Abstract
CYLD is a tumour-suppressor gene that is mutated in a benign skin tumour syndrome called cylindromatosis. The CYLD gene product is a deubiquitinating enzyme that was shown to regulate cell proliferation, cell survival and inflammatory responses, mainly through inhibiting NF-κB signalling. Here we show that CYLD controls cell growth and division at the G1/S-phase as well as cytokinesis by associating with α-tubulin and microtubules through its CAP-Gly domains. Translocation of activated CYLD to the perinuclear region of the cell is achieved by an inhibitory interaction of CYLD with histone deacetylase-6 (HDAC6) leading to an increase in the levels of acetylated α-tubulin around the nucleus. This facilitates the interaction of CYLD with Bcl-3, leading to a significant delay in the G1-to-S-phase transition. Finally, CYLD also interacts with HDAC6 in the midbody where it regulates the rate of cytokinesis in a deubiquitinase-independent manner. Altogether these results identify a mechanism by which CYLD regulates cell proliferation at distinct cell-cycle phases.
Keywords: α-tubulin, acetylation, cell cycle, CYLD, HDAC6
Introduction
Familial cylindromatosis is characterized by formation of multiple benign tumours originating from skin (Bignell et al, 2002). The tumour syndrome is caused by the loss of both CYLD alleles. The CYLD gene encodes a deubiquitinating enzyme, which removes lysine 48- or lysine 63-linked polyubiquitin chains from target proteins (Massoumi and Paus, 2007). Depending on the cellular context CYLD has been shown to negatively regulate NF-κB and/or JNK-signalling pathways resulting in suppression of cell proliferation and survival (Brummelkamp et al, 2003; Kovalenko et al, 2003; Trompouki et al, 2003; Reiley et al, 2004). The mechanism by which CYLD exerts its tumour-suppressor function in vivo has been analyzed in CYLD-null mice, which are highly susceptible to chemically induced skin tumours. The increased tumour incidence was attributed to the loss of an inhibitory interaction between CYLD and the proto-oncogene Bcl-3. The association of CYLD with Bcl-3, which results from activation and subsequent perinuclear translocation of the protein, leads to the removal of a lysine 63-linked ubiquitin chain from Bcl-3, which in turn inhibits the nuclear translocation and activity of Bcl-3 (Massoumi et al, 2006). In the absence of CYLD, Bcl-3 is able to translocate into the nucleus where it forms a complex with the NF-κB p50 and p52 isoforms. This results in activation of the cyclin-D1 promoter and increased proliferation and tumour growth (Massoumi et al, 2006). It is not clear, however, how CYLD translocates to the perinuclear region to capture Bcl-3 and whether this is the only mechanism by which CYLD regulates tumour cell proliferation.
In addition to the C-terminal ubiquitin C-terminal hydrolase (UCH) domain, which executes the removal of ubiquitin chains, CYLD contains three cytoskeleton-associated protein-glycine-conserved (CAP-Gly) domains in the N-terminal portion of the protein. The exact function of CAP-Gly domains is not known, but their presence in various microtubule (MT)-binding proteins suggests that they enable binding to MTs (Riehemann and Sorg, 1993; Pierre et al, 1994; Bateman et al, 2002). The third CAP-Gly domain (CAP-Gly3) of CYLD has recently been shown to associate directly with the proline-rich sequences of NEMO/IKKγ, a pseudokinase, which together with IKKα and IKKβ triggers TNF-receptor-mediated NF-κB activation (Bateman et al, 2002). Interestingly, this CAP-Gly domain (Saito et al, 2004) differs structurally from the first two CAP-Gly domains (CAP-Gly1 and CAP-Gly2), which have been recently shown to associate directly with tubulin and promote tubulin polymerization (Gao et al, 2008). Whether the CAP-Gly domains in CYLD have any additional function besides tubulin binding is not known.
The stability of MTs is accompanied by post-translational modifications of α-tubulin such as acetylation, which usually occurs after MT assembly (Westermann and Weber, 2003). The level of α-tubulin acetylation in cells is accurately adjusted by the activities of tubulin acetyltransferases and deacetylases, which catalyze the acetylation and deacetylation of α-tubulin, respectively. Elp3 was identified very recently as α-tubulin acetyltransferase in neuronal cells (Creppe et al, 2009), while HDAC6 (histone deacetylase-6) and SIRT2 (the Silent Information Regulator Type-2) have been shown to deacetylate α-tubulin (Hubbert et al, 2002; North et al, 2003).
HDAC6 contains two intact HDAC catalytic domains, which execute deacetylation of α-tubulin (Zhang et al, 2006; Zou et al, 2006; Boyault et al, 2007a), and a ubiquitin-binding zinc-finger domain, which enables binding to ubiquitinated proteins (Seigneurin-Berny et al, 2001; Hook et al, 2002; Boyault et al, 2006). Even though HDAC6 contains both intrinsic nuclear import and export signals, it is mainly found in the cytoplasm (Verdel et al, 2000) where it localizes around the nucleus or at the leading edge of migrating cells (Hubbert et al, 2002). HDAC6 has also been shown to have a scaffold role for lymphocyte migration independent of its activity (Cabrero et al, 2006). Furthermore, the activity of HDAC6 promotes cell motility by regulation of membrane ruffle formation, macropinocytosis, and actin remodelling (Gao et al, 2007). Besides regulating cell motility, HDAC6 is also involved in the response of cells to various types of stress stimuli (Boyault et al, 2007b; Kawaguchi et al, 2003; Kwon et al, 2007), immune synapse organization, and the antigen-specific reorientation of the microtubule organizing center (MTOC) (Serrador et al, 2004).
In this paper we addressed the role of CYLD and specifically its N-terminal CAP-Gly domains in the regulation of cell proliferation both in primary keratinocytes and malignant melanoma cells. We found that CYLD associates with MTs and colocalizes primarily with acetylated tubulin. Activation of CYLD increases the levels of acetylated α-tubulin by interaction of the N-terminal domain of CYLD with the catalytic site of HDAC6. This interaction inhibits HDAC6-mediated tubulin deacetylation, allowing CYLD to translocate to the perinuclear region. Perinuclear localized CYLD induces a delay in the G1/S transition phase of the cell cycle through a Bcl-3-mediated pathway. In addition, CYLD is found in the midbody where it also associates with HDAC6 and regulates the rate of cytokinesis.
Results
Interaction of CYLD with tubulin and MT
We have earlier shown that UV or TPA treatment of primary mouse keratinocytes triggers perinuclear accumulation of CYLD, which correlates with its ability to interact with its downstream targets (Massoumi et al, 2006). The present work aimed at understanding whether an MT-dependent mechanism was involved in the translocation of CYLD. To explore this we used primary mouse keratinocytes, primary human melanocytes, and different human malignant melanoma cells. Analysis of subcellular localization of CYLD in primary mouse keratinocytes revealed that TPA treatment caused a robust increase in perinuclear, acetylated α-tubulin (Figure 1A), and a colocalization of CYLD with acetylated MTs (Figure 1A). However, we could observe only partial colocalization of CYLD with tyrosinated α-tubulin in the cytoplasm (Supplementary Figure S1A). Interestingly, all melanoma cell lines we have tested so far lack detectable CYLD expression (Massoumi et al, 2009). Therefore, we restored CYLD expression in two different malignant melanoma cell lines, MEL and Juso melanoma cells (Rothhammer et al, 2005), by lentivirally transducing full-length, EGFP-tagged CYLD cDNA. In melanoma cells EGFP–CYLD was constitutively present in the perinuclear region where it colocalized with acetylated tubulin (Supplementary Figure S1B, lower panels), while no colocalization with peripheral, tyrosinated α-tubulin could be observed (Supplementary Figure S1B, upper panels). These results suggest that CYLD colocalizes mainly with acetylated MTs in the perinuclear region, and this localization is constitutive in CYLD-transduced melanoma cells, whereas in primary mouse keratinocytes or primary human melanocytes (Supplementary Figure S1C) it is induced after TPA treatment or exposure to UV light. A constitutive perinuclear localization of CYLD has also been reported for other tumour cell lines (Regamey et al, 2003).
Figure 1.
