1A1 Polymorphisms in OCD Probands and Controls
|
Total Samplea |
Malesb |
Femalesc |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap Frequency |
|||||||||||||||||
| rs7858819 (+) |
rs301430 (+) |
rs3087879 (+) |
Hap- Specific Test, |
P Value |
Corrected P Valued |
Hap- Specific Test, |
P Value |
Corrected P Valued |
Hap- Specific Test, |
P Value |
Corrected P Valued |
||||||
| Hap | OCD | Controls | OR | OR | OR | ||||||||||||
| H1 | C | T | G | 0.408 | 0.358 | 4.58 | .03 | .15 | 0.12 | .73 | .97 | 6.48 | .01 | .04 | |||
| H2 | C | T | C | 0.296 | 0.351 | 5.58 | .02 | .04 | 0.78 | 2.14 | .14 | .50 | 0.80 | 3.39 | .07 | .16 | 0.77 |
| H3 | T | C | G | 0.202 | 0.240 | 3.56 | .06 | .24 | 0.80 | 0.11 | .74 | >.99 | 0.94 | 4.85 | .03 | .15 | 0.71 |
| H4 | C | C | G | 0.094 | 0.051 | 11.54 | <.001 | .001 | 1.89 | 6.05 | .01 | .03 | 1.93 | 5.67 | .02 | .05 | 1.86 |
Abbreviations: Hap, haplotype; OCD, obsessive-compulsive disorder; OR, odds ratio.
Three hundred twenty-five probands; 662 controls. Omnibustest: , P<.001.
One hundred thirty-nine probands; 302 controls. Omnibus test: , P=.07.
One hundred eighty-six probands; 360 controls. Omnibus test: , P=.002.
Family-wise–corrected empirical P values as determined by permutation.
SUBPHENOTYPE ANALYSES OF OCD
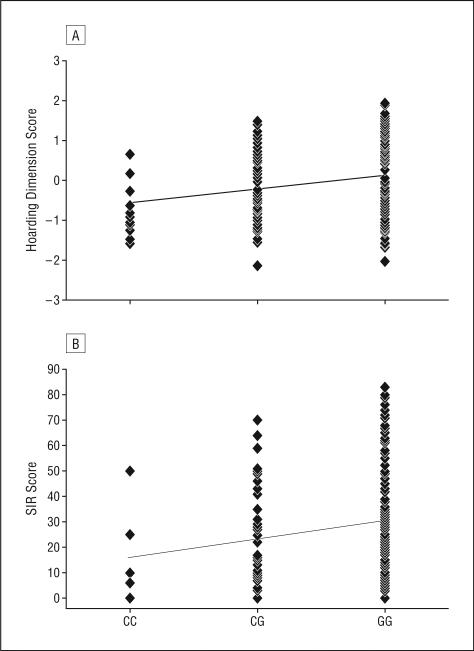
Lastly, we set out to determine whether or not SLC1A1 is also associated with differential OCD symptom presentation, given the growing appreciation of OCD as a clinically, and possibly etiologically, heterogeneous disorder.38,41-43 We performed subphenotype analyses on the basis of Y-BOCS–ascertained OCD symptom severity and the hoarding phenotype evaluated by 2 measures: (1) a Y-BOCS factor analysis–derived hoarding symptom dimension,37 and (2) the magnitude of hoarding symptoms, as indexed by the SIR.39,44 We observed a nominally significant association of 1 eQTL SNP, rs3933331, with the hoarding phenotype. Specifically, this SNP was significantly associated with hoarding as assessed by both the Y-BOCS hoarding symptom dimension (β=.21, t216=2.9, P=.005) and total SIR scores (β=.19, t198=2.6, P=.009). That is, greater scores on the hoarding dimension, as well as more severe SIR symptom scores, were strongly correlated with the number of G alleles at rs3933331 (Figure 2). The association with SIR scores appears to be largely driven by the acquisitioning subscale (β=.22, P=.002), as rs3933331 was less strongly associated with the clutter (β=.18, P=.01) and difficulty discarding (β=.115, P>.05) subscales.
COMMENT
In the present study, we analyzed the candidate gene SLC1A1, which encodes the neuronal glutamate transporter, in a large collection of OCD probands and controls in light of increasing interest in glutamatergic neurotransmission in OCD development and its treatment. We identified an SLC1A1 haplotype that was strongly associated with OCD that consists of 3 markers between intron 2 and the 3′ untranslated region of SLC1A1, 2 of which we found to be SNPs that predict gene expression levels (ie, eQTLs) in lymphoblastoid cell lines. In addition, another eQTL polymorphism, located somewhat upstream from SLC1A1, was strongly correlated with an OCD subphenotype (hoarding), as evaluated by 2 validated scales. Our data corroborate the hypothesis of dysfunctional glutamatergic neurotransmission in OCD and hoarding, mediated in part through SLC1A1.
