Abstract
A number of genome-wide linkage analyses have identified the 2q33.3-2q37.2 region as most likely to contain the genes that contribute to the susceptibility to chronic obstructive pulmonary disease (COPD). It was hypothesized that the SERPINE2 gene, which is one of the genes located at the 2q33.3-2q37.2 region, may act as a low-penetrance susceptibility gene for COPD. To test this hypothesis, the association of four SERPINE2 single nucleotide polymorphisms (SNPs; rs16865421A>G, rs7583463A>C, rs729631C>G, and rs6734100C>G) with the risk of COPD was investigated in a case-control study of 311 COPD patients and 386 controls. The SNP rs16865421 was associated with a significantly decreased risk of COPD in a dominant model for the polymorphic allele (adjusted odds ratio [OR]=0.66, 95% confidence interval [CI]=0.45-0.97, P=0.03). In haplotype analysis, the GACC haplotype carrying the polymorphic allele at the rs16865421 was associated with a significantly decreased risk of COPD when compared to the AACC haplotype (adjusted OR=0.58, 95% CI=0.38-0.89, P=0.01), and this effect was evident in younger individuals (adjusted OR=0.30, 95% CI=0.14-0.64, P=0.002). This study suggests that the SERPINE2 gene contributes to the susceptibility to COPD.
Keywords: Serpine2; Polymorphism; Pulmonary Disease, Chronic Obstructive
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is characterized by progressive irreversible airflow obstruction caused by a mixture of obstructive bronchiolitis and emphysema (1). COPD is a major health problem worldwide with a high prevalence and cost. Despite public awareness regarding the health risks of smoking, mortality from COPD is still rising (1, 2); therefore, identification of risk factors involved in COPD is urgently needed to prevent this disease. Although cigarette smoking is the most important risk factor for the development of COPD, only a fraction of chronic smokers develops symptomatic COPD (3), which suggests that genetic factors play an important role in the development of COPD (4). Based on the known or presumed pathophysiology of COPD, the genes involved in the recruitment of inflammatory cells, mucociliary clearance, protease-antiprotease imbalance, oxidants-antioxidants imbalance, apoptosis, and xenobiotic metabolism have been investigated; however, the results of such studies have been inconsistent (5).
A number of genome-wide linkage analyses have identified chromosomal regions that are likely to contain genes that contribute to the susceptibility to COPD (6-9). For example, the 2q33.3-2q37.2 region has been found to be associated with logarithm of the likelihood for linkage scores greater than 2.0 for the ratio of forced expiratory volume at one second (FEV1) to forced vital capacity (FVC) in a study of Boston Early-Onset COPD families (6). In addition, a similar region on chromosome 2q was linked to the FEV1/FVC ratio in the families studied in the Utah Genetic Reference Project (8). Furthermore, it has been suggested that the 2q33.3-2q37.2 region of linkage contains genes that contribute to COPD susceptibility through gene-by-smoking interactions (9).
Using a combination of whole genome-wide linkage analysis and microarray gene expression analysis of murine and human lung tissues, DeMeo et al. (10) isolated the SERPINE2 (serine peptidase inhibitor, clade E [nexin, plasminogen activator inhibitor type 1], member 2) gene, which is located on chromosome 2q33-35, a genomic region linked to COPD phenotypes, as a candidate gene for susceptibility to COPD. In addition, they found a significant association of multiple single nucleotide polymorphisms (SNPs) in the SERPINE2 gene with COPD-phenotypes in family-based and case-control studies (10). However, Chappell et al. (11) found no evidence of association of the five SERPINE2 SNPs with COPD in a European case-control study. Recently, Zhu et al. (12) examined the association of SERPINE2 SNPs with COPD-related phenotypes in a Norwegian population and found that six SNPs were significantly associated with COPD-related phenotypes in a family-based association study, and that five of these SNPs showed replicative associations in a case-control study. These contradictory results across studies indicate that more genetic association studies in different ethnic populations are necessary to understand the role of the SERPINE2 gene on the COPD susceptibility. Therefore, we conducted a case-control study to evaluate the association of SERPINE2 SNPs with the risk of COPD in a Korean population.