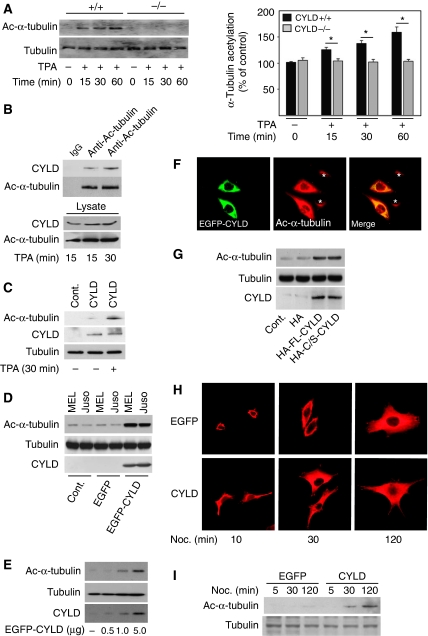
Interaction of CYLD with tubulin and microtubules. (A) The confocal plane of keratinocytes stained for acetylated α-tubulin and CYLD in the absence or presence of 100 nM TPA for 30 min. The fluorescence intensity linescan profile was generated through the merged image and demonstrates the dependence of the two fluorescent populations. (B) Immunoprecipitation of endogenous CYLD (left panels), α-tubulin (right panels), or control antibody (IgG) from human primary melanocytes, and immunoblotting against α-tubulin or CYLD. The lysate shows equal amount of protein used for immunoprecipitation. (C) Immunoprecipitation of α-tubulin from primary mouse keratinocytes in the absence or presence of nocodazole (10 μM for 60 min), and immunoblotting against α-tubulin, CYLD, or MAP4. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (D) Immunoprecipitation of endogenous CYLD from human melanocytes in the absence or presence of nocodazole (10 or 30 μM for 60 min), and immunoblotting against α-tubulin or CYLD. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (E) Microtubule co-sedimentation assay using in vitro translated, 35S-labelled CYLD (upper panel) or Bax (lower panel) in the presence or absence of taxol-stabilized microtubules. S: Supernatant and P: Pellet fraction of samples separated with SDS–PAGE after which the presence of 35S-labelled CYLD or Bax was detected by fluorography. (F) FLAG immunoprecipitates of HeLa cells transiently transfected with different FLAG-tagged deletion mutants of CYLD (1.0 μg) and analysed by immunoblotting with antibodies against α-tubulin.
Next we investigated whether CYLD, through its CAP-Gly domains, can interact with MTs. Immunoprecipitation of endogenous CYLD in melanocytes revealed an interaction with α-tubulin (Figure 1B). The interaction of CYLD with tubulin was confirmed by reverse immunoprecipitation of α-tubulin and subsequent immunoblotting against CYLD in melanocytes (Figure 1B) and in primary mouse keratinocytes (Figure 1C). As expected, immunoprecipitation of EGFP–CYLD from melanoma cells co-precipitated α-tubulin (Supplementary Figure S2A). Furthermore, exposure of keratinocytes (Figure 1C) or melanocytes (Figure 1D) to nocodazole, which depolymerises MTs, induced rapid dissociation of CYLD from MTs, suggesting that CYLD associates with polymerised tubulin. In contrast, the association of drug- and cold-resistant, MT-associated protein-4 (MAP4) (Webster and Bratcher, 2006), and tubulin was unaffected in the presence of nocodazole (Figure 1C). To confirm the association of CYLD with polymerised MTs, we performed MT-co-sedimentation assays using in vitro translated CYLD and in vitro assembled MTs. The majority of the CYLD protein was found in the heavy pellet fraction indicating that CYLD binds to polymerised MTs (Figure 1E, upper panels). To control the specificity of our assay, we used in vitro translated Bax, which does not bind MTs. As expected, Bax was exclusively found in the supernatant fraction both in the presence and absence of MTs (Figure 1E, lower panels).
To identify the interaction site on CYLD for α-tubulin binding, we transfected HeLa cells with cDNAs encoding FLAG-tagged, full-length CYLD or FLAG-tagged, CYLD-deletion mutants (Supplementary Figure S2B). FLAG pull-down assays confirmed the interaction between CYLD and α-tubulin, and further showed that the first and the second, but neither the third CAP-Gly nor the UCH, domain of CYLD interacted with α-tubulin (Figure 1F). Together these results indicate that CYLD associates preferentially with polymerised tubulin and that the first and second CAP-Gly domains of CYLD mediate the binding to MTs both in vitro and in vivo.
CYLD induces acetylation of α-tubulin and stabilization of MTs
As CYLD associates with MTs and colocalizes specifically with acetylated MTs, we hypothesized that CYLD might directly regulate α-tubulin acetylation to control its own localization. To investigate this we analysed the levels of acetylated tubulin in primary mouse keratinocytes. Both Cyld+/+ and Cyld−/− keratinocytes contained low levels of acetylated tubulin (Figure 2A). While TPA treatment did not change the total levels of α-tubulin, it increased the levels of acetylated α-tubulin in Cyld+/+ keratinocytes but not in Cyld−/− keratinocytes. (Figure 2A). This increase was accompanied by enhanced association of CYLD with acetylated α-tubulin (Figure 2B). Importantly, re-expression of CYLD in Cyld−/− keratinocytes restored the ability of these cells to respond to TPA by increasing the levels of acetylated α-tubulin (Figure 2C). Reconstitution of melanoma cells with EGFP–CYLD induced an almost sixfold increase in the levels of acetylated α-tubulin (Figure 2D). Furthermore, transient transfection of melanoma cells with increasing concentrations of the EGFP–CYLD cDNA showed direct correlation between levels of CYLD expression and acetylated α-tubulin (Figure 2E and F). These results suggest that CYLD increases the level of acetylated α-tubulin in both TPA-stimulated keratinocytes as well as in untreated melanoma cells. To determine whether the increase in α-tubulin acetylation depends on the deubiquitinase activity of CYLD, we transduced melanoma cells with catalytically inactive CYLD (CYLDC/S). CYLDC/S increased the acetylation of α-tubulin to the same extent as with full-length, wild-type CYLD (Figure 2G), indicating that the UCH domain of CYLD is not required for α-tubulin acetylation.
Figure 2.
CYLD increases the level of acetylated α-tubulin. (A) Analysis of the levels (left panels) and quantification (right panel) of acetylated α-tubulin and total tubulin in Cyld+/+ and Cyld−/− keratinocytes treated with TPA (15, 30 and 60 min). (B) Immunoprecipitation of endogenous CYLD from keratinocytes treated with 100 nM TPA and immunoblotting against acetylated-α-tubulin or CYLD. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (C) Analysis of the levels of acetylated α-tubulin and total tubulin in untreated or TPA-treated (30 min) Cyld−/− keratinocytes (control) or Cyld−/− keratinocytes transiently transfected with full-length-CYLD for 48 h. (D) Analysis of the levels of acetylated α-tubulin, total tubulin, or CYLD in two different melanoma cell lines (MEL Im and Juso) infected with mock, EGFP, or EGFP–CYLD lentivirus (1 × 105 IU) for 24 h. The medium was replaced two times before the cells were lysed and used for immunoblotting against α-tubulin, total tubulin, or CYLD. (E) Analysis of the levels of acetylated α-tubulin, total tubulin, and CYLD in melanoma cells (Juso) transiently transfected with increasing concentrations of EGFP–CYLD cDNA. (F) The confocal plane of human malignant melanoma cell line (Juso) transiently transfected with 1.0 μg EGFP–CYLD (green) for 24 h and stained for acetylated α-tubulin (red). The asterisk indicates cells lacking CYLD expression. (G) Analysis of the levels of acetylated α-tubulin and total tubulin in melanoma cells transduced with 1 × 105 IU of HA, HA–CYLD, or the catalytically inactive mutant HA–CYLDC/S lentivirus for 24 h. The medium was replaced two times before the cells were lysed and used for immunoblotting against α-tubulin, total tubulin, or CYLD. (H) Immunofluorescence staining of microtubule re-growth of EGFP and EGFP–CYLD lentivirus-infected melanoma cells after treatment with nocodazole (50 μM) for 10 min, followed by a washout and incubation in fresh culture medium at 37°C for 10, 30, or 120 min. (I) EGFP or EGFP–CYLD lentivirus-infected melanoma cells were used to analyse the levels of acetylated α-tubulin and total tubulin in untreated cells or cells treated with nocodazole (50 μM) for 10 min followed by a washout and incubation in fresh culture medium at 37°C for 10, 30, or 120 min before cell lysis. *P<0.05.
To investigate whether the association of CYLD with MTs influences their dynamic properties, we treated melanoma cells with nocodazole and monitored MT re-growth after nocodazole washout. Polymerised MT fibres in CYLD-expressing melanoma cells were evident already 10 min after the washout (Figure 2H) and acetylated α-tubulin after 30 min (Figure 2I). In EGFP-expressing melanoma cells, however, MT fibres were detected only 30 min after nocodazole washout (Figure 2H), and we failed to observe acetylation of α-tubulin even 2 h after nocodazole washout (Figure 2I). This suggests that CYLD regulates the acetylation of tubulin and also influences the polymerization rate of MTs.
