Abstract
Thyroid hormone receptors (TRs) are hormone-regulated transcription factors that control multiple aspects of physiology and development. TRs are expressed in vertebrates as a series of distinct isoforms that exert distinct biological roles. We wished to determine whether the two most widely expressed isoforms, TRα1 and TRβ1, exert their different biological effects by regulating different sets of target genes. Using stably transformed HepG2 cells and a microarray analysis, we were able to demonstrate that TRα1 and TRβ1 regulate a largely overlapping repertoire of target genes in response to T3 hormone. However, these two isoforms display very different transcriptional properties on each individual target gene, ranging from a much greater T3-mediated regulation by TRα1 than by TRβ1, to near equal regulation by both isoforms. We also identified TRα1 and TRβ1 target genes that were regulated by these receptors in a hormone-independent fashion. We suggest that it is this gene-specific, isoform-specific amplitude of transcriptional regulation that is the likely basis for the appearance and maintenance of TRα1 and TRβ1 over evolutionary time. In essence, TRα1 and TRβ1 adjust the magnitude of the transcriptional response at different target genes to different levels; by altering the ratio of these isoforms in different tissues or at different developmental times, the intensity of T3 response can be individually tailored to different physiological and developmental requirements.
Thyroid hormone receptors α1 and β1 regulate overlapping panels of target genes, yet mediate distinct levels of transcriptional activity on any given target gene.
Thyroid hormone receptors (TRs) are ligand-regulated transcription factors that play multiple roles in biology, including the control of nervous system development, cell proliferation, cardiac function, color vision, basal metabolism, lipid utilization, hearing, and metamorphosis (1,2,3,4,5). TRs typically modulate gene regulation by binding to specific DNA sequences (known as thyroid hormone response elements, or TREs) (4,6,7,8,9,10,11); consensus TREs are comprised of repeats of AGGTCA sequences in various orientations and spacings (12,13,14,15,16,17,18,19,20,21). TRs can also regulate gene transcription indirectly through protein-protein interactions with other transcription factors, as well as exert nongenomic effects (7,22,23,24).
TRs have bimodal transcription properties and can either repress or activate their target genes depending on whether hormone (T3) is bound or not bound to the TR (3,4,6,11,25,26,27,28,29,30,31,32). Positive regulatory targets of TRs refers to those genes that are repressed in the absence and activated in the presence of T3, whereas negative regulatory targets of TRs refers to the opposite: genes that are activated in the absence and repressed in the presence of T3.
TRs are encoded at two different loci, α and β, that give rise to four major isoforms by alternative mRNA splicing: TRα1, TRα2, TRβ1, and TRβ2 (1,3,11,33,34,35). Only TRα1, TRβ1, and TRβ2 are able to bind to thyroid hormone and constitute the classic thyroid hormone receptors; TRα2 may have hormone-independent function (3,11,33,34,35). TRα1 is expressed very early in development and is found in most adult tissues, whereas TRβ1 is expressed later in fetal development and is more restricted as to location in the adult (3,11,33,34,35,36). TRβ2 expression is most restrictive of all and is primarily confined to the pituitary, hypothalamus, cone cells of the retina, and the auditory cells of the cochlear (3,11,33,34,35,36,37). TRα1, TRβ1, and TRβ2 are highly homologous within their DNA- and hormone-binding domains; both bind T3 with similar affinities and can recognize common artificial consensus TREs in vitro.
TRα and TRβ appear to be the result of an ancestral gene duplication event that occurred approximately 550 million yr ago (38,39,40). The retention of these two separate loci during a half-billion years of subsequent evolution strongly suggests that the encoded TRs must exert distinct biological functions. Gene knockout mouse studies of the various TR isoforms have indeed revealed divergent functions for the different isoforms (33,35,36,41). For example, TRα knockout mice exhibit growth retardation due to delayed bone formation, as well as cardiac defects, whereas TRβ knockout mice display elevated circulating thyroid hormone levels, deafness, and defects in retina development (1,33,35,37,42,43,44,45). However these distinctions are not absolute: the two different TR loci display a significant degree of functional redundancy, and TRα and TRβ double-knockout mice exhibit more severe phenotypes than do the single-knockout mice (43,46).