MATERIALS AND METHODS
Study population
The patient group (n=311) consisted of male COPD patients who visited the respiratory center of Kyungpook National University Hospital between July 2006 and December 2006, and the details of the study population have been described elsewhere (13). Briefly, COPD was diagnosed according to the criteria established by the NHLBI/WHO Global Initiative for COPD (GOLD) (1). The criteria for COPD were as follows: chronic respiratory symptoms and signs such as cough and dyspnea; FEV1/FVC <70% and FEV1 reversibility after inhaling 200 µg salbutamol <12% of the pre-bronchodilator FEV1. Patients with bronchial asthma were excluded on the basis of the reversibility of airflow obstruction. In addition, subjects with radiographic abnormalities suggestive of other significant respiratory diseases such as bronchiectasis and pulmonary tuberculosis were also excluded. The severity of COPD was classified by the percentage predicted FEV1, according to the guidelines established by the GOLD: mild (>80%, GOLD stage I), moderate (50-80%, GOLD stage II), severe (30-50%, GOLD stage III) or very severe (<30%, GOLD stage IV). According to the GOLD classification, 97 cases (31.2%) had mild disease, 101 cases (32.5%) were moderate, 86 cases (27.6%) were severe and 27 cases (8.7%) were very severe. Control subjects (n=386) were selected from a pool of healthy men who visited the general health check-up center. The enrollment criteria for the controls were as follows: male, age >45 yr, no known disease and no history of any disease and no airflow limitation. All of the cases and controls were ethnic Koreans that resided in Daegu City or the surrounding regions. A trained interviewer completed detailed questionnaires for each patient and each control subject. This study was approved by the institutional review board of the Kyungpook National University Hospital, and written informed consent was obtained from each participant.
Genotyping
We first examined the frequencies of 6 SERPINE2 SNPs (rs16865421A>G [IVS3-1376], rs6748795G>C [IVS4-1056], rs7583463A>C [IVS5+360], rs975278A>G [IVS5-209], rs729631C>G [IVS7+109], and rs6734100C>G [IVS8+266]), which were found to be significantly associated with COPD-related phenotypes in a case-control study conducted by Zhu et al. (12), in 27 healthy Korean individuals by sequencing analysis. The primer sets used for sequencing were designed based on the GenBank reference sequence (accession no. NT_005403.16) and the sequence variants were confirmed by two independent authors. The SNPs rs6748795 and rs7583463, as well as the SNPs rs975278 and rs729631 were in complete linkage disequilibrium (LD). Therefore, four SNPs (rs16865421, rs7583463, rs729631, and rs6734100) were chosen for an association study. The genotypes of the four SNPs were determined by PCR and melting-curve analysis using fluorescence-labeled hybridization probes (Light-Cycler, Roche Diagnostic, Mannheim, Germany). A genotype success rate of greater than 96% was achieved using the LightCycler. Samples that could not be scored by the Light-Cycler were re-genotyped by direct sequencing using an ABI PRISM 3700 genetic analyzer (Applied Biosystems, Foster City, CA, U.S.A.). All genotyping analyses were performed "blind" with respect to the case/control status in order to ensure quality control. Approximately 10% of the samples were randomly selected to be genotyped again by a different investigator, and the results were 100% concordant. To determine the analytic accuracy of the melting curve-based genotyping, 10% of the samples were randomly selected using random number generated from the uniform distribution and then analyzed by DNA sequencing. The genotypes determined using the fluorescent hybridization probe melting curve analysis were 100% concordant with those determined by DNA sequencing. Information regarding all SNPs, SNP IDs and allele frequencies was obtained from the NCBI homepage (http://www.ncbi.nlm.nih.gov/SNP).