Next, we analysed the effect of CYLD on MT depolymerisation by treating EGFP–CYLD- or EGFP-expressing melanoma cells with increasing concentrations of nocodazole and found that in the presence of CYLD, MTs began to partially depolymerise at 5-μM concentrations (with some MT fibres still detectable) (Supplementary Figure S3A). Total depolymerisation was achieved with 50 μM nocodazole. In sharp contrast, already 1 μM nocodazole induced partial depolymerisation of MT fibres in EGFP-expressing cells and 5 μM nocodazole was sufficient to induce complete depolymerisation (Supplementary Figure S3A). Furthermore, treatment with 1 μM nocodazole did not change the level of acetylated tubulin in EGFP–CYLD-expressing melanoma cells (Supplementary Figure S3B), while treatment with 50 μM of nocodazole resulted in almost complete deacetylation of α-tubulin (Supplementary Figure S3B). These results indicate that CYLD stabilizes the MT network and protects the MT network from nocodazole-induced depolymerisation.
CYLD binding to HDAC6 increases α-tubulin acetylation
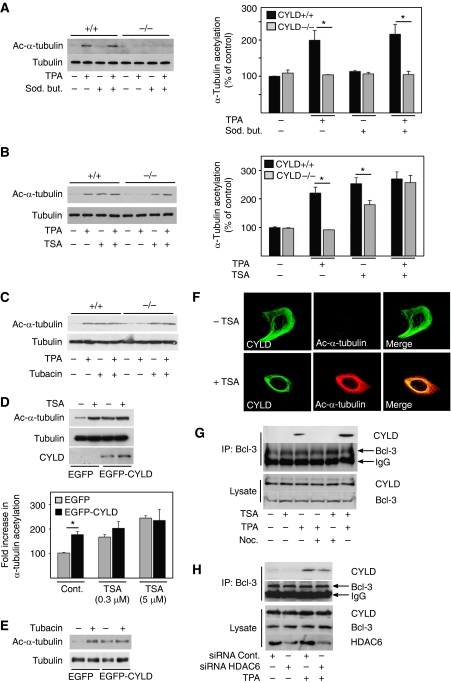
To analyse whether CYLD regulates tubulin acetylation by inhibiting HDAC6, we treated keratinocytes and melanoma cells with different HDAC inhibitors: tubacin, a small-molecule inhibitor of HDAC6; trichostatin-A (TSA), a chemical inhibitor of class-I and II HDACs, including HDAC6; and sodium butyrate, a potent inhibitor of class-I and II HDACs except HDAC6. Sodium butyrate treatment did not affect the levels of acetylated tubulin either in keratinocytes or in melanoma cells (Figure 3A and Supplementary Figure S3C). In addition, TPA stimulation of Cyld+/+ keratinocytes led to an increase in acetylated α-tubulin levels both in the presence and absence of sodium butyrate (Figure 3A). In contrast, treatment of Cyld+/+ keratinocytes with either TPA or TSA alone increased acetylated α-tubulin levels to a similar extent, while simultaneous treatment with both the compounds failed to induce a synergistic effect (Figure 3B). Moreover, TSA treatment of Cyld−/− keratinocytes increased the level of acetylated α-tubulin, while TPA treatment had no effect (Figure 3B). A similar effect was observed in keratinocytes treated with tubacin, which is a selective inhibitor for HDAC6 (Figure 3C). Furthermore, TSA or tubacin treatment increased the levels of acetylated α-tubulin in EGFP-expressing melanoma cells (Figure 3D and E), while treatment of EGFP–CYLD-expressing melanoma cells with TSA or tubacin failed to increase the levels of acetylated α-tubulin (Figure 3D and E). Taken together, these findings suggest that CYLD, TSA, and tubacin act within the same pathway to inhibit HDAC6, which results in elevated levels of acetylated α-tubulin.
Figure 3.
CYLD inhibits the activity of HDAC6 and tubulin deacetylation. (A) Analysis of the levels (left panels) and quantification (right panel) of acetylated α-tubulin and total tubulin in Cyld+/+ and Cyld−/− keratinocytes treated with 1 mM sodium butyrate (4 h) before stimulation with 100 nM TPA (30 min). (B) Analysis of the levels (left panels) and quantification (right panel) of acetylated α-tubulin and total tubulin in Cyld+/+ and Cyld−/− keratinocytes treated with 0.3 μM TSA (60 min) before stimulation with 100 nM TPA (30 min). (C) Analysis of the levels of acetylated α-tubulin and total tubulin in Cyld+/+ and Cyld−/− keratinocytes treated with 2 μM tubacin (4 h) before stimulation with 100 nM TPA (30 min). (D) Analysis of the levels (left panels) and quantification (right panel) of acetylated α-tubulin and total tubulin in untreated or TSA (0.3 or 5 μM for 60 min)-treated, EGFP or EGFP–CYLD lentivirus-infected melanoma cells. (E) Analysis of the levels of acetylated α-tubulin and total tubulin in untreated or tubacin (2 μM for 4 h)-treated, EGFP or EGFP–CYLD lentivirus-infected melanoma cells. (F) The confocal plane of primary mouse keratinocytes untreated or treated with 0.3 μM TSA (15 min) and stained for acetylated α-tubulin (red) and CYLD (green). (G) Untreated, TPA- (100 nM for 30 min), nocodazole- (10 μM for 60 min), or TSA (0.3 μM for 60 min)-treated Cyld+/+ keratinocytes immunoprecipitated with antibodies against Bcl-3 and immunoblotted against CYLD. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (H) Cyld+/+ keratinocytes were transiently transfected with control or HDAC6 siRNA (24 h) in the absence or presence of TPA (100 nM for 30 min), followed by immunoprecipitation of endogenous Bcl-3 and immunoblotting against CYLD. The lysate shows equal amount of protein used for immunoprecipitation and knockdown efficiency of HDAC6. *P<0.05.
Next we investigated whether levels of acetylated tubulin regulate the subcellular localization of CYLD. To analyse this, we inhibited HDAC6 in Cyld+/+ keratinocytes using TSA or by downregulating HDAC6 using siRNA. In the absence of TSA, CYLD localized throughout the cytoplasm (Figure 3F), while in the presence of TSA or in HDAC6-depleted cells, the levels of acetylated tubulin increased and CYLD translocated to the perinuclear region where it strongly colocalized with acetylated MTs (Figure 3F and Supplementary Figure S3D). TPA treatment of HDAC6-depleted cells induced a similar affect as HDAC6 siRNA alone (Supplementary Figure S3D). In addition, treatment of keratinocytes with TSA caused an increase in the interaction of CYLD with tubulin as compared with that in untreated cells (Supplementary Figure S3E).
Since CYLD regulates tumour cell proliferation through direct association with Bcl-3, which in turn inhibits the nuclear translocation and activity of Bcl-3 (Massoumi et al, 2006), we investigated whether intact MTs are required for this interaction. As expected, TPA induced interaction of CYLD with Bcl-3, while this interaction (Figure 3G), as well as perinuclear localization of CYLD (Supplementary Figure S4A), was prevented in the presence of nocodazole. Furthermore, TSA together with TPA, but not in combination with nocodazole, was able to rescue this interaction (Figure 3G). However, TSA treatment or HDAC6 depletion alone did not induce interaction of CYLD with Bcl-3 (Figure 3G and H).These results suggest that inhibition of HDAC6 and subsequent accumulation of acetylated MTs facilitates the translocation of CYLD to the perinuclear region, while binding to Bcl-3 requires both an intact network of acetylated MTs and an additional signal induced by TPA.
