Abstract
Preadipocyte factor-1 [Pref-1; also called Dlk1 (Delta-like protein 1)] is made as an epidermal growth factor-repeat containing transmembrane protein that produces a biologically active soluble form by TNF-α-converting enzyme (TACE)-mediated cleavage. Soluble Pref-1 activates the MAPK kinase/ERK pathway. In adipose tissue, Pref-1 is specifically expressed in preadipocytes but not in adipocytes and thus is used as a preadipocyte marker. Inhibition of adipogenesis by Pref-1 has been well established in vitro as well as in vivo by ablation and overexpression of Pref-1. SRY (sex determining region Y)-box 9 (Sox9), a transcription factor expressed in preadipocytes to suppress CCAAT enhancer binding protein β and (C/EBP) δ expression, is required to be down-regulated before adipocyte differentiation can proceed. By activating MAPK kinase/ERK, Pref-1 prevents down-regulation of Sox9, resulting in inhibition of adipogenesis. Furthermore, by inducing Sox9, Pref-1 promotes chondrogenic induction of mesenchymal cells but prevents chondrocyte maturation as well as osteoblast differentiation. Thus, Pref-1 directs multipotent mesenchymal cells toward the chondrogenic lineage but inhibits differentiation into adipocytes as well as osteoblasts and chondrocytes. Pref-1, encoded by an imprinted gene, has also been detected in progenitor cells in various tissues during regeneration and therefore may have a more general role in maintaining cells in an undifferentiated state.
Pref-1 activates MEK/ERK and induces Sox9 expression thereby causing inhibition of adipocyte differentiation, promotion of chondrogenic commitment, inhibition of chondrocyte maturation and osteoblast differentiation.
Excess adipose tissue leading to obesity has become a severe public health threat. Adipose tissue development can be affected by genetic background, hormonal balance, diet, and physical activity. Adipose tissue mass can increase when fat cells are increased in size due to higher triacylglycerol accumulation. In addition, an increase in fat cell number, arising from differentiation of precursor cells into adipocytes, can also occur even in adults as observed in severe human obesity and in rodents fed a high-carbohydrate or high-fat diet (1,2,3).
Adipose tissue includes, not only adipocytes, but also stromal vascular cells representing preadipocytes and monocytes/macrophages as well as endothelial and other cell types. Adipocytes specifically are thought to arise from mesenchymal cells that undergo the commitment and differentiation process, adipogenesis (4,5). A recent report suggests that, in adipose tissue, precursor fat cells reside in the mural compartment of vasculature (6). During the last two decades, transcriptional events leading to preadipocyte differentiation into adipocytes have been extensively studied. Preadipocyte cell lines, including 3T3-L1 and 3T3-F442A, which can undergo adipocyte differentiation upon treatment with adipogenic agents comprised of synthetic glucocorticoid, dexamethasone (DEX), isobutylmethylxanthine (IBMX), and insulin, have been valuable in these studies (7,8,9). Peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT enhancer-binding protein (C/EBP) family of transcription factors have been firmly established to play critical roles in adipocyte differentiation (10,11,12,13). Early during adipocyte differentiation, C/EBPβ and C/EBPδ are induced by DEX and IBMX, respectively, which together then induce PPARγ and C/EBPα to activate various adipocyte markers that are required for adipocyte function (12,14). Thus, thiazolidinediones, synthetic ligands of PPARγ, can enhance adipocyte differentiation in culture. Other transcription factors have also been reported to either positive or negatively regulate adipogenesis. Recently, the author’s laboratory reported that SRY (sex determining region Y)-box 9 (Sox9) present in mesenchymal cells and preadipocytes suppresses expression of C/EBPδ and C/EBPβ. Hence, suppression of Sox9 is necessary before preadipocytes can undergo the differentiation process to become adipocytes (15). Moreover, various growth factors/hormones can affect adipocyte differentiation by regulating expression of adipogenic transcription factors. In fact, in addition to being the main site for energy storage in mammals by storing triacyglycerol and releasing fatty acids in times of need, adipose tissue secretes a wide array of molecules that are involved in diverse physiological processes including immune response, vascular function, and energy homeostasis (16,17). Among these factors are leptin and adiponectin that are secreted from adipocytes. Cytokines such as TNF-α and IL-6 are secreted from adipocytes as well as macrophages, whereas preadipocyte factor-1 [Pref-1; also called Dlk1 (Delta-like protein 1)] is secreted from preadipocytes. Some of these factors may also affect growth and development of adipose tissue by autocrine/paracrine action.
This review focuses on the current knowledge of the role of Pref-1 in the regulation of adipocyte differentiation and mesenchymal cell fate and its signaling mechanism, as well as its effect on pathological disorders such as obesity and diabetes.
Pref-1 Structure
Pref-1 is synthesized as a protein of 385 amino acids containing a signal sequence at the N terminus and a single membrane-spanning domain of amino acids 300-322. The most striking structural feature of Pref-1 is in the extracellular domain presence of six tandem epidermal growth factor (EGF)-like repeats maintaining both the conserved spacing of six cysteines for three disulfide bonding as well as the other amino acids characteristic of EGF-like repeat motif-containing proteins (Fig. 1) (18). The EGF-like motif was originally described for EGF and other growth factors, which, by binding to EGF receptor, act as signals for cell proliferation and differentiation. However, Pref-1 does not contain the conserved amino acid residues that are required for EGF receptor binding. Rather, Pref-1 shares structural characteristics with another class of EGF-like repeat-containing signaling proteins, the Notch/Delta/Serrate family, that are involved in cell signaling and cell fate determination (19). However, Pref-1 lacks the DSL (Delta/Serrate/LAG-2) domain that is conserved in all classic Notch ligands to mediate receptor-ligand interaction for Notch (20,21). Instead, the N-terminal tandem EGF repeats of Pref-1 represent a DOS (Delta and OSM-11)-motif, recently defined specialized tandem EGF repeats found in classic Notch ligands as well as in other soluble and membrane-bound proteins reported to function in the presence of DSL containing Notch ligands in this pathway (19,22).