Statistical analysis
The cases and controls were compared using the Student's t-test for continuous variables and a chi-suquare test for categorical variables. Each of the SNPs was analyzed for Hardy-Weinberg equilibrium using the goodness-of-fit test, as implemented through SAS Genetics, version 9.1.3 (SAS institute, Gary, NC, U.S.A.). The LD structure among SNPs was examined using HaploView 4.0 (14), which is available at http://www.broad.mit.edu/mpg/haploview. LD blocks were inferred from the definition proposed by Gabriel et al. (15). Haplotype frequencies were estimated based on a Bayesian algorithm using PHASE 2.1 (16). Unconditional logistic regression analyses were used to calculate the odds ratio (OR) and 95% confidence interval (CI), with adjustment for possible confounders (age and pack-years of smoking as continuous variables). In addition to the overall association analysis, we performed a stratified analysis according to age, smoking status and severity of COPD to further explore the association between genotypes/haplotypes and the risk of COPD in each stratum. To correct for multiple testing, we applied the Bonferroni correction based on the effective number of independent SNPs (Meff) for genotype analysis and the Benjamini and Hochberg's method for haplotype analysis (17, 18). To assess the potential interaction between genotype/haplotype and smoking (smoking status or pack-years of smoking), we first included the interaction term in the logistic model. The interaction between genotype/haplotype and smoking was not statistically significant, and was therefore removed from the model. All of the analyses were performed using Statistical Analysis Software for Windows, version 9.1.3 (SAS institute).
RESULTS
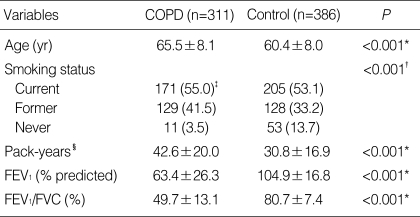
The baseline characteristics of the cases and controls enrolled in this study are shown in Table 1. The FEV1 and FEV1/FVC ratio were significantly lower in the COPD group than in the control group.
Table 1.
Characteristics of the study population
*t-test; †χ2 test; ‡Numbers in parenthesis, column percentage; §In current and former smokers.
FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity.
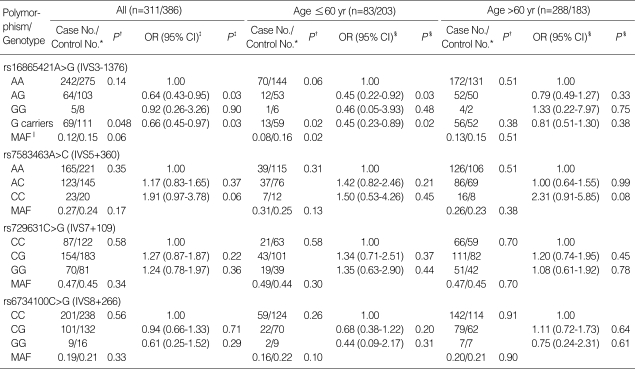
The genotype and polymorphic allele frequencies of the four SERPINE2 SNPs (rs16865421A>G, rs7583463A>C, rs729631C>G, and rs6734100C>G) in the cases and controls are shown in Table 2. The genotype distributions of the four SNPs in the controls were in Hardy-Weinberg equilibrium. The frequency of the rs16865421G allele was a borderline significantly lower in the cases than in the control (P=0.06), and the presence of at least one rs16865421G allele was associated with a significantly decreased risk of COPD when compared to the homozygous wild-type genotype (adjusted OR=0.66, 95% CI=0.45-0.97, P=0.03, corrected P [Pc] for multiple testing by Meff=0.11). When the subjects were stratified by median age (≤60 yr/>60 yr), the presence of at least one rs16865421G allele was associated with a significantly decreased risk of COPD when compared to the AA genotype in the younger-aged group (adjusted OR=0.45, 95% CI=0.23-0.89, P=0.02, Pc=0.08), but not in older individuals (adjusted OR=0.83, 95% CI=0.51-1.30, P=0.38). The genotype and allele frequencies of the other three SNPs examined were not significantly different between the cases and controls.