Do TRα and TRβ mediate their different biological roles by regulating distinct sets of genes? To address this question, we examined global transcriptional regulation in HepG2 human liver cells engineered to highly express either TRα1 or TRβ1. Using microarray analysis, we determined that TRα1 and TRβ1 regulate a mostly shared panel of genes in response to T3. Despite this general overlap in target gene recognition, it was nonetheless apparent that the ability of TRα1 and TRβ1 to activate any given target gene in response to T3 displayed substantial isoform specificity. Although TRα1 displayed stronger transcriptional activation than did TRβ1 on several genes in these cells, the relative ability of TRβ1 to activate a given target gene in response to T3 ranged from virtually undetectable to slightly greater than that of TRα1. We propose that it is this TR isoform-specific control of the amplitude of transcriptional regulation that is the likely basis for the appearance and maintenance of TRα1 and TRβ1 over evolutionary time. By altering the ratio of TRα1 to TRβ1 in different tissues or at different developmental times, the magnitude of the T3 response can be individually tailored for different target genes, allowing the organism to customize the hormone response under different conditions to different physiological needs. We also identified an additional set of target genes that are repressed or activated by TRα1 and TRβ1 irrespective of T3 status, and which also appear likely to contribute to the divergent biological roles mediated by TRα1 and TRβ1.
Results
Stable ectopic expression of TRα1 or TRβ1 in HepG2 cells can dictate the T3 response in HepG2 cells
We wished to identify TRα and TRβ target genes in a cell type, such as hepatocytes, naturally responsive to thyroid hormone (5,10,47). We chose for our studies HepG2 cells, a well-characterized cell line derived from a human hepatocellular carcinoma that retains many liver-specific functions. HepG2 cells display a detectable response to thyroid hormone (e.g. Refs. 48,49,50,51,52), confirming that the necessary auxiliary machinery exists in these cells (53). Using quantitative RT-PCR (qRT-PCR) and suitable primers, we determined that TRα1 and TRβ1 are both expressed endogenously in these cells at modest, but near equal, levels (288 ± 15 vs. 294 ± 10 copies per ng RNA), consistent with their near equal expression in intact liver (54). To identify the specific target genes regulated by TRα1 or TRβ1, we chose a strategy of overexpressing one or the other TR isoform to create a TRα1- or TRβ1-dominant environment. We used a neomysin-resistance cotransformation protocol to isolate HepG2 cell lines containing an integrated ectopic human TRα1 construct, an integrated ectopic human TRβ1 construct, or an integrated empty expression plasmid (as a negative control).
Multiple independent HepG2 cell transformant clones were obtained for each construct and were analyzed for TR integration and expression, after which appropriate positive clones were pooled (at least six for each construct). We employed this strategy to minimize any possible clone to clone variation in the T3 response among the transformant population. We also confirmed the legitimacy of this pooling strategy by analysis of individual clones (described below).
Using qRT-PCR, we determined that overall TRα1 expression was elevated more than 4-fold in the TRα1 transformants relative to the empty vector control cells (Fig. 1A). Similarly, overall TRβ1 expression was elevated approximately 5-fold in the TRβ1 transformant pool (Fig. 1A). Ectopic overexpression of a given isoform did not detectably alter expression of the other, endogenous isoform (Fig. 1A). Other than the actual receptor-coding sequences, our TRα1 and TRβ1 expression constructs possess identical backbones, 5′- and 3′-untranslated regions, and Kozak sequences, and are expected to yield comparable translational efficiency/protein expression.
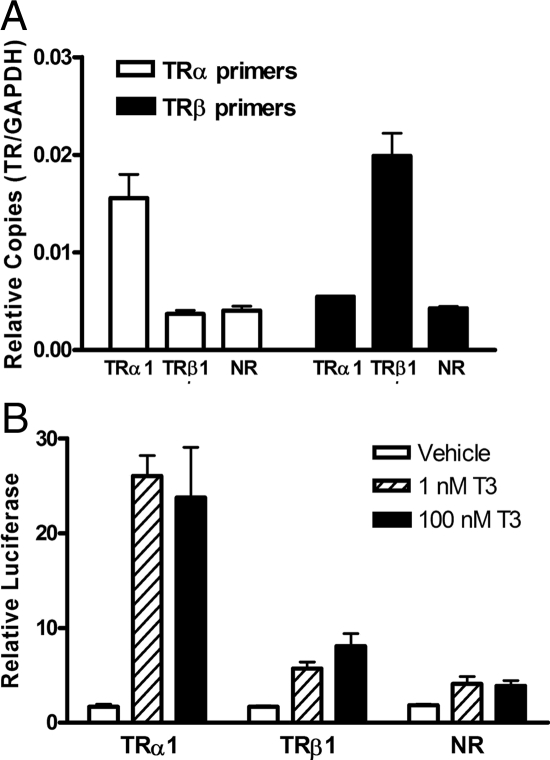
Figure 1.