Figure 1.
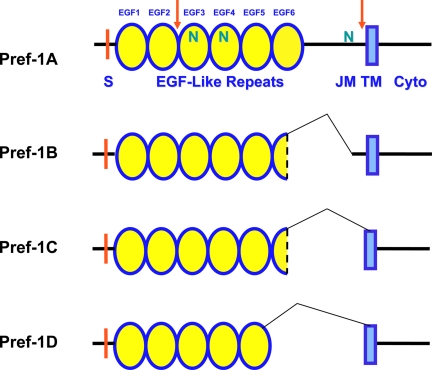
Domain structure of Pref-1 isoforms. EGF (EGF-like repeat), S (signal sequence), JM (juxtamembrane), TM (transmembrane domain), Cyto (cytoplasmic region), and N (N-linked glycosylation sites). Cleavage sites are marked orange.
In preadipocytes, multiple transmembrane forms of Pref-1, ranging from 50 to 60 kDa, are found in the cell membrane due, in part, to posttranslational modifications containing N-linked oligosaccharides as well as sialic acids in the extracellular domain. Moreover, there are four major alternative splicing products of Pref-1 (Pref-1A–D) (Fig. 1) (23). In addition to the largest full-length Pref-1 form, alternate splicing generates three major shorter forms of Pref-1, each containing in-frame deletions in the extracellular juxtamembrane region or part of the sixth EGF-like repeat domain. The relative abundance of the different splice forms varies depending on the tissue or developmental stage investigated (24).
Pref-1 is proteolytically cleaved at the extracellular domain at two sites to generate soluble forms of Pref-1 (25). Thus, the two larger alternate splice forms of Pref-1, Pref-1A and Pref-1B, are cleaved at a juxtamembrane site as well as at a site closer to the N terminus to generate a larger 50-kDa and a smaller 25-kDa soluble form, respectively. The smaller Pref-1C and Pref-1D, due to the larger deletions that include the juxtamembrane processing site, are cleaved only at the N-terminal site to generate only the smaller 25-kDa soluble form but not the larger 50-kDa form. In this regard, fetal antigen 1, the protein in fetal circulation, likely corresponds to the larger soluble form of human Pref-1 (26,27,28). Detection of the cleaved form in circulation clearly validates Pref-1 processing and release shown in cultured cells. Using various inhibitors and in vitro approaches, the author’s laboratory reported that one of the ADAM family members, TNF-α-converting enzyme (TACE, ADAM 17), can cleave Pref-1 at the juxtamembrane region to generate the large soluble form of Pref-1 (29). Both basal and stimulated cleavage was inhibited by the broad metalloproteinase inhibitor GM6001, also suggesting that the cleavage of membrane Pref-1 is dependent on a metalloproteinase. In addition, lentivirus-mediated overexpression of TACE increased Pref-1 cleavage, which produces the large soluble form. Conversely, small interfering (siRNA)-mediated knockdown of TACE decreased the release of the large soluble form from the membrane form (29). Moreover, this cleavage was not detectable or was markedly decreased in cells bearing mutated TACE or in cells transfected with TACE siRNA. These data clearly demonstrate TACE-mediated cleavage at the juxtamembrane in generating the large 50-kDa soluble form of Pref-1. The release of the 50-kDa full extracellular domain can be markedly enhanced by phorbol ester treatment in a dose- and time-dependent manner, indicating that increased Pref-1 cleavage is regulated by protein kinase C activation (25). It is plausible that Pref-1 activity may be regulated at the proteolytic cleavage step to generate the active form.
Inhibition of Adipocyte Differentiation by Pref-1
Pref-1 is highly expressed in preadipocytes but decreases during differentiation and is absent in mature adipocytes (18,30). In fact, the author’s laboratory originally cloned Pref-1 from a 3T3-L1 preadipocyte cDNA library employing the selection criterion based on preadipocyte-specific expression. Pref-1 reflects the degree of adipocyte differentiation in vitro and in vivo and thus is used as a preadipocyte marker (6,30,31,32,33,34,35,36,37). Pref-1 expression is down-regulated specifically by a component of adipogenic agents, DEX. There is a close correlation between DEX-mediated down-regulation of Pref-1 and efficacy of DEX on differentiation of 3T3-L1 preadipocytes (38,39). Thus, in addition to inducing C/EBPδ, DEX may promote adipocyte differentiation by suppressing Pref-1 expression. Pantoja et al. (40) recently reported that DEX may drive preadipocytes to an intermediate state that can then undergo differentiation by IBMX treatment and, thus, Pref-1, which is suppressed by DEX, may be a cell fate determinant.
Studies utilizing various in vitro approaches have clearly demonstrated the inhibitory role of Pref-1 in adipocyte differentiation (18,30). Constitutive expression of Pref-1 in 3T3-L1 cells by stable transfection markedly lowers the degree of adipocyte differentiation. Pref-1 prevents lipid accumulation and expression of adipocyte transcription factors such as PPARγ and C/EBPα as well as other late adipocyte markers, including fatty acid synthase, stearoyl-coenzyme A desaturase, and FABP4/aP2. Conversely, decreasing Pref-1 levels by transfection of antisense sequence greatly enhances adipogenesis. In this regard, the antiadipogenic activity of GH has been attributed to the enhanced expression of Pref-1 through FoxA2 activation (41). Overall, these studies demonstrate that Pref-1 expression inhibits adipocyte differentiation and that down-regulation of Pref-1 is a necessary step in adipocyte differentiation.