Three recently published, family-based association studies have suggested SLC1A1 to be a candidate gene for OCD.16-18 However, no given SNP within SLC1A1 in these 3 studies was consistently positively associated with OCD across the 3 studies when tested individually, with the exception of rs3780412, which was considered significant in 2 studies: the overall and males-only study by Dickel and coworkers17 and in the males-only sample by Stewart and colleagues.18 However, our results do not support a specific involvement of this marker in OCD, nor did we observe any preferential or stronger association of single variants or haplotypes in males compared with females in our case-control sample (Table 1 and Table 2). Speculatively, this observation might be related to the increased power of our case-control design compared with the family-based studies published previously.16-18 Clearly, additional studies are warranted to clarify the discrepancy for preferential association in males observed in prior family-based studies,16-18 though the overall finding of involvement of SLC1A1 in OCD now appears more clearly replicable.
One of the most important common findings in these 3 previous SLC1A1 studies was a haplotypic association in the 3′ region of the gene.16-18 Our results are in line with this observation, as 2 of the 3 SNPs of the associated haplotype we identified are located in exon 10 and the 3′ untranslated region. The third SNP of this haplotype, however, lies somewhat upstream (intron 2) of this region, and although all 3 SNPs (rs7858819, rs301430, and rs3087879) were in considerable linkage disequilibrium with each other (Table 1), we speculatively interpret this observation as evidence for more than 1 functional locus within SLC1A1, as has been suggested previously.17,18 Additional support for this tentative conclusion stems from another SNP, rs3933331, located farther upstream and clearly not in linkage disequilibrium with the previously mentioned haplotype markers that we found to be associated with gene expression functionality and that predicted an OCD hoarding subphenotype in our sample (Figure 1 and Figure 2). The possibility of 2 or more functional (possibly causal) variants of differential effect size provides 1 plausible explanation for the molecular discrepancies between present and published SLC1A1 genotyping studies in OCD, especially if these were found to be interacting from within different haplotype blocks. This possibility might be addressed by genotyping markers at higher density or by resequencing large genomic regions.
We have made extensive use of publicly accessible data with the aim of better understanding 1 facet of SLC1A1 functionality—gene expression. Some of the most extensive gene expression data sets currently available were obtained from lymphoblastoid cell lines, and we used these data to better understand how genetic SLC1A1 variation might affect its gene expression levels. To this end, it was first necessary to determine whether or not SLC1A1 is expressed in lymphoblastoid cell lines at sufficiently high levels for quantitative analyses and then to identify the degree to which (if at all) SLC1A1 gene expression in lymphoblastoid cell lines constitutes a heritable trait. The additive heritability (h2) value of 0.23 determined herein, while highly significant, is quite modest compared with some other genes expressed in lymphoblastoid cell lines, whose heritabilities ranged between 0.18 and 1, with a median of 0.34.45 In light of the relatively modest additive heritability for SLC1A1 expression now determined in our analysis, it is not surprising that the eQTL markers identified herein also had a small effect size (Figure 1). Nevertheless, our approach proved valuable, not only because it identified a previously OCD-associated SNP, rs301430, as an eQTL, but also because (1) the 3-marker haplotype that was strongly associated with OCD contained 2 eQTLs, and (2) another eQTL marker farther upstream correlated with an OCD hoarding subphenotype.
Given the modest eQTL significance values and effect sizes from our analyses in lymphoblastoid cell lines that would not withstand multiple testing correction (Figure 1), we conducted additional studies to further assess functionality. We were able to show that 2 of the 3 initial eQTLs, rs7858819 and rs301430, are also associated with SLC1A1 messenger RNA expression in the brain with similar significance levels and the same effect direction (Figure 1 and Figure 2). These results not only strengthen the relevance of these 2 markers for SLC1A1 genetics research, but also suggest that regulation of gene expression in lymphoblastoid cell lines resembles brain regulation for this gene. This would make this readily obtainable cell line suitable for future functional and population-based analyses of SLC1A1 in a manner that has been used extensively for a related molecule, the serotonin transporter (SLC6A4).46,47 It also calls for a systematic comparison of gene expression genetics between brain tissue and lymphoblastoid cell lines now that suitable brain data sets have become available.48
The second complementary study, ie, luciferase reporter gene constructs, was aimed at direct functional assessment of individual SNPs. While association analyses with tissue samples or cell lines have the advantage of more closely resembling in vivo conditions, the eQTLs do not necessarily have to directly confer functionality, as they may merely be in linkage disequilibrium with the actual functional polymorphism. We addressed this uncertainty by examining individual polymorphisms in vitro with reporter gene assays. Our luciferase data for rs301430 suggest that this variant is indeed directly functional. Additional studies are now needed to elucidate the molecular mechanisms through which rs301430 affects gene expression.