Table 2.
SERPINE2 genotypes of COPD cases and controls, and their association with the risk of COPD
*Number of cases/number of controls; †Two-sided χ2 test for either genotype distributions or allele frequencies between cases and controls; ‡Odds ratios (ORs), 95% confidence intervals (95% CIs) and P values were calculated by unconditional logistic regression analysis, adjusted for age and pack-years of smoking; §ORs (95% CIs) and P values were calculated by unconditional logistic regression analysis, adjusted for pack-years of smoking; ∥Minor allele frequency.
Threshold P value for a significant association=0.014, based on Bonferroni correction for the effective number of independent tests (Meff=3.59).
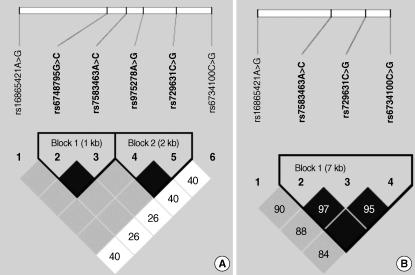
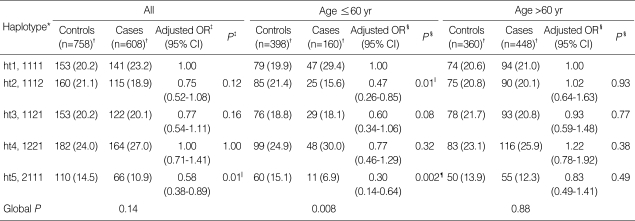
We next examined the association between the haplotypes of the four SERPINE2 SNPs and the risk of COPD. The four SERPINE2 SNPs were in strong LD (|D'| value: range from 0.90 to 1.00; Fig. 1). Among the 10 haplotypes observed, five had a frequency of less than 1% and were excluded from the analysis. The remaining five haplotypes accounted for 98.0% of the chromosomes obtained from the 697 subjects (cases: 98.2% and controls: 97.8%). Consistent with the results of the genotyping analysis, haplotype 5 (ht5, rs16865421G/rs7583463A/rs729631C/rs6734100C), carrying the rs168-65421G allele, was associated with a significantly decreased risk of COPD when compared to the ht1 (rs16865421A/rs7583463A/rs729631C/rs6734100C; adjusted OR=0.58, 95% CI=0.38-0.89, P=0.01, Pc for multiple testing by Hochberg and Benjamini's method=0.04, Table 3). When the subjects were stratified by median age, the distribution of the inferred haplotypes of the cases was significantly different from that of the controls in the younger age group (Pdf=5=0.008), but not in the older age group. Haplotype 5 was associated with a significantly decreased risk of COPD when compared to the ht1 in the younger individuals (adjusted OR=0.30, 95% CI=0.14-0.64, P=0.002, Pc=0.02), but not in the older individuals (adjusted OR=0.83, 95% CI=0.49-1.41, P=0.49).
Fig. 1.
Linkage disequilibrium (LD) blocks between SERPINE2 polymorphisms in 27 healthy Koreans (A) and the subjects (n=697) of a case-control study (B). The LD blocks were generated by the Haploview program. Black boxes indicate complete LD (|D'|=1.0 and r2=1.0). Dark gray boxes indicate strong evidence of LD (confidence interval [CI] minima for strong LD: upper 0.98, low 0.7; fraction of strong LD in informative comparisons must be at least 0.95). White boxes indicate strong recombination (upper CI ≤0.9), and the gray boxes indicate uninformative findings. Triangles indicate haplotype blocks. Numbers in squares are |D'| (×100) values.
Table 3.