Because soluble forms of Pref-1 can be generated by cleavage of the extracellular domain, the results obtained from transfection of full-length Pref-1 could not distinguish the effect of the membrane form of Pref-1 from soluble Pref-1 in regulating adipocyte differentiation. However, addition of the large soluble form purified from mammalian cells transfected with the 50-kDa full-length Pref-1 extracellular domain (referred to as “soluble Pref-1”) fused to human Ig γ constant (hFc) as well as Pref-1 extracellular domain fused to glutathione-S transferase expressed in Escherichia coli can effectively inhibit adipocyte differentiation. Moreover, unlike Pref-1A and Pref-1B that can generate the 50-kDa soluble Pref-1, Pref-1C and Pref-1D (that can produce only the 25-kDa cleavage product, but not 50-kDa form) cannot inhibit adipocyte differentiation (24). These observations clearly show the inhibitory effect of the large soluble 50-kDa protein, but not the full-length membrane form, on adipocyte differentiation. More definitive evidence was provided by transfection of a noncleavable transmembrane form of Pref-1A, constructed by the deletion of the 22 amino acids containing the putative TACE cleavage site at the juxtamembrane region, confirming that the membrane form of Pref-1 is not capable of inhibiting adipocyte differentiation (24). Although further studies are needed, because the smaller 25-kDa cleavage product is not effective in inhibiting adipocyte differentiation, the additional N-terminal cleavage can be a means to inactivate the biologically active 50-kDa soluble Pref-1. As described below, the similar developmental defects described in Pref-1 bac transgenic mice (42) to that of transgenic mice expressing only the 50-kDa soluble Pref-1 further argue that the large soluble form is the biologically active form. Therefore, it can be predicted that Pref-1 acts by binding to a yet to be identified interacting protein/receptor that may contain EGF-repeats that are presumably involved in protein-protein interaction. As mentioned above, Pref-1 does not contain a DSL domain, which is required for interaction with Notch, predicting that Pref-1 is not a Notch ligand. In this regard, inhibition of Notch activation by Pref-1 has been reported (43). In addition, expression of full-length Pref-1 by using Gal4/UAS system in Drosophila showed, contrary to mammalian Delta, inhibitory effects on Notch function in addition to other unrelated phenotypes (44). On the other hand, a recent study showed that Pref-1 could substitute for OSM-11 in Caenorhabditis elegans development, suggesting not only conservation of function of the DOS-motif, but its potential coactivating role in Notch/DSL ligand signaling (22). These contradictory results of inhibition and stimulation by Pref-1 on DSL ligand-induced Notch activation will require further investigation. Moreover, Pref-1 effect on expression of Hes-1, a downstream target of Notch, has also been controversial at best (46,47). Regardless, direct interaction of biologically active Pref-1 with Notch has not been demonstrated (43,45). In fact, the author’s laboratory could detect neither the changes in Hes-1 expression by Pref-1 nor the binding of Pref-1 to Notch (Wang, Y., and H. S. Sul, unpublished results). Identification of the Pref-1 interacting protein/receptor and biochemical and functional evidence for its interaction with Pref-1 are needed in elucidating Pref-1 action.
Pref-1 Activates the MAPK Kinase (MEK)/ERK Pathway
Mouse embryo fibroblasts (MEFs) in culture can be differentiated into adipocytes and have been employed by researchers to investigate adipocyte differentiation. MEFs isolated from Pref-1 knockout mice provide a system to examine the signaling pathway for Pref-1 inhibition of adipogenesis. Although there are reports that suggest Pref-1 affects other growth factor/cytokine secretion and function, its specific signaling pathway was largely unknown (48,49). Addition of the large soluble Pref-1- to Pref-1-null MEFs increases phosphorylation of ERK in a time- and dose-dependent manner (50). Pref-1-null MEFs, as compared with wild-type MEFs, show significantly higher degree of adipose conversion: compared with the 50% differentiation of wild-type MEFs into adipocytes, 90% of Pref-1-null MEFs can be differentiated into adipocytes. Moreover, infection of lentivirus containing the large soluble form of Pref-1 into Pref-1-null MEFs decreases the degree of differentiation, confirming the inhibitory effect of Pref-1 on adipogenesis. An initial transient burst of ERK phosphorylation upon addition of adipogenic agents has been found to be required for induction of adipocyte differentiation (51,52). However, wild-type MEFs also have a low but significant second increase in ERK phosphorylation peaking at d 2 that parallels the expression level of Pref-1 in these cells. This second peak in ERK phosphorylation is absent in Pref-1-null MEFs. Furthermore, a specific MEK inhibitor or siRNA-mediated ERK depletion, after the first burst but before the second peak of ERK activation, can prevent the second ERK phosphorylation and also enhance adipocyte differentiation of wild-type MEFs. Treatment of Pref-1-null MEFs with the large soluble Pref-1 restores the second ERK phosphorylation, resulting in inhibition of adipocyte differentiation (50). These studies clearly demonstrate that Pref-1 inhibition of adipogenesis is through activation of the MEK/ERK pathway during differentiation (Fig. 2). Because the soluble Pref-1 corresponding to the cleaved ectodomain containing the six EGF-like repeats is biologically active, Pref-1 must interact with a yet to be identified Pref-1 interacting protein/receptor, in order to activate MEK/ERK pathway to inhibit adipocyte differentiation. The identification of the putative Pref-1 interacting protein/receptor is critical for understanding Pref-1 activation of MEK/ERK as well as its inhibition of adipocyte differentiation.
Figure 2.