The H4 haplotype, as detailed in Table 2, which was almost twice as common in those with OCD as in controls (odds ratio, 1.89), represents the strongest haplotypic association of SLC1A1 with OCD published to date.16-18 It illustrates the usefulness of our complementary expression–mining approach, because 2 of its markers were eQTLs, and conditional haplotypic association testing with the remaining markers failed to yield positive results (see the “Methods” section).28,32 The individual alleles in H4, however, are not fully compatible with the notion of increased or decreased SLC1A1 gene expression in OCD, as the C allele at rs7858819 was the lower-expressing allele in the regression analyses (Figure 1B), while the C allele at rs301430 in H4 correlated with increased expression (Table 2 and Figure 1). On the other hand, the mildly protective haplotype H2 in Table 2 (odds ratio, 0.78), which narrowly survived correction for multiple testing in individual haplotype analysis, contained the lower-expressing allele at both eQTLs. Here, too, it will be essential to conduct gene expression analyses in larger samples and with more densely spaced markers.
The major limitation of our study is the potential susceptibility of the case-control design to population stratification, especially because our controls originated from 3 different groups that did not closely correspond in their geographic origin to our proband sample. While we addressed this issue by matching self-reported ethnicities of probands and controls, spurious effects in case-control studies (which are regarded by some authors as relatively uncommon49) can be ruled out by genotyping multiple ancestry-informative markers, which was not feasible for this work. Dense genetic information on SLC1A1 markers from different ethnicities and geographic locations will increasingly become available from genome-wide association studies. Such data will allow a better judgment of how strongly (and if at all) population stratification is relevant to the SLC1A1 markers analyzed herein.
Another aspect of genetic studies such as ours with several investigated phenotypes that requires discussion is the multiple comparison problem. In these studies, the multiple statistical tests are clearly not independent of one another given linkage disequilibrium between variants (Table 1) and the high correlation between subphenotypes (hoarding in this case) and the general diagnosis of OCD. Therefore, methods that correct by the total number of tests (eg, the Bonferroni procedure) would be more conservative than appropriate in our view. We conducted empirical family-wise error correction for the categorical single-marker and haplotype analyses by permutation and determined that 2 haplotypes remain significant (H4 and H2) (Table 2). The significance values for our subphenotype analyses are nominally robust (for association of rs3933331 with Y-BOCS score, P=.005, and with SIR-assessed hoarding, P=.009) (Figure 4). They would, however, not withstand Bonferroni correction and therefore need to be interpreted with caution and observed in larger samples in future studies.
Figure 4.
Two linear regression analyses of rs3933331 for the hoarding subphenotype, both factor analysis–derived (A) and on the basis of the Saving Inventory–Revised (SIR) score (B), which were nominally significant. A, P=.005. B, P=.009.
In summary, we have identified a strong association of OCD with a 3-marker haplotype composed of eQTLs for SLC1A1, which in turn were determined by exploiting expression and genotype data repositories and further validated with brain expression studies and reporter assays. Our complementary expression analysis approach might prove to be of use in future genetic studies for genes expressed at quantifiable levels in lymphoblastoid cell lines and other population-based tissue collections. Our haplotypic association results call for continued appreciation of glutamatergic neurotransmission abnormalities in OCD research.
Funding/Support
This research was supported by the Intramural Research Program of the National Institute of Mental Health, National Institutes of Health, and by a NARSAD 2007 Young Investigator Award and a National Institute of Mental Health Division of Intramural Research Programs Julius Axelrod Memorial Fellowship Training Award to Dr Wendland.
Footnotes
Financial Disclosure: None reported.
Additional Contributions: We are indebted to all the participants in this study. Diane Kazuba, BS, and Brenda Justement, RN, BSN, MA, conducted patient interviews; Teresa Tolliver, MS, provided technical assistance in DNA extraction; and James Nagle, MS, and Debbie Kauffman, BS, of the National Institute of Neurological Disorders and Stroke DNA sequencing facility provided technical assistance. Postmortem brain tissue was supplied by the Stanley Medical Research Institute.