Association of CYLD and HDAC6 in the presence of TPA
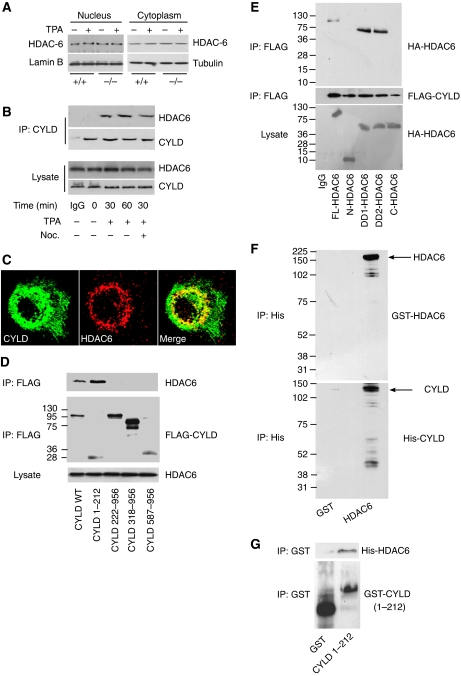
To investigate how CYLD regulates HDCA6 function we analysed whether CYLD regulates the expression of HDAC6. Western blot analyses excluded reduced HDAC6 expression as the cause for diminished HDAC6 function in keratinocytes or melanoma cell lines (Figure 4A and Supplementary Figure S4B). Furthermore, similar levels of HDAC6 were found in nuclear and cytoplasmic extracts from TPA-treated Cyld+/+ and Cyld−/− keratinocytes, as well as from melanoma cells before and after EGFP–CYLD transduction (Figure 4A and Supplementary Figure S4B), indicating that CYLD does not interfere with the subcellular distribution of HDAC6. We also analysed SIRT2, another α-tubulin deacetylase, and did not detect SIRT expression in keratinocytes (data not shown), suggesting that HDAC6 is the major α-tubulin deacetylase in keratinocytes.
Figure 4.
Interaction of CYLD with HDAC6. (A) Cytoplasmic and nuclear extracts of keratinocytes (Cyld+/+ and Cyld−/−) in the presence or absence of TPA (100 nM for 30 min), immunoblotted with antibodies against HDAC6, lamin-B, and tubulin. (B) Immunoprecipitation of CYLD and immunoblotting against HDAC6 and CYLD untreated, TPA- (100 nM for 30 min) and/or nocodazole (10 μM for 60 min)-treated Cyld+/+ keratinocytes. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (C) The confocal plane of Cyld+/+ keratinocytes stimulated for 30 min with 100 nM TPA and stained for HDAC6 (red) and CYLD (green). (D) HeLa cells were transiently transfected with FLAG-tagged, wild-type or different deletion mutants of CYLD, incubated for 24 h, lysed, immunoprecipitated using α-FLAG M2 agarose beads, and immunoblotted against endogenous HDAC6 or FLAG. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (E) HeLa cells were transiently transfected with HA-tagged, full-length or different deletion mutants of HDAC6 and FLAG-tagged, full-length CYLD. After transfection (24 h) the cells were lysed, immunoprecipitated using α-FLAG M2 agarose beads, and immunoblotted against HA or FLAG. The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (F) His-tagged CYLD was purified from COS cells transiently transfected with His–CYLD (1.0 μg for 48 h) and incubated with recombinant N-terminal GST–HDAC6 for 30 min followed by precipitation of His-tagged protein complexes using Ni–NTA agarose beads. The bound proteins were recovered from the beads and analysed by immunoblotting against GST to detect GST–HDAC6 or His to detect His–CYLD. (G) Purified GST or GST–CYLD1–212 protein was incubated with His–HDAC6 in vitro for 30 min followed by GST pull down using glutathione–agarose beads. The complex was recovered from the beads and analysed by immunoblotting against His to detect His–HDAC6 or GST to detect GST–CYLD1–212.
To analyse whether CYLD directly influences HDAC6 activity, we first performed co-immunoprecipitation and immunofluorescence experiments using primary keratinocytes. We observed both co-precipitation (Figure 4B) and colocalization (Figure 4C) of the proteins after TPA treatment, indicating that they form a protein complex in response to CYLD activation. The interaction between CYLD and HDAC6 was retained in the presence of nocodazole (Figure 4B), indicating that the interaction between CYLD and HDAC6 does not require intact MTs. Similarly, HDAC6 co-precipitated with EGFP–CYLD in melanoma cells, and this interaction was also retained in the presence of nocodazole (Supplementary Figure S4C). Furthermore, FLAG pull-down assays with a series of in vitro translated, FLAG-tagged CYLD-deletion mutants (Supplementary Figure S2B) showed that the N-terminal region of CYLD (CYLD1–212) harboured the ability to interact with purified HDAC6 protein (Figure 4D).
To define which domain(s) in HDAC6 is required for the interaction with CYLD, a series of HA-tagged HDAC6-deletion mutants (Zhang et al, 2003) were transiently transfected together with full-length, FLAG-tagged CYLD into HeLa cells. FLAG pull-down assays of CYLD demonstrated that the deletion mutants containing the HDAC6 domain DD1 or DD2 interacted with CYLD (Figure 4E). To confirm this interaction we expressed recombinant GST–HDAC6, His–CYLD, and GST alone as control (Supplementary Figure S5A). HDAC6 co-precipitated with His–CYLD-bound Ni–NTA agarose, indicating association of these two proteins (Figure 4F). Furthermore, GST pull-down assays using the N-terminal region of CYLD (GST–CYLD1–212) and the purified form of His-tagged, full-length HDAC6 confirmed an interaction between CYLD and HDAC6 (Figure 4G).
CYLD inhibits HDAC6 activity by binding to its catalytic domains
As it was shown earlier that DD1 and DD2 are critical for the catalytic activity of HDAC6 (Zhang et al, 2006), we hypothesized that binding of CYLD to these domains leads to HDAC6 inactivation. To test this hypothesis, we performed in vitro tubulin deacetylation assays. We incubated endogenous HDAC6 and/or EGFP–CYLD immunoprecipitates from melanoma cells in the absence or presence of TSA together with MAP-enriched polymerised MTs purified from bovine brain. Whereas HDAC6 alone induced deacetylation of α-tubulin, CYLD or TSA treatment could maintain acetylated α-tubulin even in the presence of HDAC6 (Figure 5A). To confirm the specificity of this effect, we transiently transfected COS cells with increasing concentrations of FLAG–CYLD or HA–HDAC6 cDNA. The polymerised MTs were then incubated with CYLD and/or HDAC6 immunoprecipitates after peptide elution. We found a direct correlation between the levels of CYLD and acetylated α-tubulin in vitro (Figure 5B). To identify the domain of CYLD responsible for inhibition of HDAC6, we transiently transfected COS cells with different fragments of CYLD (Supplementary Figure S2B) or HDAC6. The polymerised MTs were then incubated with immunoprecipitates of these fragments together with HDAC6. As expected, only the CYLD1–212 fragment, which mediates the association of CYLD with HDAC6 (Figure 4D), was able to inhibit HDAC6 activity and subsequent α-tubulin deacetylation (Figure 5C).
Figure 5.
The N-terminal domain of CYLD is responsible for inhibition of HDAC6 activity. (A) MAP-enriched bovine tubulin (cytoskeleton) was polymerised into microtubules in the absence of taxol or glycerol by incubation for 30 min at 35°C. The polymerised microtubules were incubated with endogenous HDAC6 and/or EGFP–CYLD immunoprecipitates from melanoma cells in the absence or presence of TSA (0.5 μM) at 37°C for 2 h. Samples were placed on ice for 15 min and the supernatant was collected by centrifugation and analysed by immunoblotting. (B) COS cells were transiently transfected with different concentrations of FLAG–CYLD (0.5, 1.0 and 3.0 μg) or HA–HDAC6 (1.0 μg) for 24 h. Polymerised microtubules were incubated with CYLD and/or HDAC6 eluted from the corresponding immunoprecipitates at 37°C for 2 h. Samples were placed on ice for 15 min and the supernatant was collected by centrifugation and analysed by immunoblotting. (C) COS cells were transiently transfected with FLAG-tagged, wild-type or truncation mutants of CYLD or HA-tagged HDAC6 constructs (1.0 μg of each for 24 h). Polymerised microtubules were then incubated with CYLD and/or HDAC6 eluted from the corresponding immunoprecipitates at 37°C for 2 h. Samples were placed on ice for 15 min and the supernatant was collected by centrifugation and analysed by immunoblotting. (D) Immunoprecipitation of endogenous HDAC6 from Cyld+/+ and Cyld−/− keratinocytes in the absence or presence of TPA (100 nM for 30 min) and immunoblotting against tubulin and HDAC6 in the absence or presence of TPA (100 nM for 30 min). The lysate (lower panel) shows equal amount of protein used for immunoprecipitation. (E) Immunoblot analysis of HDAC6, acetylated α-tubulin, and total tubulin in EGFP- or EGFP–CYLD-expressing melanoma cells before or after 24 h of transient transfection with HDAC6 siRNAs (0.2 μM). (F) Immunoblot analysis of HDAC6, acetylated α-tubulin, and total tubulin in untreated or TPA (100 nM for 30 min)-treated Cyld−/− keratinocytes before or after transient transfection with HDAC6 siRNAs.