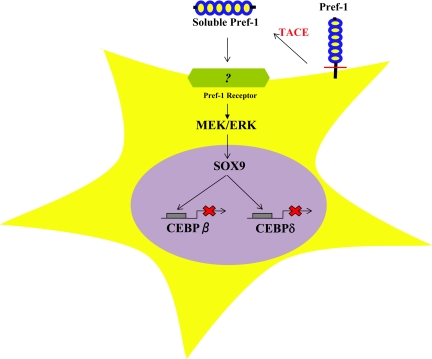
Pref-1 inhibition of adipogenesis. Membrane-bound Pref-1A and Pref-1B generate 50-kDa soluble Pref-1 by TACE-mediated cleavage at the juxtamembrane region. Binding of the 50-kDa soluble Pref-1 to the putative Pref-1 receptor at the preadipocyte membrane activates MEK/ERK which, in turn, increases Sox9 expression. Sox9 binds to its binding sites at the C/EBPβ and C/EBPδ promoter regions to suppress their transcription, resulting in inhibition of adipocyte differentiation.
Pref-1 Induces Sox9 to Inhibit Adipocyte Differentiation
Although expression of C/EBPα and PPARγ is suppressed upon inhibition of adipogenesis by Pref-1, the specific target of Pref-1 action for inhibition of adipocyte differentiation was not known. During the search for Pref-1 targets by microarray analysis and other candidate approaches, Sox9 was identified as a downstream target of Pref-1. Pref-1 treatment causes a rapid increase in Sox9 expression. Sox9, a member of the large Sox gene family, belongs to the high-mobility group-box class of transcription factors and is responsible for human campomelic dysplasia, an autosomal-dominant condition of skeletal malformation in which most XY patients have male to female sex reversal (53,54). Sox9 plays an important role during embryogenesis and cellular differentiation of various tissues, including testogenesis, chondrogenesis, and osteoblastogenesis. However, the role of Sox9 in adipocyte commitment/differentiation was not known. Recently, it has been shown that, when treated with adipogenic agents (DEX/IBMX/insulin), Sox9 expression decreases early during 3T3-L1 differentiation in a pattern similar to Pref-1 (15). Down-regulation of Sox9 during adipocyte differentiation coincides with the early induction of C/EBPβ and C/EBPδ. Overexpression and short hairpin RNA-mediated knockdown of Sox9 clearly demonstrated that Sox9 negatively regulates adipocyte differentiation (15). It has also been revealed that Sox9 directly binds its binding sites at the promoter regions of C/EBPβ and C/EBPδ genes, suppressing their transcription. In this regard, sc implantation in mice of 3T3-F442A cells that stably overexpress Sox9 does not form adipocytes efficiently, whereas cells expressing Sox9 short hairpin RNA can form a greater number of adipocytes (15). This clearly provided in vivo evidence for the inhibitory role of Sox9 in adipogenesis. Sox9 may maintain cells in the preadipocyte stage through suppressing C/EBPβ and C/EBPδ, and Sox9 suppression is an early event in adipocyte differentiation (Fig. 2). More importantly, it has also been demonstrated that Pref-1 inhibits adipocyte differentiation by preventing this down-regulation of Sox9 that normally occurs during differentiation (15). The fact that Sox9 is induced by ERK activation, and that inhibition of ERK activation by Pref-1 prevents Sox9 induction further support the notion that Sox9 is a downstream target of Pref-1. Indeed, studies employing MEK inhibitors as well as siRNA approaches also provided evidence that, by activating MEK/ERK, Pref-1 up-regulates Sox9 and thereby inhibits adipocyte differentiation (15).
In Vivo Effect of Pref-1 on Adipogenesis and Glucose/Insulin Homeostasis
The role of Pref-1 in adipogenesis has been clearly shown by ablation of the Pref-1 gene in mice (55) as well as by overexpression of Pref-1 in adipose tissue of transgenic mice (56,57). Both male and female Pref-1-null mice weighed significantly less than wild-type mice at weaning (55). However, Pref-1-null mice gained body weight more rapidly. The weight of major fat depots (inguinal, retroperitoneal, and gonadal) was significantly higher in Pref-1-null mice, indicating that the accelerated body weight gain in Pref-1-null mice was due to an increase in adipose tissue mass. Histological analysis of fat depots revealed that adipocytes from Pref-1-null mice were larger than those from wild-type littermates. Moreover, mRNA levels of late markers of adipocyte differentiation, stearoyl-coenzyme A desaturase and fatty acid synthase, were significantly higher in adipose tissue from Pref-1-null mice, indicating enhanced adipogenesis. Pref-1-null mice also showed an enlarged fatty liver as well as increased circulating levels of triglycerides, cholesterol, and free fatty acids, characteristics usually associated with obesity. Pref-1-null mice fed a high-fat diet develop impaired insulin resistance and glucose intolerance when compared with wild-type mice on the same diet (Wang, Y., and H. S. Sul, unpublished results). Thus, studies of Pref-1 ablation in mice demonstrate that ablation of Pref-1 enhances adipogenesis in vivo and support the proposed role of Pref-1 as a negative regulator of the adipogenesis.
Transgenic mice overexpressing the large soluble form of Pref-1 as an hFc-fusion protein in adipose tissue have been generated using the FABP4/aP2 promoter. These transgenic mice showed a marked decrease in adipose tissue mass and reduced expression of adipocyte markers and adipocyte-secreted factors (56,57). With decreased adipose tissue development, as observed in lipodystrophy mouse models, these mice suffered from hypertriglyceridemia, decreased glucose tolerance, and lower insulin sensitivity. Mice ectopically expressing the Pref-1/hFc exclusively in liver under the control of the albumin promoter also showed a decrease in adipose mass and adipocyte marker expression, suggesting a potential endocrine mode of action of Pref-1 (56). Data from these in vivo studies with Pref-1 knockout and transgenic mice models are consistent with in vitro studies, strongly demonstrating the inhibitory effect of Pref-1 on adipogenesis. The findings that both Pref-1-null and Pref-1-overexpressing transgenic mice develop insulin resistance and glucose intolerance also suggest that proper development of adipose tissue and adipose tissue function are critical for maintenance of glucose/insulin homeostasis.