REFERENCES
- 1.Rosenberg DR, Keshavan MS. A.E. Bennett Research Award: toward a neurodevelopmental model of obsessive-compulsive disorder. Biol Psychiatry. 1998;43(9):623–640. doi: 10.1016/s0006-3223(97)00443-5. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg DR, MacMaster FP, Keshavan MS, Fitzgerald KD, Stewart CM, Moore GJ. Decrease in caudate glutamatergic concentrations in pediatric obsessive-compulsive disorder patients taking paroxetine. J Am Acad Child Adolesc Psychiatry. 2000;39(9):1096–1103. doi: 10.1097/00004583-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 3.McGrath MJ, Campbell KM, Parks CR, Burton FH. Glutamatergic drugs exacerbate symptomatic behavior in a transgenic model of comorbid Tourette's syndrome and obsessive-compulsive disorder. Brain Res. 2000;877(1):23–30. doi: 10.1016/s0006-8993(00)02646-9. [DOI] [PubMed] [Google Scholar]
- 4.Nordstrom EJ, Burton FH. A transgenic model of comorbid Tourette's syndrome and obsessive-compulsive disorder circuitry. Mol Psychiatry. 2002;7(6):617–625. doi: 10.1038/sj.mp.4001144. [DOI] [PubMed] [Google Scholar]
- 5.Pittenger C, Krystal JH, Coric V. Glutamate-modulating drugs as novel pharmacotherapeutic agents in the treatment of obsessive-compulsive disorder. NeuroRx. 2006;3(1):69–81. doi: 10.1016/j.nurx.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarty K, Bhattacharyya S, Christopher R, Khanna S. Glutamatergic dysfunction in OCD. Neuropsychopharmacology. 2005;30(9):1735–1740. doi: 10.1038/sj.npp.1300733. [DOI] [PubMed] [Google Scholar]
- 7.Nestadt G, Samuels J, Riddle M, Bienvenu OJ, III, Liang KY, LaBuda M, Walkup J, Grados M, Hoehn-Saric R. A family study of obsessive-compulsive disorder. Arch Gen Psychiatry. 2000;57(4):358–363. doi: 10.1001/archpsyc.57.4.358. [DOI] [PubMed] [Google Scholar]
- 8.Pauls DL, Alsobrook JP, II, Goodman W, Rasmussen S, Leckman JF. A family study of obsessive-compulsive disorder. Am J Psychiatry. 1995;152(1):76–84. doi: 10.1176/ajp.152.1.76. [DOI] [PubMed] [Google Scholar]
- 9.Arnold PD, Rosenberg DR, Mundo E, Tharmalingam S, Kennedy JL, Richter MA. Association of a glutamate (NMDA) subunit receptor gene (GRIN2B) with obsessive-compulsive disorder: a preliminary study. Psychopharmacology (Berl) 2004;174(4):530–538. doi: 10.1007/s00213-004-1847-1. [DOI] [PubMed] [Google Scholar]
- 10.Delorme R, Krebs MO, Chabane N, Roy I, Millet B, Mouren-Simeoni MC, Maier W, Bourgeron T, Leboyer M. Frequency and transmission of glutamate receptors GRIK2 and GRIK3 polymorphisms in patients with obsessive compulsive disorder. Neuroreport. 2004;15(4):699–702. doi: 10.1097/00001756-200403220-00025. [DOI] [PubMed] [Google Scholar]
- 11.Hanna GL, Veenstra-VanderWeele J, Cox NJ, Boehnke M, Himle JA, Curtis GC, Leventhal BL, Cook EH., Jr Genome-wide linkage analysis of families with obsessive-compulsive disorder ascertained through pediatric probands. Am J Med Genet. 2002;114(5):541–552. doi: 10.1002/ajmg.10519. [DOI] [PubMed] [Google Scholar]
- 12.Willour VL, Yao Shugart Y, Samuels J, Grados M, Cullen B, Bienvenu OJ, III, Wang Y, Liang KY, Valle D, Hoehn-Saric R, Riddle M, Nestadt G. Replication study supports evidence for linkage to 9p24 in obsessive-compulsive disorder. Am J Hum Genet. 2004;75(3):508–513. doi: 10.1086/423899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoyama K, Suh SW, Hamby AM, Liu J, Chan WY, Chen Y, Swanson RA. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat Neurosci. 2006;9(1):119–126. doi: 10.1038/nn1609. [DOI] [PubMed] [Google Scholar]
- 14.Rothstein JD, Martin L, Levey AI, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl RW. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13(3):713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 15.Veenstra-VanderWeele J, Kim SJ, Gonen D, Hanna GL, Leventhal BL, Cook EH., Jr Genomic organization of the SLC1A1/EAAC1 gene and mutation screening in early-onset obsessive-compulsive disorder. Mol Psychiatry. 2001;6(2):160–167. doi: 10.1038/sj.mp.4000806. [DOI] [PubMed] [Google Scholar]