Next we tested whether TPA-mediated CYLD activation inhibits HDAC6 activity by regulating its interaction with tubulin. To this end, we immunoprecipitated HDAC6 from Cyld+/+ and Cyld−/− keratinocytes and found that TPA treatment, which was found to induce the interaction of CYLD and HDAC6, did not affect the association of HDAC6 with α-tubulin (Figure 5D). Similarly, EGFP–CYLD did not affect the association of HDAC6 with α-tubulin in the melanoma cells (Supplementary Figure S5B).
If loss of CYLD was directly responsible for elevated HDAC6 activity and increased deacetylation of α-tubulin, siRNA-mediated depletion of HDAC6 in EGFP-expressing melanoma cells or Cyld−/− keratinocytes should restore α-tubulin acetylation. In line with this hypothesis, the levels of acetylated α-tubulin increased in melanoma cells upon HDAC6 depletion (Figure 5E). Furthermore, HDAC6-depleted Cyld−/− keratinocytes showed significant increase in α-tubulin acetylation irrespective of whether they were untreated or treated with TPA (Figure 5F).
Delay in the G1/S-phase of the cell cycle and cytokinesis induced by CYLD
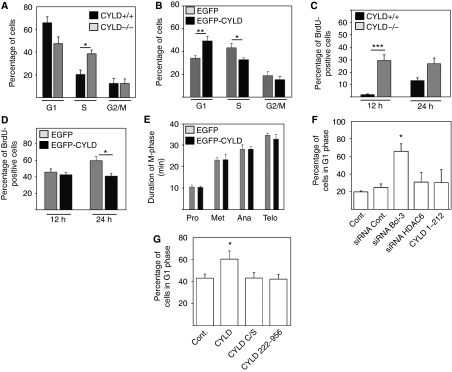
To determine whether CYLD, through its effects on MTs, can regulate the duration of the cell cycle, we used serum starvation in combination with contact inhibition to arrest keratinocytes in G1-phase and nocodazole treatment to synchronize melanoma cells in G2/M-phase, and then analysed cell-cycle progression using flow cytometry (Supplementary Figures S6 and 7). The release of synchronized, TPA-treated keratinocytes caused a delay in G1-to-S phase transition in Cyld+/+ but not in Cyld−/− keratinocytes (Figure 6A and Supplementary Figure S6). Similarly, release of synchronized, nocodazole-treated melanoma cells revealed a significant delay in G1-to-S phase progression and an accumulation in S-phase of EGFP–CYLD-expressing cells as compared with EGFP-expressing control cells (Figure 6B and Supplementary Figure S7). To further corroborate this finding, we performed BrdU pulse labelling of TPA-stimulated keratinocytes as well as melanoma cells. TPA-stimulated Cyld+/+ keratinocytes and EGFP–CYLD-expressing melanoma cells showed reduced levels of BrdU-positive nuclei as an indication of delayed S-phase progression in the presence of activated CYLD (Figure 6C and D). To exclude the possibility that the delay in the G1/S-phase transition in CYLD-expressing cells is due to differences in mitosis, we quantified mitotic intervals by live-cell imaging. The experiments revealed that EGFP–CYLD-expressing melanoma cells and EGFP-expressing control cells displayed similar rates of mitosis (Figure 6E).
Figure 6.
CYLD induces a delay in the G1/S phase of the cell cycle. (A) Cell-cycle distribution of Cyld+/+ and Cyld−/− keratinocytes after synchronization by serum starvation and contact inhibition for 24 h and stimulation with 100 nM TPA for 24 h. (B) Cell-cycle distribution of EGFP or EGFP–CYLD-infected melanoma cells 16 h after synchronization with 30 μM nocodazole. (C) BrdU pulse labelling of keratinocytes synchronized by serum starvation and contact inhibition followed by BrdU exposure (10 μM) for 12 or 24 h after the release. (D) BrdU pulse labelling of EGFP and EGFP–CYLD lentivirus-infected melanoma cells after arrest with with nocodazole (30 μM for 16 h) and exposure to BrdU (10 μM) for 12 or 24 h after the release. (E) Duration of mitosis in EGFP- or EGFP–CYLD-infected melanoma cells after synchronization by a double thymidine (2 mM) block. (F) The G1 cell-cycle phase of melanoma cells transiently transfected with EGFP (control), CYLD1–212, or/and Bcl-3 or HDAC6 siRNA and synchronized with nocodazole. (G) The G1 cell-cycle phase of melanoma cells transiently transfected with mock (control), CYLD, CYLDC/S, or CYLD222–956 and synchronized with nocodazole. *P<0.05, **P<0.01, ***P<0.001.
We have reported earlier that CYLD decreases the proliferation of keratinocytes (Massoumi et al, 2006) and melanoma cells (Massoumi et al, 2009) by retaining Bcl-3 in the cytoplasm and thereby reducing the expression of cyclin-D1. Interestingly, depletion of Bcl-3 in EGFP-expressing or control melanoma cells increased the number of cells in G1-phase (Figure 6F), while expression of CYLD1–212 or siRNA-mediated depletion of HDAC6 had no significant effect (Figure 6F). To further assess whether the catalytic activity and/or HDAC6 and MT binding of CYLD are necessary to induce a delay in the cell cycle, we analysed the duration of the cell cycle in melanoma cells expressing full-length CYLD, catalytically inactive CYLDC/S, and a deletion mutant that lacks the first CAP-Gly domains (CYLD222–956) and does not bind HDAC6. Full-length CYLD induced a delay in G1/S transition, whereas the catalytically inactive CYLDC/S or CYLD222–956 had no effect. Together, these results suggest that both deubiquitinase activity as well as HDAC6 binding are required for CYLD to regulate the cell cycle (Figure 6G).
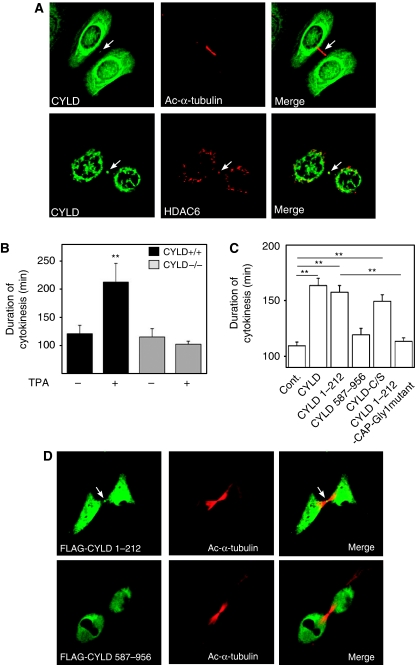
It has been suggested earlier that the localization of HDAC6 in the midbody might regulate mitosis by affecting MT dynamics during cytokinesis (Zhang et al, 2003). This correlates with the notion that the cytoplasmic bridge connecting the two daughter cells contains high levels of acetylated MTs (Piperno and Fuller, 1985; Schatten et al, 1988; Chu and Klymkowsky, 1989). We observed that CYLD localizes to the midbody during cytokinesis (Figure 7A, upper panels). The localization of CYLD to the midbody has also previously been reported for dividing HeLa cells overexpressing CYLD (Stegmeier et al, 2007). In addition, we observed that CYLD colocalized with HDAC6 in the midbody of both non-stimulated and TPA-stimulated primary Cyld+/+ keratinocytes (Figure 7A, lower panels).
Figure 7.
The N-terminal part of CYLD induces a delay in the cytokinesis rate. (A) The confocal plane of keratinocytes stained for acetylated α-tubulin, CYLD, or HDAC6 as indicated. The arrows indicate the localization of CYLD and HDAC6 in the midbody. (B) Duration of cytokinesis of Cyld+/+ and Cyld−/− cells after TPA (100 nM) treatment. (C) Duration of cytokinesis in melanoma cells (Juso) transiently transfected with mock (control), CYLD, CYLD1–212, CYLD587–956, CYLDC/S, or CYLD1–212–CAP1-Gly mutant. (D) The confocal plane of melanoma (Juso) transfected with expression vectors FLAG-CYLD1–212, or FLAG–CYLD587–956 and stained for acetylated α-tubulin and FLAG. The arrow indicates the localization of CYLD1–212 in the midbody. **P<0.01.