Pref-1 Controls Mesenchymal Cell Fate
Pref-1-null mice display greater than 50% perinatal lethality, and the surviving animals show growth retardation, skeletal malformation, and other defects in addition to increased adiposity (55). These phenotypes suggest that Pref-1 may function in other developmental processes including skeletal formation. MEFs and bone marrow mesenchymal stem cells can differentiate not only into adipocytes but other mesenchymal derived cell types, including chondrocytes and osteoblasts. Thus, these cells provide an excellent system to examine mesenchymal cell commitment and differentiation in vitro. In these cells, Pref-1 promotes chondrogenic induction but prevents chondrocyte maturation to become hypertrophic chondrocytes. Pref-1 also inhibits osteoblast differentiation of MEFs and bone marrow mesenchymal cells. Both Pref-1 ablation and Pref-1 overexpression in mice cause bone malformation with smaller skeletons (55,56). In fact, this potential role of Pref-1 can be predicted because Pref-1 up-regulates Sox9. It is well recognized that Sox9 plays an essential role in recruiting mesenchymal cells to undergo chondrogenic commitment and early chondrogenesis but inhibits maturation into hypertrophic chondrocytes (58,59,60,61,62). In this regard, inactivation of Sox9 before mesenchymal condensation in mice results in complete prevention of condensation and cartilage formation. Pref-1 has similar but not additive effects to Sox9 on MEF commitment and early differentiation into immature chondrocytes (15). Moreover, in the absence of Sox9, Pref-1 cannot bring chondrogenic induction in micromass culture, evidence that Sox9 mediates the Pref-1 effect of chondrogenic induction. It is noteworthy that Pref-1-null and Pref-1-transgenic mice have smaller skeletons as seen in Sox9 deficiency and Sox9 overexpression in mice. Pref-1-null mice appear to have, albeit less severe, a phenotype in skeletal development that resembles that found in Sox9 knockdown or mutated mice, whereas the phenotype of Pref-1-transgenic mice has similarities to the Sox9 transgenic mouse model. In this regard, deficiencies in some of the growth factors/hormones that affect skeletogenesis, at least in part, through Sox9 regulation, also have less severe phenotypes (61,62). Pref-1 prevents Sox9 down-regulation and thereby inhibits osteoblast differentiation and the expression of the osteogenic transcription factor Runx2. Progenitor cells in early mesenchymal condensations have dual differentiation potentials as they coexpress Sox9 and Runx2. Runx2 is known to accelerate chondrocyte maturation to become hypertrophic chondrocytes and promote osteogenesis (63). Because Runx2 exerts its function when Sox9 is down-regulated in hypertrophic chondrocytes or in osteoblasts, Pref-1 may inhibit osteoblast differentiation by preventing down-regulation of Sox9. Similar inhibition of osteoblast differentiation of bone marrow mesenchymal cells by Pref-1 has also been observed (64). Overall, Sox9 is a Pref-1 target that directs multipotent mesenchymal cells to the chondrogenic lineage but inhibits differentiation into mature chondrocytes and osteoblasts as well as adipocytes.
Pref-1 is encoded by the imprinted gene (dlk1) and, along with meg8, dat, gtl2, peg11, and antipeg11, is located in an imprinted syntenic region of mouse chromosome 12, human chromosome 14, and sheep chromosome 18 (65,66,67). Paternal monoallelic expression of the Pref-1 gene, due to differential methylation, is documented and evidenced in Pref-1-ablated heterozygous mice (55). Interestingly, Pref-1-null and transgenic mice show distinct defects similar to maternal uniparental disomy (UPD)12 and paternal UPD12 in mice, respectively, and syntenic maternal and paternal UPD14 syndromes in humans. Calipyge sheep with a mutation in chromosome 18 show decreased adiposity with elevated Pref-1 levels (68). It appears that observed UPD syndromes are due, at least in part, to altered expression of Pref-1. Although a significant level of Pref-1 expression is detected in adults, only in preadipocytes and in certain neuroendocrine types of cells, Pref-1 is widely expressed in multiple mouse embryonic tissues, such as liver, lung, tongue, pituitary, and developing vertebrae (18) and placenta. Circulating Pref-1 is defected in maternal serum in concentrations that correlate with the number of fetuses in utero (26). Thus, the human homolog of Pref-1, Dlk1, was purified as fetal antigen 1 (27) from fetal circulation and was shown to be expressed in a wide array of human embryonic tissues (28). However, after birth, expression of Pref-1 is rapidly abolished in most tissues and becomes restricted to certain type of cells that include preadipocytes (18) and thymic stromal cells (47) as well as some neuroendocrine types of cells such as pancreatic islet β-cells (9), adrenal glands (10), and pituitary. Given the role of imprinted genes in fetal growth and development in general, and the expression of Pref-1 during embryonic development in a variety of tissues, Pref-1 undoubtedly has a function beyond the regulation of adipogenesis or even mesenchymal cell fate (69). In this regard, Pref-1 was reported recently to be found in progenitor cell compartments in many other cell types, specifically during regeneration, including bone marrow stem cells and hepatoblasts, as well as in muscle satellite cells (70,71,72,73,74,75,76,77,78,79,80,81). Thus, as in adipocyte and mesenchymal cell differentiation, Pref-1 may function as a soluble factor, maintaining proliferating cells in an undifferentiated state during development.