Stable ectopic expression of TRα1 and TRβ1 in HepG2 cells results in enhanced T3-dependent gene activation. A, TRα1 and TRβ1 mRNA levels in HepG2 cells stably transformed with TRα1, TRβ1, or empty [no receptor (NR)] expression plasmids. Total RNA from each pool of stable HepG2 cells was isolated, and TR mRNA levels were determined by quantitative RT-PCR. Relative copy number was calculated as a ratio of TR to GAPDH expression used as an internal normalization control (mean + sem; n = 3). B, Enhanced T3-dependent reporter activation in TRα1 and TRβ1 transformants. HepG2 cells stably integrated with TRα1, TRβ1, or empty plasmid control (NR) were transiently transfected with a DR4-TK-luciferase reporter and a pCH110 construct as an internal control. The cells were treated with or without T3 24 h later as indicated and harvested 48 h after transfection, and the luciferase activity was determined relative to β-galactosidase activity (mean + sem; n = 3).
To examine the overall T3 response in our stable HepG2 transformants, each was transiently transfected with a thymidine kinase promoter-luciferase reporter bearing a generic DR-4 TR response element/binding site. The empty plasmid control transformants displayed a low basal level of luciferase expression that was induced 2- to 3-fold higher in the presence of 1 nm T3; no further activation was observed at 100 nm treatment (Fig. 1B). Both the TRα1 and the TRβ1 transformants displayed levels of basal luciferase expression approximately equal to that of the empty plasmid control (repression below basal level is not always observed with this reporter), but a much greater response to T3 (Fig. 1B). Notably the activation of luciferase expression by T3 in the TRβ1 transformants (∼6- to 8-fold over that seen in the absence of T3) was less than that observed for the TRα1 transformants (∼24- to 28-fold over that seen in the absence of T3) (Fig. 1B). This appears to reflect a generally somewhat higher transcriptional activity of the TRα1 isoform over the TRβ1 isoform (see below and Discussion). Our results, taken collectively, indicate that ectopic expression of TRα1 or TRβ1 significantly enhances the overall T3 response of our HepG2 transformants beyond that observed for the endogenous TRs.
TRα1 and TRβ1 regulate primarily overlapping sets of T3-responsive genes
To identify the specific cellular target genes regulated by TRα1 or TRβ1, our stable transformants expressing TRα1, TRβ1, or the empty plasmid control were treated with either ethanol vehicle alone, or ethanol plus 100 nm T3 for 6 h (a time period designed to primarily focus on direct, rather than secondary, targets of T3 regulation). RNAs were isolated from each transformant after the treatment period, were converted to cDNA probes, and were analyzed on Affymetrix GeneChip Human Gene 1.0 ST microarrays. Using R or dChip statistics packages and an unadjusted P value <0.05, 59–100 genes were regulated by T3 in the control transformants (presumably reflecting the activity of the endogenous TRs in these cells), and more than double that number in the ectopic TRα1 or TRβ1 transformants (data not shown). To minimize false positives and also focus on the genes that reflect the actions of the ectopically introduced TR isoforms, we further applied a Benjamini-Hochberg analysis to yield an adjusted P value of < 0.05. The genes that displayed a statistically significant induction by T3 in each cell population by these criteria are listed in Table 1 and are presented schematically in Fig. 2.
Table 1.