Distribution of the SERPINE2 haplotypes in the cases and controls
*Wild-type allele is denoted by 1 and polymorphic allele, by 2. The order of the polymorphisms is as follows; rs16865421A>G, rs7583463A>C, rs729631C>G and rs6734100C>G; †Five haplotypes that had a frequency of less than 1% were excluded from analysis; controls, 14 (age ≤60, 8; and age >60, 6) and cases, 14 (age ≤60, 6; and age >60, 8); ‡OR (95% CI) and corresponding P values were calculated by unconditional logistic regression analysis, adjusted for age and pack-years of smoking; §OR (95% CI) and corresponding P values were calculated by unconditional logistic regression analysis, adjusted for pack-years of smoking. Corrected P value (Pc) by Hochberg and Benjamini's method; ∥Pc=0.04 and ¶Pc=0.02.
The association of the SERPINE2 SNPs and haplotypes were further examined after stratifying the subjects according to smoking status, smoking exposure level, and quantitative (FEV1 and FEV1/FVC) and qualitative (severity of COPD) spirometric phenotypes of COPD. There were no effect modification by smoking status and smoking exposure level. In addition, the SNPs and haplotypes were not significantly associated with the quantitative and qualitative spirometric phenotypes of COPD (data not shown).
DISCUSSION
We investigated the association between SNPs in the SERPINE2 gene, which is a positional candidate gene for COPD susceptibility, and the risk of COPD in a Korean population. In the present study, the SNP rs16865421 and haplotypes composed of the four SNPs rs16865421, rs7583463, rs729631, and rs6734100, were significantly associated with the risk of COPD, particularly in younger individuals. This study is an important addition to previous studies that have shown an association between SERPINE2 SNPs and COPD-related phenotypes. In addition, this is the first case-control study investigating an association of SERPINE2 SNPs with the risk of COPD in an Asian population.
In the present study, the polymorphic allele frequencies of the SERPINE2 SNPs rs16865421A>G, rs7583463A>C, rs729631C>G, and rs6734100C>G among healthy Koreans were 0.15, 0.24, 0.45, and 0.21, respectively, which were similar to those of Asians quoted in the NCBI database. However, the frequencies were significantly different from those of Norwegians (0.08, 0.82, 0.84, and 0.13 respectively) (12), Europeans (0.04, 0.83, 0.16, and 0.13, respectively) and Africans (0.17, 0.59, 0.32, and 0.03, respectively) reported in the NCBI database.
To date, three studies investigating the association of SERPINE2 SNPs and COPD-related phenotypes have been reported; however, the results have been inconsistent. DeMeo et al. (10) evaluated the associations of 48 SERPINE2 SNPs with COPD phenotypes in both family and case-control cohorts, and found that 18 SNPs were significantly associated with quantitative and/or qualitative spirometric phenotypes in the family study of severe early-onset COPD pedigrees, and that eight SNPs (rs1438831, rs7579646, rs840088, rs7562213, rs920251, rs3795877, rs6747096, and rs3795879), including 5 SNPs (underlined), that were significant in the family-based study demonstrated a significant association with severe COPD (FEV1 of ≤45% predicted) in a case-control study of American Caucasians. In contrast, Chappell et al. (11) reported that five SNPs (rs1438831, rs920251, rs6747096, rs3795879, ss49785625) that were significantly associated with COPD phenotypes in the family-based or case-control study conducted by DeMeo et al. (10) were not significantly associated with the risk of COPD in a European case-control study. Furthermore, Zhu et al. (12) investigated the association of 25 SERPINE2 SNPs with COPD phenotypes in a family-based study and a case-control study in a Norwegian population. They reported that five SNPs (rs6748795, rs7583463, rs975278, rs729631, and rs6734100) were significantly associated with the risk of COPD in the family-based study. However, in the case-control study, only the SNP rs16865421 was associated with a significantly increased risk of COPD, while the five SNPs (rs6748795, rs7583463, rs975278, rs729631, and rs6734100) were only associated with a reduction of FEV1 and/or FEV1/VC in the COPD patients. In the current study, we examined the six SERPINE2 SNPs that were significantly associated with COPD-related phenotypes in the study conducted by Zhu et al. (12). In concordance with their study, we found that the SNP rs16865421 was significantly associated with the risk of COPD in the present case-control study performed in a Korean population; however, the other five SNPs were not associated with the quantitative spirometric phenotypes. In addition, in contrast to the Norwegian study (12), the SNP rs16865421 was found to be associated with a significantly decreased risk of COPD in this study.