Conclusion
Both obesity and lipodystrophy are commonly associated with pathologies including diabetes and cardiovascular diseases. It is now recognized that adipose tissue is an endocrine organ that secretes a wide variety of factors, and dysregulated secretion affects adiopogenesis as well as whole-body glucose/insulin homeostasis. Here, current knowledge on the function of the preadipocyte secreted protein Pref-1 is discussed. Pref-1 is synthesized and released from preadipocytes in both rodents and humans. The inhibitory role of Pref-1 on adipogenesis, as well as its effect on glucose/insulin homeostasis, has been demonstrated in vivo by Pref-1-null and Pref-1-overexpressing transgenic mice. A variety of experimental approaches using 3T3-L1 cells and Pref-1-null MEFs clearly showed that Pref-1 is processed by TACE to generate a biologically active soluble form containing the full ectodomain, and the soluble form prevents differentiation into adipocytes through activation of the MEK/ERK pathway. Recently, the molecular events underlying the inhibition of adipogenesis by Pref-1 have been revealed; Pref-1, via MEK/ERK activation, maintains preadipocyte expression of Sox9, which, by direct binding to the promoter regions, suppresses C/EBPβ and C/EBPδ expression. Furthermore, by regulating Sox9 expression, Pref-1 promotes multipotent mesenchymal stem cells into the chondrogenic lineage but inhibits chondrocyte and osteoblast as well as adipocyte differentiation. Therefore, both ablation and overexpression of Pref-1 in mice cause skeletal malformation. Although Pref-1 expression is restricted to preadipocytes and a few neuroendocrine types of cells in adults, Pref-1 is present in variety of embryonic tissues. Pref-1, encoded by an imprinted gene expressed by the paternal copy through differential methylation, may affect embryonic development. In this regard, Pref-1 is detected in progenitor cells of different tissues especially during regeneration, probably maintaining cells in an undifferentiated state. Clearly, identification of the interacting protein/receptor through which Pref-1 signaling occurs is critical and would represent a major advance in understanding the molecular mechanism underlying the function of Pref-1.
Footnotes
This work was supported by National Institutes of Health Grant DK50828 (to H.S.S.).
Disclosure Summary: The author has nothing to disclose.
First Published Online June 18, 2009
Abbreviations: C/EBP, CCAAT/enhancer binding protein; DEX, dexamethasone; Dlk1, Delta-like protein 1; DOS, (Delta and OSM-11)-motif; FABP4/aP2, fatty acid binding protein 4; hFc, human Ig γ constant; MEK, MAPK kinase; IBMX, isobutylmethylxanthine; MEF, mouse embryo fibroblast; Pref-1, preadipocyte factor-1; PPARγ, peroxisome proliferator-activated receptor γ; Sox9, SRY (sex determining region Y)-box 9; TACE, TNF-α cleavage enzyme; UPD, uniparental disomy.
References
- Faust IM, Miller Jr WH, Sclafani A, Aravich PF, Triscari J, Sullivan AC 1984 Diet-dependent hyperplastic growth of adipose tissue in hypothalamic obese rats. Am J Physiol 247:R1038–R1046 [DOI] [PubMed] [Google Scholar]
- Miller Jr WH, Faust IM, Hirsch J 1984 Demonstration of de novo production of adipocytes in adult rats by biochemical and radioautographic techniques. J Lipid Res 25:336–347 [PubMed] [Google Scholar]
- Kirkland JL, Hollenberg CH, Gillon WS 1990 Age, anatomic site, and the replication and differentiation of adipocyte precursors. Am J Physiol 258:C206–C210 [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR 2007 Developmental origin of fat: tracking obesity to its source. Cell 131:242–256 [DOI] [PubMed] [Google Scholar]
- Otto TC, Lane MD 2005 Adipose development: from stem cell to adipocyte. Crit Rev Biochem Mol Biol 40:229–242 [DOI] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM 2008 White fat progenitor cells reside in the adipose vasculature. Science 322:583–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O 1976 Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell 7:105–113 [DOI] [PubMed] [Google Scholar]
- Green H, Meuth M 1974 An established pre-adipose cell line and its differentiation in culture. Cell 3:127–133 [DOI] [PubMed] [Google Scholar]
- Rubin CS, Lai E, Rosen OM 1977 Acquisition of increased hormone sensitivity during in vitro adipocyte development. J Biol Chem 252:3554–3357 [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA 2006 Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896 [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, Sul HS 1998 Understanding adipocyte differentiation. Physiol Rev 78:783–809 [DOI] [PubMed] [Google Scholar]
- Farmer SR 2006 Transcriptional control of adipocyte formation. Cell Metab 4:263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM 2008 Fat and beyond: the diverse biology of PPARγ. Annu Rev Biochem 77:289–312 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL 1995 Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9:168–181 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sul HS 2009 Pref-1 regulates mesenchymal cell commitment and differentiation through Sox9. Cell Metab 9:287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahima RS 2006 Adipose tissue as an endocrine organ. Obesity 14:242S–249S [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Scherer PE 2006 Adipose tissue-derived factors: impact on health and disease. Endocr Rev 27:762–778 [DOI] [PubMed] [Google Scholar]
- Smas CM, Sul HS 1993 Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73:725–734 [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG 2009 The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax FE, Yeargers JJ, Thomas JH 1994 Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368:150–154 [DOI] [PubMed] [Google Scholar]
- Gordon WR, Arnett KL, Blacklow SC 2008 The molecular logic of Notch signaling—a structural and biochemical perspective. J Cell Sci 121:3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, Dionne HM, White JQ, Wani K, Boxem M, Hart AC 2008 OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol 8:e196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smas CM, Green D, Sul HS 1994 Structural characterization and alternate splicing of the gene encoding the preadipocyte EGF-like protein pref-1. Biochemistry 33:9257–9265 [DOI] [PubMed] [Google Scholar]