- 16.Arnold PD, Sicard T, Burroughs E, Richter MA, Kennedy JL. Glutamate transporter gene SLC1A1 associated with obsessive-compulsive disorder. Arch Gen Psychiatry. 2006;63(7):769–776. doi: 10.1001/archpsyc.63.7.769. [DOI] [PubMed] [Google Scholar]
- 17.Dickel DE, Veenstra-VanderWeele J, Cox NJ, Wu X, Fischer DJ, Van Etten-Lee M, Himle JA, Leventhal BL, Cook EH, Jr, Hanna GL. Association testing of the positional and functional candidate gene SLC1A1/EAAC1 in early-onset obsessive-compulsive disorder. Arch Gen Psychiatry. 2006;63(7):778–785. doi: 10.1001/archpsyc.63.7.778. [DOI] [PubMed] [Google Scholar]
- 18.Stewart SE, Fagerness JA, Platko J, Smoller JW, Scharf JM, Illmann C, Jenike E, Chabane N, Leboyer M, Delorme R, Jenike MA, Pauls DL. Association of the SLC1A1 glutamate transporter gene and obsessive-compulsive disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(8):1027–1033. doi: 10.1002/ajmg.b.30533. [DOI] [PubMed] [Google Scholar]
- 19.Samuels J, Shugart YY, Grados MA, Willour VL, Bienvenu OJ, Greenberg BD, Knowles JA, McCracken JT, Rauch SL, Murphy DL, Wang Y, Pinto A, Fyer AJ, Piacentini J, Pauls DL, Cullen B, Rasmussen SA, Hoehn-Saric R, Valle D, Liang KY, Riddle MA, Nestadt G. Significant linkage to compulsive hoarding on chromosome 14 in families with obsessive-compulsive disorder: results from the OCD Collaborative Genetics Study. Am J Psychiatry. 2007;164(3):493–499. doi: 10.1176/ajp.2007.164.3.493. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Leckman JF, Pauls DL, Tsai CP, Kidd KK, Campos MR, Tourette Syndrome Association International Consortium for Genetics Genomewide scan of hoarding in sib pairs in which both sibs have Gilles de la Tourette syndrome. Am J Hum Genet. 2002;70(4):896–904. doi: 10.1086/339520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang KY, Wang Y, Shugart YY, Grados M, Fyer AJ, Rauch S, Murphy D, McCracken J, Rasmussen S, Cullen B, Hoehn-Saric R, Greenberg B, Pinto A, Knowles J, Piacentini J, Pauls D, Bienvenu O, Riddle M, Samuels J, Nestadt G. Evidence for potential relationship between SLC1A1 and a putative genetic linkage region on chromosome 14q to obsessive-compulsive disorder with compulsive hoarding. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):1000–1002. doi: 10.1002/ajmg.b.30713. [DOI] [PubMed] [Google Scholar]
- 22.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430(7001):743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39(2):226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavaré S, Deloukas P, Dermitzakis ET. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendland JR, Moya PR, Kruse MR, Ren-Patterson RF, Jensen CL, Timpano KR, Murphy DL. A novel, putative gain-of-function haplotype at SLC6A4 associates with obsessive-compulsive disorder. Hum Mol Genet. 2008;17(5):717–723. doi: 10.1093/hmg/ddm343. [DOI] [PubMed] [Google Scholar]
- 29.LaSalle VH, Cromer KR, Nelson KN, Kazuba D, Justement L, Murphy DL. Diagnostic interview assessed neuropsychiatric disorder comorbidity in 334 individuals with obsessive-compulsive disorder. Depress Anxiety. 2004;19(3):163–173. doi: 10.1002/da.20009. [DOI] [PubMed] [Google Scholar]
- 30.Wendland JR, Kruse MR, Cromer KR, Murphy DL. A large case-control study of common functional SLC6A4 and BDNF variants in obsessive-compulsive disorder. Neuropsychopharmacology. 2007;32(12):2543–2551. doi: 10.1038/sj.npp.1301394. [erratum published in Neuropsychopharmacology. 2008;33(6):1476. Cromer, Kiara C (corrected to Cromer, Kiara R)] [DOI] [PubMed] [Google Scholar]