Although it is hard to decipher the reasons for the different results across studies, particularly the conflicting association of the SNP rs16865421 with COPD, the different genetic backgrounds and environmental factors in the study populations may have been responsible for these differences. The allele and genotype frequencies of the SERPINE2 SNPs, as well as the status of LD between the SNPs, vary greatly between ethnic groups (10-12, 19). Therefore, the genetic effects of the SERPINE2 SNPs and haplotypes on the susceptibility to COPD may differ by ethnicity. In a complex polygenic disease such as COPD, it is likely that multiple genes are involved in its pathogenesis, which would result in the genetic susceptibility being dependent on the coincidence of several polymorphisms acting together. Therefore, a protective genotype in one population may increase the risk of COPD in another population due to other linked polymorphisms that exhibit stronger effects on the susceptibility to COPD. These discrepancies may be related to differences in the distribution of COPD phenotypes across studies. COPD is a heterogeneous disorder that includes obstructive bronchiolitis and emphysema. These COPD phenotypes share some similar pathophysiology, and likely share some similar genetic determinants; however, there may also be divergent underlying pathways involved in their development (20, 21). Therefore, it is possible that the effects of the SERPINE2 SNPs on the risk of COPD will differ according to the phenotype of COPD (19-21). In addition, differences in the severity of the disease in COPD cases across different studies should be also considered. Furthermore, the SERPINE2 SNPs and haplotypes were only associated with the risk of COPD in younger individuals in the present study. This may indicate that the negative association observed in the study conducted by Chappell et al. (11) occurred due to their study primarily consisting of older-aged patients. The inadequacies in the study design, such as nonrandom sampling, limited sample sizes and the pitfalls arising from unknown confounders, also need be considered.
The physiologic function of SERPINE2 and its role in the pathogenesis of COPD remains to be elucidated. One of the most prevailing hypotheses for the pathogenesis of COPD is the inflammatory theory, which suggests that cigarette smoke and other inhaled irritants stimulates inflammatory cells such as neutrophils, macrophages and CD8+ T cells to release a variety of mediators, oxidants and proteases. These inflammatory events then induce airways remodeling and parenchymal destruction. Based on this theory, disturbance of the balance between proteases and their inhibitors by an excess of protease or lack of inhibitor is believed to allow structural damage to the lung to occur (22, 23). SERPINE2 is a 43-kDa member of the serine protease inhibitor (SERPIN) superfamily that has been shown to inhibit several serine proteases, including thrombin, urokinase, and plasmin, which play important roles in inflammation and wound repair following tissue injury (24, 25). In addition, it has been shown that SERPINE2 gene expression is up-regulated by cytokines such as interleukin-1, tumor necrosis factor-α, and transforming growth factor-β, which play important roles in the development of COPD (26, 27). Taken together, these findings suggest that SERPINE2 may play a role in the pathogenesis of COPD by mediating the maintenance of the protease-antiprotease balance (10, 12). However, further studies must be conducted to confirm this.
In disease-gene association studies, analysis of haplotypes composed of multiple SNPs can increase the power to detect disease associations when compared with single SNP analysis due to the higher heterozygosity and tighter LD with the disease-causative variants. In addition, these analyses allow for the possibility of an ungenotyped functional variant to be in LD with the genotyped SNPs (28, 29). Because none of the SERPINE2 SNPs examined in the present study were located in potentially functional regions of the gene, it is likely that the association between the SNPs and COPD susceptibility may be due to LD with other functional SERPINE2 variant(s) rather than to direct functional effects of the SNPs examined. Therefore, our investigation was extended to an analysis of the SERPINE2 haplotypes composed of the four SNPs. In the haplotype analysis, ht5 was associated with a significantly decreased risk of COPD when compared to ht1 among the four haplotypes carrying the rs16865421A allele.