- Mei B, Zhao L, Chen L, Sul HS 2002 Only the large soluble form of preadipocyte factor-1 (Pref-1), but not the small soluble and membrane forms, inhibits adipocyte differentiation: role of alternative splicing. Biochem J 364:137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smas CM, Chen L, Sul HS 1997 Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol Cell Biol 17:977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann E, Krogh TN, Højrup P, Skjødt K, Teisner B 1996 Mouse fetal antigen 1 (mFA1), the circulating gene product of mdlk, pref-1 and SCP-1: isolation, characterization and biology. J Reprod Fertil 107:279–285 [DOI] [PubMed] [Google Scholar]
- Jensen CH, Krogh TN, Højrup P, Clausen PP, Skjødt K, Larsson LI, Enghild JJ, Teisner B 1994 Protein structure of fetal antigen 1 (FA1). A novel circulating human epidermal-growth-factor-like protein expressed in neuroendocrine tumors and its relation to the gene products of dlk and pG2. Eur J Biochem 225:83–92 [DOI] [PubMed] [Google Scholar]
- Floridon C, Jensen CH, Thorsen P, Nielsen O, Sunde L, Westergaard JG, Thomsen SG, Teisner B 2000 Does fetal antigen 1 (FA1) identify cells with regenerative, endocrine and neuroendocrine potentials? A study of FA1 in embryonic, fetal, and placental tissue and in maternal circulation. Differentiation 66:49–59 [DOI] [PubMed] [Google Scholar]
- Wang Y, Sul HS 2006 Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor α converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol Cell Biol 26:5421–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim KA, Kim JH, Sul HS 2006 Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J Nutr 136:2953–2956 [DOI] [PubMed] [Google Scholar]
- Wang MY, Grayburn P, Chen S, Ravazzola M, Orci L, Unger RH 2008 Adipogenic capacity and the susceptibility to type 2 diabetes and metabolic syndrome. Proc Natl Acad Sci USA 105:6139–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox KE, Fankell DM, Erickson PF, Majka SM, Crossno Jr JT, Klemm DJ 2006 Depletion of cAMP-response element-binding protein/ATF1 inhibits adipogenic conversion of 3T3-L1 cells ectopically expressing CCAAT/enhancer-binding protein (C/EBP)α, C/EBPβ, or PPARγ2. J Biol Chem 281:40341–40353 [DOI] [PubMed] [Google Scholar]
- Tseng YH, Butte AJ, Kokkotou E, Yechoor VK, Taniguchi CM, Kriauciunas KM, Cypess AM, Niinobe M, Yoshikawa K, Patti ME, Kahn CR 2005 Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat Cell Biol 7:601–611 [DOI] [PubMed] [Google Scholar]
- Zhou YT, Wang ZW, Higa M, Newgard CB, Unger RH 1999 Reversing adipocyte differentiation: implications for treatment of obesity. Proc Natl Acad Sci USA 96:2391–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS 1998 Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev 12:3182–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson B, Smith U 2006 Cytokines promote Wnt signaling and inflammation and impair the normal differentiation and lipid accumulation in 3T3-L1 preadipocytes. J Biol Chem 281:9507–9516 [DOI] [PubMed] [Google Scholar]
- Rodheheffer MS, Birsoy K, Friedman JM 2008 Identification of white adipocyte progenitor cells in vivo. Cell 135:240–249 [DOI] [PubMed] [Google Scholar]
- Smas CM, Kachinskas D, Liu CM, Xie X, Dircks LK, Sul HS 1998 Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J Biol Chem 273:31751–31758 [DOI] [PubMed] [Google Scholar]
- Smas CM, Chen L, Zhao L, Latasa MJ, Sul HS 1999 Transcriptional repression of pref-1 by glucocorticoids promotes 3T3-L1 adipocyte differentiation. J Biol Chem 274:12632–12641 [DOI] [PubMed] [Google Scholar]
- Pantoja C, Huff JT, Yamamoto KR 2008 Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol Biol Cell 19:4032–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Shih DQ, Kuwajima S, Norris AW, Kahn CR, Stoffel M 2003 Role of Foxa-2 in adipocyte metabolism and differentiation. J Clin Invest 112:345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Charalambous M, Lin SP, Gutteridge I, Ito Y, Gray D, Dean W, Ferguson-Smith AC 2009 Gene dosage effects of the imprinted Delta-like homologue 1(dlk1/pref1) in development: implications for the evolution of imprinting. PLoS Genet 5:e1000392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baladrón V, Ruiz-Hidalgo MJ, Nueda ML, Díaz-Guerra MJ, García-Ramírez JJ, Bonvini E, Gubina E, Laborda J 2005 dlk Acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res 303:343–359 [DOI] [PubMed] [Google Scholar]
- Bray SJ, Takada S, Harrison E, Shen SC, Ferguson-Smith AC 2008 2008 The atypical mammalian ligand delta-like homologue 1 (dlk1) can regulate Notch signaling in Drosophila. BMC Dev Biol 8:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza B, Miyamoto A, Weinmaster G 2008 The many facets of Notch ligands. Oncogene 27:5148–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DA, Rao PK, Kadesch T 2004 Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol Cell Biol 24:3505–3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneta M, Osawa M, Sudo K, Nakauchi H, Farr AG, Takahama Y 2000 A role for pref-1 and HES-1 in thymocyte development. J Immunol 164:256–264 [DOI] [PubMed] [Google Scholar]
- Zhang H, N¿ohr J, Jensen CH, Petersen RK, Bachmann E, Teisner B, Larsen LK, Mandrup S, Kristiansen K 2003 Insulin-like growth factor-1/insulin bypasses Pref-1/FA1-mediated inhibition of adipocyte differentiation J Biol Chem 278 20906–21014 [DOI] [PubMed] [Google Scholar]
- Abdallah BM, Boissy P, Tan Q, Dahlgaard J, Traustadottir GA, Kupisiewicz K, Laborda J, Delaisse JM, Kassem M 2007 dlk1/FA1 regulates the function of human bone marrow mesenchymal stem cells by modulating gene expression of pro-inflammatory cytokines and immune response-related factors. J Biol Chem 282:7339–7351 [DOI] [PubMed] [Google Scholar]