To analyse whether the observed localization of CYLD in the midbody could affect cytokinesis, we determined the duration of cytokinesis using live-cell imaging. The experiments revealed no differences in the duration of cytokinesis in non-stimulated Cyld+/+ and Cyld−/− keratinocytes (Figure 7B), while treatment of cells with TPA induced a significant delay in cytokinetic rate of Cyld+/+ as compared with that of Cyld−/− keratinocytes (Figure 7B). This phenotype could also be detected in EGFP–CYLD-expressing melanoma cells as compared with EGFP-expressing control cells (Figure 7C). Furthermore, the N-terminal but not the C-terminal domain of CYLD localized to the midbody of melanoma cells (Figure 7D) and induced a delay in cytokinesis (Figure 7C). However, expressing CYLD1–212-carrying mutations in the CAP-Gly1 domain (Supplementary Figure S2B), to disrupt MT binding (Feierbach et al, 1999), failed to induce a delay in cytokinesis, whereas the catalytically inactive mutant CYLDC/S retained this ability (Figure 7C). Altogether these results indicate that inhibition of Bcl-3 by the deubiquitinase activity of CYLD induces a delay in the G1/S transition, while the N-terminal domain of CYLD, which binds MTs and inhibits HDAC6 activity, regulates the perinuclear localization of CYLD and reduces the rate of cytokinesis.
Discussion
CYLD is a deubiquitination enzyme, which acts primarily as a tumour-suppressor gene. Mutations in the CYLD gene were discovered in patients suffering from cylindromatosis, which is a benign tumour derived from the cells of the skin (Bignell et al, 2002). We have recently shown that CYLD expression is very low in melanoma cells due to transcriptional suppression by high levels of Snail (Massoumi et al, 2009). Interestingly, gene mutation studies on cylindroma patients showed that the known mutations can cause either loss of expression or expression of C-terminally truncated versions of CYLD. The C-terminal part of CYLD harbours the UCH domain, which is responsible for the deubiquitinase activity of CYLD (Brummelkamp et al, 2003; Kovalenko et al, 2003; Trompouki et al, 2003; Massoumi et al, 2006). So far, no cylindroma patients with mutations giving rise to N-terminal truncations of CYLD have been identified (Massoumi and Paus, 2007). It was speculated that this could be due to the ability of this region to delay tumour growth and thus be associated with lower disease penetrance. In the present study we investigated the role of the N-terminal CAP-Gly domains of CYLD in the growth and proliferation of both primary and transformed cells derived from the skin.
The N-terminal region of CYLD contains three CAP-Gly motifs. We found that two of them (CAP-Gly1 and CAP-Gly2) associated with tubulin and inhibited tubulin deacetylation. In addition, we found that TPA treatment of Cyld+/+ keratinocytes caused redistribution of CYLD and induced its accumulation in the perinuclear region, which has been previously shown to be critical in mediating the interaction with its downstream substrate Bcl-3 (Massoumi et al, 2006). In addition, TPA treatment lead to a marked increase in the levels of acetylated MTs in this area, and CYLD was found to extensively colocalize and associate with acetylated MTs. These observations suggest that CYLD colocalizes preferentially with acetylated MTs, and that the TPA-mediated increase of acetylated MTs in the perinuclear region might in fact regulate the translocation of CYLD to this area. This hypothesis is supported by the observation that inhibition of tubulin deacetylation by downregulation of HDAC6 or inhibition of its activity by TSA induced a similar translocation of CYLD to the perinuclear area where it colocalized with the abundant acetylated MTs, whereas depolymerisation of MTs inhibited the translocation.
HDAC6, which has been shown to operate as a α-tubulin-specific deacetylase (Hubbert et al, 2002), contains two intact catalytic domains in the central region of the protein. Mutations in the catalytic domains or treatment of cells with the HDAC6 inhibitor TSA abrogates the catalytic activity (Zhang et al, 2006) and increases the pool of acetylated tubulin. Several lines of evidence suggest that CYLD regulates the levels of acetylated tubulin by acting as an endogenous inhibitor of HDAC6. First, inhibition of HDAC6 by TSA or tubacin treatment failed to induce an additive effect on acetylated tubulin in the presence of CYLD, suggesting that TSA, tubacin, and CYLD act within the same pathway. In addition, CYLD colocalized with HDAC6 in the perinuclear region, and the endogenous proteins were found in the same protein complex. Finally, we observed that CYLD binds to the catalytic domains of this enzyme and inhibits its tubulin deacetylase activity in vitro.
What are the functional consequences of CYLD-mediated inhibition of HDAC6 and the increased levels of acetylated MTs? We observed that CYLD induces a delay in the G1/S transition of the cell cycle, both in melanoma cells as well as in keratinocytes. This is in contrast to an earlier study where downregulation of CYLD by siRNA in HeLa cells induced a delay in G2/M transition of the cell cycle (Stegmeier et al, 2007). This discrepancy could be explained by different functions of CYLD depending on the tissue/cell type in combination with the requirement of a specific stimulus such as TPA and/or by siRNA-mediated downregulation of CYLD versus complete absence of the protein. We have previously shown that CYLD, through deubiquitination of Bcl-3, prevents its nuclear translocation. This in turn inhibits the transcriptional activity of NF-κB p50/p52 and reduces the expression of cyclin-D1 (Massoumi et al, 2006). Since expression of cyclin-D1 is critical for the G1/S transition of the cell cycle, it is very likely that CYLD regulates the G1/S transition by inhibiting the expression of cyclin-D1. Interestingly, depletion of HDAC6 or expression of the N-terminal domain of CYLD, which interacts and inhibits HDAC6, was unable to increase the number of G1-phase cells, suggesting that the inactivation of HDAC6 and hyperacetylation of MTs alone is not the cause of the G1/S cell-cycle delay. This is supported by the finding that depletion of HDAC6 was not sufficient to induce the interaction of CYLD with Bcl-3, but TPA-mediated activation of CYLD is additionally required. However, a deletion mutant of CYLD, which is unable to bind HDAC6, but contains an intact catalytical domain, did not induce a delay in cytokinesis. Thus, we suggest that the association of CYLD with acetylated MTs leads to its perinuclear accumulation, which when followed by an additional TPA-mediated signal prevents nuclear accumulation of Bcl-3, leading to decreased cyclin-D1 expression and delay in G1/S transition. It is interesting to note that a direct interaction between HDAC6 and Bcl-3 has been reported earlier in other cell types (Viatour et al, 2004), suggesting that formation of these protein complexes could be spatiotemporally linked.
The finding that CYLD colocalizes with HDAC6 in the midbody between bundles of acetylated MTs in both keratinocytes and melanoma cells, prompted us to investigate the rate of cytokinesis in these cells. Although CYLD is present in the midbody of primary keratinocytes both before and after TPA treatment, CYLD was found to induce a delay in cytokinesis only after TPA treatment. We could also detect a reduction in the rate of cytokinesis in melanoma cells expressing EGFP–CYLD, and in line with the effect of EGFP–CYLD on G1/S-phase transition of melanoma cells, also this effect did not require an additional stimulus. Interestingly, the N-terminal domain, which binds to tubulin and HDAC6, but not the UCH-containing C-terminal domain of CYLD, was able to localize to the midbody and induce a delay in cytokinesis. In addition, mutations in the CAP-Gly1 domain, which impair MT binding, abolished this effect, whereas a catalytically inactive CYLD had no effect. This suggests that localization of CYLD to the midbody is important for its effect on cytokinesis, whereas deubiquitinase activity of CYLD is not required for this function. The reason why CYLD does not have an effect on cytokinesis in the absence of TPA treatment in the primary keratinocytes, despite its presence in the midbody, is unclear. It is possible that TPA induces additional changes in CYLD itself or in unidentified inhibitory proteins, which are necessary for the function of CYLD. This hypothesis is supported by our finding that CYLD is unable to bind HDAC6 in the absence of TPA treatment. The molecular details of this process need to be addressed in future studies.