It is important to note that this study had several limitations. Because this study was designed to evaluate the effects of SERPINE2 SNPs on the risk of overall COPD, the stratification analyses according to age, smoking status and spirometric phenotypes might have a type I error (due to multiple comparisons) and/or a type II error (due to the small number of subjects in the subgroups). Therefore, additional studies with a larger sample sizes should be conducted to confirm our findings. Second, among several candidate SNPs in the SERPINE2 gene, only 6 intronic SNPs were examined, which obviously limit the ability to elucidate the entire effect of the SERPINE2 gene on the susceptibility to COPD. Therefore, a comprehensive evaluation of SERPINE2 SNPs and identification of functional variants are needed to better understand the role of SERPINE2 in the development of COPD.
In conclusion, this case-control study demonstrates a significant association of SERPINE2 SNPs and haplotypes with decreased risk of COPD, particularly among younger individuals. However, because this is the first case-control study investigating the associations of SERPINE2 SNPs with the risk of COPD in an Asian population, additional studies are required to confirm our findings. Moreover, further study is needed to identify functional variants of the SERPINE2 gene.
Footnotes
This study was supported by a grant of the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (A060165), and in part by the Brain Korea 21 Project in 2006.
References
- 1.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, Zielinski J. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 2.Murray CJ, Lopez AD. Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study. Lancet. 1997;349:1498–1504. doi: 10.1016/S0140-6736(96)07492-2. [DOI] [PubMed] [Google Scholar]
- 3.Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J. Developing COPD: a 25 year follow up study of the general population. Thorax. 2006;61:935–939. doi: 10.1136/thx.2006.062802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampsonas F, Karkoulias K, Kaparianos A, Spiropoulos K. Genetics of chronic obstructive pulmonary disease, beyond α1-antitrypsin deficiency. Cur Med Chem. 2006;13:2857–2873. doi: 10.2174/092986706778521922. [DOI] [PubMed] [Google Scholar]
- 5.Hersh CP, DeMeo DL, Lange C, Litonjua AA, Reilly JJ, Kwiatkowski D, Laird N, Sylvia JS, Sparrow D, Speizer FE, Weiss ST, Silverman EK. Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am J Respir Cell Mol Biol. 2005;33:71–78. doi: 10.1165/rcmb.2005-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman EK, Palmer LJ, Mosley JD, Barth M, Senter JM, Brown A, Drazen JM, Kwiatkowski DJ, Chapman HA, Campbell EJ, Province MA, Rao DC, Reilly JJ, Ginns LC, Speizer FE, Weiss ST. Genomewide linkage analysis of quantitative spirometric phenotypes in severe early-onset chronic obstructive pulmonary disease. Am J Hum Genet. 2002;70:1229–1239. doi: 10.1086/340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer LJ, Celedon JC, Chapman HA, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage analysis of bronchodilator responsiveness and post-bronchodilator spirometric phenotypes in chronic obstructive pulmonary disease. Hum Mol Genet. 2003;12:1199–1210. doi: 10.1093/hmg/ddg125. [DOI] [PubMed] [Google Scholar]
- 8.Malhotra A, Peiffer AP, Ryujin DT, Elsner T, Kanner RE, Leppert MF, Hasstedt SJ. Further evidence for the role of genes on chromosome 2 and chromosome 5 in the inheritance of pulmonary function. Am J Respir Crit Care Med. 2003;168:556–561. doi: 10.1164/rccm.200303-410OC. [DOI] [PubMed] [Google Scholar]