- Kim KA, Kim JH, Wang Y, Sul HS 2007 Pref-1 (preadipocyte factor-1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol 27:2294–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font de Mora J, Porras A, Ahn N, Santos E 1997 Mitogen activated protein kinase activation is not necessary for but antagonizes 3T3-L1 adipocyte differentiation. Mol Cell Biol 17:6068–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty D, Park BH, Davis KE, Farmer SR 2002 Activation of MEK/ERK signaling promotes adipogenesis by enhancing PPARγ and C/EBPα gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem 277:46226–46232 [DOI] [PubMed] [Google Scholar]
- Akiyama H 2008 Control of chondrogenesis by the transcription factor Sox9. Mod Rheumatol 18:213–219 [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R 2009 Sex determination and SRY: down to a wink and a nudge? Trends Genet 25:19–29 [DOI] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS 2002 Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol 22:5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Villena JA, Moon YS, Kim KH, Lee S, Kang C, Sul HS 2003 Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J Clin Invest 111:453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA, Choi CS, Wang Y, Kim S, Hwang YJ, Kim YB, Cline G, Shulman GI, Sul HS 2008 Resistance to high-fat diet-induced obesity but exacerbated insulin resistance in mice overexpressing preadipocyte factor-1 (Pref-1): a new model of partial lipodystrophy. Diabetes 57:3258–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B 2001 Haploinsufficiency of Sox9 results in defective cartilage primordial and premature skeletal mineralization. Proc Natl Acad Sci USA 98:6698–6703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Kan M, McKeehan WL, de Crombrugghe B 2000 Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA 97:1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Bonnamy JP, Owen MJ, Ducy P, Karsenty G 2001 Continuous expression of Cbfa1 in nonhypertrophic condrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa 1-deficient mice. Genes Dev 15:467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, Mulligan RC 1994 Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev 8:277–289 [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM 2002 Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev 16:859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G 2008 Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet 9:183–196 [DOI] [PubMed] [Google Scholar]
- Abdallah BM, Jensen CH, Gutierrez G, Leslie RG, Jensen TG, Kassem M 2004 Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res 19:841–852 [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM 2000 The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev 14:1997–2002 [PMC free article] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC 2000 Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol 10:1135–1138 [DOI] [PubMed] [Google Scholar]
- Wylie AA, Murphy SK, Orton TC, Jirtle RL 2000 Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res 10:1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Segers K, Karim L, Shay T, Gyapay G, Cockett N, Georges M 2001 The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet 4:367–369 [DOI] [PubMed] [Google Scholar]
- Lui JC, Finkielstain GP, Barnes KM, Baron J 2008 An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol 295:R189–R196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakajiri S, O'kelly J, Yin D, Miller CW, Hofmann WK, Oshimi K, Shih LY, Kim KH, Sul HS, Jensen CH, Teisner B, Kawamata N, Koeffler HP 2005 Dlk1 in normal and abnormal hematopoiesis. Leukemia 19:1404–1410 [DOI] [PubMed] [Google Scholar]
- Carlsson C, Tornehave D, Lindberg K, Galante P, Billestrup N, Michelsen B, Larsson LI, Nielsen JH 1997 Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/Delta-like protein in pancreatic islets: molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology 138:3940–3948 [DOI] [PubMed] [Google Scholar]
- Halder SK, Takemori H, Hatano O, Nonaka Y, Wada A, Okamoto M 1998 Cloning of a membrane-spanning protein with epidermal growth factor-like repeat motifs from adrenal glomerulosa cells. Endocrinology 139:3316–3328 [DOI] [PubMed] [Google Scholar]
- Jensen CH, Teisner B, Højrup P, Rasmussen HB, Madsen OD, Nielsen B, Skjødt K 1993 Studies on the isolation, structural analysis and tissue localization of fetal antigen 1 and its relation to a human adrenal-specific cDNA, pG2. Hum Reprod 8:635–641 [DOI] [PubMed] [Google Scholar]
- Laborda J, Sausville EA, Hoffman T, Notario V 1993 dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem 268:3817–3820 [PubMed] [Google Scholar]
- Moore KA, Pytowski B, Witte L, Hicklin D, Lemischka IR 1997 Hematopoietic activity of a stromal cell transmembrane protein containing epidermal growth factor-like repeat motifs. Proc Natl Acad Sci USA 94:4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen DC, Petersson SJ, Jorgensen LH, Bollen P, Jensen PB, Teisner B, Schroeder HK, Jensen CH 2009 Characterization of dlk1+ cells emerging during skeletal muscle remodeling in response to myositis, myopathies and acute injury. Stem Cells 27:898–908 [DOI] [PubMed] [Google Scholar]
- Jensen CH, Jauho EI, Santoni-Rugiu E, Holmskov U, Teisner B, Tygstrup N, Bisgaard HC 2004 Transit-amplifying ductular (oval) cells and their hepatocytic progeny are characterized by a novel and distinctive expression of Delta-like protein/preadipocyte factor 1/fetal antigen 1. Am J Pathol 164:1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Tsujimura T, Takahide K, Kodama T, Nakamura K, Miyajima A 2004 Expression of Dlk/Pref-1 defines a subpopulation in the oval cell compartment of rat liver. Gene Expr Patterns 5:209–218 [DOI] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa M, Saito H, Tsujimura T, Miyajima A 2003 Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J Cell Sci 116:1775–1786 [DOI] [PubMed] [Google Scholar]
- Terrace JD, Currie IS, Hay DC, Masson NM, Anderson RA, Forbes SJ, Parks RW, Ross JA 2007 Progenitor cell characterization and location in the developing human liver. Stem Cells Dev 16:771–778 [DOI] [PubMed] [Google Scholar]
- Raghunandan R, Ruiz-Hidalgo M, Jia Y, Ettinger R, Rudikoff E, Riggins P, Farnsworth R, Tesfaye A, Laborda J, Bauer SR 2008 Dlk1 influences differentiation and function of B lymphocytes. Stem Cells Dev 17:495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]