The precise role of acetylated MTs and tubulin acetylation in the regulation of the cell cycle and mitosis is not understood, although it is known that the cytoplasmic bridge connecting the two daughter cells during cytokinesis is highly enriched in acetylated tubulin in many cell types (Piperno and Fuller, 1985; Schatten et al, 1988; Chu and Klymkowsky, 1989). Previous studies have shown that the degree of tubulin acetylation reflects the dynamics of MTs (reviewed by Westermann and Weber, 2003), and that stable, nocodazole-resistant MTs are essential for recruitment of myosin-II and RhoA to allow contraction of the cleavage furrow during cytokinesis (Bement et al, 2005). In addition, tubulin acetylation itself regulates the recruitment and dynamics of the MT motor protein kinesin-1, which regulates long-distance intracellular delivery (Reed et al, 2006). Interestingly, HDAC6-mediated tubulin acetylation was recently shown to regulate intracellular transport in neurons (Dompierre et al, 2007). It is, therefore, possible that CYLD, through its effect on HDAC6 or a direct effect on the MTs, regulates the dynamics of MTs and/or recruitment of proteins required for successful execution of cytokinesis. Interestingly, other tumour suppressors have previously been shown to use tubulin acetylation as mechanism to regulate the cell cycle. Loss of the tumour suppressor LAPSER1, which also localizes to the midbody, leads to a decrease in acetylated tubulin accompanied by an increase in the rate of cytokinesis and proliferation (Sudo and Maru, 2007). The tumour suppressor CHFR also localizes to the midbody and regulates the levels of acetylated α-tubulin in the mitotic spindle during cytokinesis (Privette et al, 2008).
Taken together, CYLD simultaneously employs two functional properties to negatively regulate the proliferation rates of keratinocytes and melanoma cells. The abnormal cell-cycle progression through G1 phases and cytokinesis are both hallmarks of cancer cell proliferation. Since many tumour types and cancer cell lines lack or have low CYLD expression (Hellerbrand et al, 2007; Massoumi et al, 2009), our findings provide a mechanistic explanation for their elevated proliferation rates.
Materials and methods
Cell culture and transfection
The melanoma cell lines Mel Im and Mel Juso were derived from metastases of malignant melanomas (Rothhammer et al, 2005). Isolation and culture of primary normal human epidermal melanocytes (NHEM) were performed as previously described by Rothhammer et al (2005). Primary keratinocytes were isolated from back skin of 8-week-old mice and cultured as described by Romero et al (1999).
Viral infections
Stable expression of EGFP, EGFP–CYLD, HA–CYLD, and HA–CYLDC/S in melanoma cells was achieved by lentiviral gene transfer. In brief, CMV promoter-driven lentiviral constructs were obtained by cloning EGFP, EGFP–CYLD, HA–CYLD, and HA–CYLDC/S into a lentiviral vector (p156rrlsinPPTCMV). The titre of the lentiviral particles was determined by measuring the amount of HIV p24 gag antigen by ELISA (Alliance; NEN). To calculate the amount of infectious units (IU), the p24 titre was correlated to the biological activity of a similar virus carrying a green-fluorescent protein (GFP) cassette by using serial dilutions of the GFP virus to transduce 293T cells (1 ng of p24=1 × 105 IU). Melanoma cells were infected with 1 × 105 IU for 24 h using the calcium phosphate method and the medium was replaced two times before the cells were used in different assays.
Transient transfections
CYLD truncation mutants were amplified by PCR and cloned into CMV promoter-driven pCS2 construct. The CAP-Gly1 mutant was generated by mutating the GFTDG sequence in the CYLD1–212 construct into AFADA, Transient transfection assays were performed in six-well plates at 80% confluence using the Polyfect transfection reagent (Qiagen) and 1.0 or 3.0 μg of DNA, and 10 or 30 μl of Polyfect were suspended in cell growth medium containing no serum or antibiotics. The mixture was incubated for 10 min at room temperature to allow complex formation. Cells were washed once with phosphate-buffered saline (PBS) before adding 1 ml of fresh cell growth medium containing serum and antibiotics, after which the transfection mixture was added to the cells. Experiments were performed 24 or 48 h after transfection. All transfection assays were repeated three times and performed in triplicates.
Depletion of Bcl-3 and HDAC6
SiRNAs (control, Bcl-3, and HDAC6) were purchased from Santa Cruz. SiRNA duplexes (0.2 μM) were mixed with Lipofectamine (Invitrogen) transfection reagent (10 μl) and incubated at room temperature for 15 min. For each transfection, 0.8 ml siRNA transfection mixture was added to 80% confluent cells in six-well tissue culture plate two times within 24 h. Experiments were performed 24 h after transfection.
Immunoblots and immunoprecipitation
For immunobloting, lysates from whole cells or cytosolic and nuclear extracts were resolved in SDS±PAGE gels and transferred to PVDF membranes, followed by incubation with primary antibodies anti-total α-tubulin; anti-tyrosinated α-tubulin; anti-acetylated α-tubulin; anti HA and α-FLAG M2 agarose beads (all from Sigma); anti-rat monoclonal tubulin (from Abcam); anti-HDAC6; anti-lamin-B and anti-Bcl-3 (from Santa Cruz). Rabbit polyclonal Cyld antibody has been described earlier (Massoumi et al, 2006).
For immunoprecipitations lysates were obtained from untreated or TPA-treated (100 nM) primary keratinocytes, human primary melanocytes (NHEM), or human malignant melanoma cell lines. The samples were centrifuged at 10 000 g for 20 min. For anti-CYLD immunoprecipitation the lysates were pre-cleared for 30 min at 4°C. The protein content was determined and compensated for equal content in all supernatants and used for immunoprecipitation. FLAG immunoprecipitation was performed by using anti-FLAG M2 affinity gel and eluted using FLAG-peptide purchased from Sigma. HA immunoprecipitation was performed by using anti-HA antibody and eluted using HA-peptide purchased from Sigma. EGFP immunoprecipitation was performed by using anti-GFP microBeads purchased from Miltenyi Biotech and used according to the manufacturer's instructions. Immunoblots were developed with the ECL-plus reagent (Amersham Biosciences) according to the manufacturer's instructions.
Immunofluorescence
Cells were grown on glass coverslips and fixed with ice-cold 100% methanol. After rehydration with PBS, unspecific binding was blocked with PBS containing 3% bovine serum albumin (BSA) and 5% goat serum, followed by incubation with primary antibodies against CYLD (Massoumi et al, 2006), tyrosinated tubulin (Chemicon), FLAG (Sigma), acetylated tubulin (Sigma), and HDAC6 (Santa Cruz), and fluorescence labelled secondary antibodies (Alexa, Invitrogen). Finally, the coverslips were washed and mounted on glass slides using Vectashield (Vector Laboratories). The fluorescence images were collected by laser-scanning confocal microscopy (DMIRE2; Leica) using Leica Confocal Software version 2.5 Build 1227 with × 100 oil-immersion objectives. All images were collected at room temperature.
Colocalization was measured by linescans from immunofluorescence images using the MetaMorph software.
In vitro tubulin deacetylation assay
Tubulin deacetylation assays were performed as described earlier (Hubbert et al, 2002). Briefly, MAP-enriched bovine tubulin (cytoskeleton) was polymerised into microtubules in the absence of paclitaxel or glycerol by incubation for 30 min at 35°C. The polymerised microtubules were incubated at 37°C for 2 h with HA–HDAC6 and/or FLAG–CYLD immunoprecipitates (after elution using HA respective FLAG peptide; Sigma) in the absence or presence of TSA (0.5 μM). Samples were then placed on ice for 15 min. The supernatant was subsequently collected by centrifugation and analysed by immunoblotting with antibodies against acetylated tubulin and total tubulin.
Flow cytometry and BrdU labelling
Malignant melanoma cells were synchronized by treatment with 100 ng/ml nocodazole for 16 h. Primary keratinocytes were synchronized by a combination of contact inhibition and serum starvation for 24 h, after which they were released into complete growth medium containing 1 μM TPA. After release the cells were harvested at the indicated time points and fixed with 70% ethanol. Cells were then stained with propidium iodide and analysed by flow cytometry (FACSCalibur, Becton Dickinson, Franklin Lakes, NY, USA) using FlowJo software (Tree Star Inc., Ashland, OR, USA).
BrdU pulse labelling was performed by arresting cells as described above. After release, the cells were exposed to BrdU (10 μM) for 12 or 24 h. The cells were then fixed with 4% paraformaldehyde and subsequently with 100% methanol. DNA was denatured using 1.5 M HCl and BrdU labelling was detected using anti-BrdU antibodies (Sigma) and fluorescent secondary antibodies.
Quantification of mitotic phases and cytokinesis
Malignant melanoma cells were plated on plastic culture dishes and synchronized using a double thymidine block. Briefly, cells were treated with 2 mM thymidine for 19 h. After a 5-h release in complete culture medium, the cells were treated with thymidine for another 16 h. After release, images of live cells were recorded with a Zeiss Axiovert 200M (Zeiss, Germany) with a cooled CCD camera (Roper Scientific, Princeton, NJ, USA). Image acquisition was performed with the MetaMorph software (Molecular Devices, Downington, PA, USA). For quantification of mitotic intervals, the duration of mitotic phases were defined as follows: prometaphase, from disassembly of nuclear membrane and nucleoli to equatorial arrangement of chromatin; metaphase, from the beginning to the end of equatorial arrangement of chromatin; anaphase, from the start of sister chromatid separation to reformation of nuclear envelopes and nucleoli; telophase, from the start of nuclear envelope and nucleolar reformation to formation of two daughter cells.