- 31.Wendland JR, Kruse MR, Murphy DL. Functional SLITRK1 var321, varCDfs and SLC6A4 G56A variants and susceptibility to obsessive-compulsive disorder. Mol Psychiatry. 2006;11(9):802–804. doi: 10.1038/sj.mp.4001848. [DOI] [PubMed] [Google Scholar]
- 32.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23(2):255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 35.Goodman WK, Price LH, Rasmussen SA, Mazure C, Fleischmann RL, Hill CL, Heninger GR, Charney DS. The Yale-Brown Obsessive Compulsive Scale, I: development, use, and reliability. Arch Gen Psychiatry. 1989;46(11):1006–1011. doi: 10.1001/archpsyc.1989.01810110048007. [DOI] [PubMed] [Google Scholar]
- 36.Hasler G, Pinto A, Greenberg BD, Samuels J, Fyer AJ, Pauls D, Knowles JA, McCracken JT, Piacentini J, Riddle MA, Rauch SL, Rasmussen SA, Willour VL, Grados MA, Cullen B, Bienvenu OJ, Shugart YY, Liang KY, Hoehn-Saric R, Wang Y, Ronquillo J, Nestadt G, Murphy DL, OCD Collaborative Genetics Study Familiality of factor analysis-derived YBOCS dimensions in OCD-affected sibling pairs from the OCD Collaborative Genetics Study. Biol Psychiatry. 2007;61(5):617–625. doi: 10.1016/j.biopsych.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 37.Hasler G, LaSalle-Ricci VH, Ronquillo JG, Crawley SA, Cochran LW, Kazuba D, Greenberg BD, Murphy DL. Obsessive-compulsive disorder symptom dimensions show specific relationships to psychiatric comorbidity. Psychiatry Res. 2005;135(2):121–132. doi: 10.1016/j.psychres.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Leckman JF, Grice DE, Boardman J, Zhang H, Vitale A, Bondi C, Alsobrook J, Peterson BS, Cohen DJ, Rasmussen SA, Goodman WK, McDougle CJ, Pauls DL. Symptoms of obsessive-compulsive disorder. Am J Psychiatry. 1997;154(7):911–917. doi: 10.1176/ajp.154.7.911. [DOI] [PubMed] [Google Scholar]
- 39.Frost RO, Steketee G, Grisham J. Measurement of compulsive hoarding: saving inventory-revised. Behav Res Ther. 2004;42(10):1163–1182. doi: 10.1016/j.brat.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 40.Cromer KR, Schmidt NB, Murphy DL. Do traumatic events influence the clinical expression of compulsive hoarding? Behav Res Ther. 2007;45(11):2581–2592. doi: 10.1016/j.brat.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Pato MT, Pato CN, Pauls DL. Recent findings in the genetics of OCD. J Clin Psychiatry. 2002;63(suppl 6):30–33. [PubMed] [Google Scholar]
- 42.Baer L. Factor analysis of symptom subtypes of obsessive compulsive disorder and their relation to personality and tic disorders. J Clin Psychiatry. 1994;55(suppl):18–23. [PubMed] [Google Scholar]
- 43.Mataix-Cols D, Rosario-Campos MC, Leckman JF. A multidimensional model of obsessive-compulsive disorder. Am J Psychiatry. 2005;162(2):228–238. doi: 10.1176/appi.ajp.162.2.228. [DOI] [PubMed] [Google Scholar]
- 44.Wheaton M, Timpano KR, Lasalle-Ricci VH, Murphy D. Characterizing the hoarding phenotype in individuals with OCD: associations with comorbidity, severity and gender. J Anxiety Disord. 2008;22(2):243–252. doi: 10.1016/j.janxdis.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75(6):1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Müller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274(5292):1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 47.Prasad HC, Zhu CB, McCauley JL, Samuvel DJ, Ramamoorthy S, Shelton RC, Hewlett WA, Sutcliffe JS, Blakely RD. Human serotonin transporter variants display altered sensitivity to protein kinase G and p38 mitogen-activated protein kinase. Proc Natl Acad Sci U S A. 2005;102(32):11545–11550. doi: 10.1073/pnas.0501432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39(12):1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 49.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361(9357):598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]