- 9.DeMeo DL, Celedon JC, Lange C, Reilly JJ, Chapman HA, Sylvia JS, Speizer FE, Weiss ST, Silverman EK. Genome-wide linkage of forced mid-expiratory flow in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170:1294–1301. doi: 10.1164/rccm.200404-524OC. [DOI] [PubMed] [Google Scholar]
- 10.DeMeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, Haley KJ, Sylvia JS, Sparrow D, Spira AE, Beane J, Pinto-Plata V, Speizer FE, Shapiro SD, Weiss ST, Silverman EK. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet. 2006;78:253–264. doi: 10.1086/499828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chappell S, Daly L, Morgan K, Baranes TG, Roca J, Rabinovich R, Millar A, Donnelly SC, Keatings V, MacNee W, Stolk J, Hiemstra PS, Miniati M, Monti S, O'Connor CM, Kalsheker N. The SERPINE2 gene and chronic obstructive pulmonary disease. Am J Hum Genet. 2006;79:184–186. doi: 10.1086/505268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, Anderson WH, Lomas DA, Silverman EK, Pilai SG. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176:167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 13.Kim KM, Park SH, Kim JS, Lee WK, Cha SI, Kim CH, Kang YM, Jung TH, Kim IS, Park JY. Polymorphisms in the Type IV collagen alpha 3 gene and the risk of chronic obstructive pulmonary disease. Eur Respir J. 2008;32:35–41. doi: 10.1183/09031936.00076207. [DOI] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 16.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 19.DeMeo DL. Reply to Chappell et al. Am J Hum Genet. 2006;79:186–187. [Google Scholar]
- 20.Litonjua A, Weiss ST. Phenotypes for human respiratory genetics. In: Silverman EK, Shapiro SD, Lomas DA, Weiss ST, editors. Respiratory Genetics. London: Edward Arnold Publishers; 2005. pp. 15–26. [Google Scholar]
- 21.Hersh CP, DeMeo DL, Silverman EK. Chronic obstructive pulmonary disease. In: Silverman EK, Shapiro SD, Lomas DA, Weiss ST, editors. Respiratory Genetics. London: Edward Arnold Publishers; 2005. pp. 253–296. [Google Scholar]
- 22.Snider GL. Chronic obstructive pulmonary disease: risk factors, pathophysiology, and pathogenesis. Annu Rev Med. 1989;40:411–429. doi: 10.1146/annurev.me.40.020189.002211. [DOI] [PubMed] [Google Scholar]
- 23.Groneberg DA, Chung KF. Models of chronic obstructive pulmonary disease. Respir Res. 2004;5:18. doi: 10.1186/1465-9921-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker JB, Low DA, Simmer RL, Cunningham DD. Protease-nexin: A cellular component that links thrombin and plasminogen activator and mediates their binding to cells. Cell. 1980;21:37–45. doi: 10.1016/0092-8674(80)90112-9. [DOI] [PubMed] [Google Scholar]
- 25.Scott RW, Bergman BL, Bajpai A, Hersh RT, Rodrigues H, Jones BN, Barreda C, Watts S, Baker JB. Protease nexin. Properties and a modified purification procedure. J Biol Chem. 1985;260:7029–7034. [PubMed] [Google Scholar]
- 26.Vaughan PJ, Cunningham DD. Regulation of protease nexin-1 synthesis and secretion in cultured brain cells by injury-related factors. J Biol Chem. 1993;268:3720–3727. [PubMed] [Google Scholar]
- 27.Mbebi C, Hantai D, Jandrot-Perrus M, Doyennette MG, Verdiere-Sahuque M. Protease nexin 1 expression is upregulated in human skeletal muscle by injury-related factors. J Cell Physiol. 1999;179:305–314. doi: 10.1002/(SICI)1097-4652(199906)179:3<305::AID-JCP8>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Judson R, Stephens JC. Notes from the SNP vs. haplotype front. Pharmacogenomics. 2001;2:7–10. doi: 10.1517/14622416.2.1.7. [DOI] [PubMed] [Google Scholar]
- 29.Rebbeck TR, Ambrosone CB, Bell DA, Chanock SJ, Hayes RB, Kadlubar FF, Thomas DC. SNPs, haplotypes, and cancer: applications in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2004;13:681–687. [PubMed] [Google Scholar]