For quantification of cytokinesis cells were plated on plastic culture dishes in complete culture medium and stimulated with 100 nM TPA, when indicated. Images of live cells were recorded as described above. The duration of cytokinesis was quantified from the formation of two daughter cells to abscission.
Statistical analysis
Data are presented as mean±s.e.m. Statistical comparisons were assessed with analysis of variance (ANOVA) or Student's t-test after data were confirmed to fulfil the criteria of normal distribution and equal variance. If overall ANOVA tests were significant, we performed a post hoc Tukey test. P<0.05 was considered significant.
Supplementary Material
Supplementary Figures S1–S7
Review Process File
Acknowledgments
We thank Dr Anja Bosserhoff for discussion and for the melanoma cell lines Mel Im, Mel Juso, and primary NHEMs; Drs Ludger Hengst, Jillian Howlin, Stephan Geley, and Karin Sadoul for critically reading the paper; Dr Alexander Pfeifer and Bodo Haas for retro- or lentiviral gene transfer; and Dr Stuart L Schreiber for tubacin. This work was supported by the Swedish Society for Medical Research, Swedish Cancer Foundation, Swedish Medical Research Council, Crafoordska Foundation, Royal Physiographic Society in Lund, U-MAS Research Foundations (to RM), the Sigrid Juselius Foundation and the Finnish Cultural Foundation (to SAW), and the Max Planck Society (to RF).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Benink HA, von Dassow GJ (2005) A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol 170: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, Jones C, Hansen J, Blair E, Hofmann B, Siebert R, Turner G, Evans DG, Schrander-Stumpel C, Beemer FA, van Den Ouweland A et al. (2002) Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet 25: 160–165 [DOI] [PubMed] [Google Scholar]
- Boyault C, Gilquin B, Zhang Y, Rybin V, Garman E, Meyer-Klaucke W, Matthias P, Müller CW, Khochbin S (2006) HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J 25: 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault C, Sadoul K, Pabion M, Khochbin S (2007a) HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene 26: 5468–5476 [DOI] [PubMed] [Google Scholar]
- Boyault C, Zhang Y, Fritah S, Caron C, Gilquin B, Kwon SH, Garrido C, Yao TP, Vourc'h C, Matthias P, Khochbin S (2007b) HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev 21: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R (2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kB. Nature 424: 797–801 [DOI] [PubMed] [Google Scholar]
- Cabrero JR, Serrador JM, Barreiro O, Mittelbrunn M, Naranjo-Suárez S, Martín-Cófreces N, Vicente-Manzanares M, Mazitschek R, Bradner JE, Avila J, Valenzuela-Fernández A, Sánchez-Madrid F (2006) Lymphocyte chemotaxis is regulated by histone deacetylase 6, independently of its deacetylase activity. Mol Biol Cell 17: 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DT, Klymkowsky MW (1989) The appearance of acetylated alpha-tubulin during early development and cellular differentiation in Xenopus. Dev Biol 136: 104–117 [DOI] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136: 551–564 [DOI] [PubMed] [Google Scholar]
- Dompierre JP, Godin JD, Charrin BC, Cordelières FP, King SJ, Humbert S, Saudou F (2007) Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J Neurosci 28: 3571–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feierbach B, Nogales E, Downing KH, Stearns TJ (1999) Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with alpha-tubulin. J Cell Biol 144: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Huo L, Sun X, Liu M, Li D, Dong JT, Zhou J (2008) The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem 283: 8802–8809 [DOI] [PubMed] [Google Scholar]
- Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP (2007) Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol Cell Biol 27: 8637–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK (2007) Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 28: 21–27 [DOI] [PubMed] [Google Scholar]
- Hook SS, Orian A, Cowley SM, Eisenman RN (2002) Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proc Natl Acad Sci USA 15: 13425–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G (2003) The tumour suppressor CYLD negatively regulates NF-kB signalling by deubiquitination. Nature 424: 801–805 [DOI] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P (2007) The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21: 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fässler R (2006) Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125: 665–677 [DOI] [PubMed] [Google Scholar]
- Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fässler R, Bosserhoff AK (2009) Downregulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med 206: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R, Paus R (2007) Cylindromatosis and the CYLD gene: new lessons on the molecular principles of epithelial growth control. Bioessays 29: 1203–1214 [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444 [DOI] [PubMed] [Google Scholar]
- Pierre P, Pepperkok R, Kreis TE (1994) Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci 107: 1909–1920 [DOI] [PubMed] [Google Scholar]
- Piperno G, Fuller MT (1985) Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol 101: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privette LM, Weier JF, Nguyen HN, Yu X, Petty EM (2008) Loss of CHFR in human mammary epithelial cells causes genomic instability by disrupting the mitotic spindle assembly checkpoint. Neoplasia 10: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regamey A, Hohl D, Liu JW, Roger T, Kogerman P, Toftgard R, Huber M (2003) The tumor suppressor CYLD interacts with TRIP and regulates negatively nuclear factor kappaB activation by tumor necrosis factor. J Exp Med 198: 1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ (2006) Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol 7: 2166–2172 [DOI] [PubMed] [Google Scholar]
- Reiley WW, Zhang M, Sun SC (2004) Negative regulation of JNK signaling by the tumor suppressor CYLD. J Biol Chem 279: 55161–55167 [DOI] [PubMed] [Google Scholar]
- Riehemann K, Sorg C (1993) Sequence homologies between four cytoskeleton-associated proteins. Trends Biochem Sci 18: 82–83 [DOI] [PubMed] [Google Scholar]
- Romero MR, Carroll JM, Watt FM (1999) Analysis of cultured keratinocytes from a transgenic mouse model of psoriasis: effects of suprabasal integrin expression on keratinocyte adhesion, proliferation and terminal differentiation. Exp Dermatol 8: 53–67 [DOI] [PubMed] [Google Scholar]
- Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK (2005) Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res 65: 448–456 [PubMed] [Google Scholar]
- Saito K, Kigawa T, Koshiba S, Sato K, Matsuo Y, Sakamoto A, Takagi T, Shirouzu M, Yabuki T, Nunokawa E, Seki E, Matsuda T, Aoki M, Miyata Y, Hirakawa N, Inoue M, Terada T, Nagase T, Kikuno R, Nakayama M et al. (2004) The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma. Structure 12: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Schatten G, Simerly C, Asai J, Szoke E, Cooke P, Schatten H (1988) Acetylated alpha-tubulin in microtubules during mouse fertilization and early development. Dev Biol 130: 74–86 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol Cell Biol 21: 8035–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrador JM, Cabrero JR, Sancho D, Mittelbrunn M, Urzainqui A, Sánchez-Madrid F (2004) HDAC6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity 20: 417–428 [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ (2007) The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci USA 104: 8869–8874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo H, Maru Y (2007) LAPSER1 is a putative cytokinetic tumor suppressor that shows the same centrosome and midbody subcellular localization pattern as p80 katanin. FASEB J 21: 2086–2100 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-kB activation by TNFR family members. Nature 424: 793–796 [DOI] [PubMed] [Google Scholar]
- Verdel A, Curtet S, Brocard MP, Rousseaux S, Lemercier C, Yoshida M, Khochbin S (2000) Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr Biol 10: 747–749 [DOI] [PubMed] [Google Scholar]
- Viatour P, Dejardin E, Warnier M, Lair F, Claudio E, Bureau F, Marine JC, Merville MP, Maurer U, Green D, Piette J, Siebenlist U, Bours V, Chariot A (2004) GSK3-mediated BCL-3 phosphorylation modulates its degradation and its oncogenicity. Mol Cell 16: 35–45 [DOI] [PubMed] [Google Scholar]
- Webster DR, Bratcher JM (2006) Developmental regulation of cardiac MAP4 protein expression. Cell Motil Cytoskeleton 63: 512–522 [DOI] [PubMed] [Google Scholar]
- Westermann S, Weber K (2003) Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol 4: 938–947 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gilquin B, Khochbin S, Matthias P (2006) Two catalytic domains are required for protein deacetylation. J Biol Chem 281: 2401–2404 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li N, Caron C, Matthias G, Hess D, Khochbin S, Matthias P (2003) HDAC6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J 3: 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Wu Y, Navre M, Sang BC (2006) Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun 341: 45–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S7
Review Process File