List of genes regulated by TRα1, TRβ1, and/or control after treatment with 100 nm T3 for 6 h compared with the absence of T3
| Regulated by | Symbol | Gene | Biological process | T3 responsea (fold, adj. P value)
|
||
|---|---|---|---|---|---|---|
| TRα1 | TRβ1 | |||||
| TRα1 | ANGPTL2 | Angiopoietin-like 2 | Signal transduction | 1.6 (0.036) | 1.4 (0.437) | |
| CLMN | Calmin | 1.7 (0.020) | 1.4 (1.000) | |||
| FLJ21963 | Metabolic process | 1.4 (0.043) | 1.3 (0.165) | |||
| GBE1 | Glucan (1,4-α-), branching enzyme 1 | Carbohydrate metabolic process | 2.5 (0.043) | 2.0 (0.468) | ||
| GRHPR | Glyoxylate reductase/hydroxypyruvate reductase | Electron transport/excretion | 1.5 (0.043) | 1.2 (1.000) | ||
| HERPUD1 | Homocysteine-inducible, ER stress-inducible, ubiquitin-like domain member 1 | Protein modification process | 1.3 (0.029) | 1.3 (0.107) | ||
| IRF1 | Interferon-regulatory factor 1 | Transcription/immune response/cell cycle | 2.1 (0.003) | 1.4 (1.000) | ||
| KCNJ10 | Potassium inwardly rectifying channel, subfamily j, member 10 | Potassium ion transport | 2.7 (1E-4) | 1.5 (0.787) | ||
| MMP11 | Matrix metallopeptidase 11 | Proteolysis | 1.7 (0.043) | 1.5 (0.486) | ||
| MPV17Lb | mpv17-like protein type 2 | 2.1 (0.002) | 1.8 (0.075) | |||
| RHPN2 | Rhophilin, ρ gtpase binding protein 2 | Signal transduction | 1.6 (0.029) | 1.5 (0.141) | ||
| SALL2 | sal-like 2 | Transcription | 1.7 (0.032) | 1.4 (0.914) | ||
| SLC38A3 | Solute carrier family 38, member 3 | Sodium ion transport/amino acid transport | 1.5 (0.029) | 1.4 (0.157) | ||
| TMEFF1 | Transmembrane protein with EGF-like and two FSH-like domains 1 | Multicellular organismal development | 1.5 (0.029) | 1.2 (1.000) | ||
| ZNF165 | Zinc finger protein 165 | Transcription | 1.8 (0.047) | 1.7 (0.156) | ||
| TRβ1 | ACSM2Bb | Hypothetical protein loc123876 | Metabolic process | 2.1 (0.081) | 2.2 (0.047) | |
| HIF1Ab | Hypoxia-inducible factor 1, α subunit | Response to hypoxia/signal transduction | 1.5 (0.068) | 1.5 (0.049) | ||
| MASP1b | Mannan-binding lectin serine peptidase 1 | Complement activation/proteolysis | 1.5 (0.090) | 1.7 (0.022) | ||
| PCK1 | Phosphoenolpyruvate carboxykinase 1 | Gluconeogenesis/lipid metabolic process | 2.0 (0.255) | 3.1 (0.004) | ||
| ZMIZ1b | Retinoic acid induced 17 | Transcription | 1.4 (0.084) | 1.5 (0.043) | ||
| TRα1/TRβ1 | ARSB | Arylsulfatase b | Lysosomal transport/glycosaminoglycan metabolic process | 1.7 (0.029) | 1.7 (0.023) | |
| C18orf17 | Chromosome 18 open reading frame 17 | 2.0 (1E-4) | 1.9 (0.001) | |||
| C1orf115 | Chromosome 1 open reading frame 115 | 1.5 (0.032) | 1.5 (0.023) | |||
| CABLES2 | cdk5 and abl enzyme substrate 2 | Cell cycle/regulation of cell division | 2.0 (0.002) | 1.8 (0.022) | ||
| CD14 | cd14 antigen | Apoptosis/immune response/signal transduction | 1.9 (0.002) | 1.7 (0.019) | ||
| CPT1A | Carnitine palmitoyltransferase 1a | Lipid/fatty acid metabolic process | 2.1 (0.003) | 2.0 (0.014) | ||
| CYP24A1 | Cytochrome p450, family 24, subfamily a, polypeptide 1 | Electron transport | 3.6 (0.017) | 3.6 (0.023) | ||
| (Continued) | ||||||
Table 1A.
Continued
| Regulated by | Symbol | Gene | Biological process | T3 responsea (fold, adj. P value)
|
||
|---|---|---|---|---|---|---|
| TRα1 | TRβ1 | |||||
| ENG | Endoglin | Cell adhesion/regulation of transcription | 2.6 (6E-5) | 1.7 (0.041) | ||
| FYN | fyn oncogene related to src, fgr, yes | T cell receptor signaling pathway | 3.5 (3E-8) | 2.6 (3E-6) | ||
| HIVEP2 | HIV type 1 enhancer binding protein 2 | Transcription | 2.3 (4E-5) | 2.4 (3E-5) | ||
| LDLRAD1 | Low-density lipoprotein receptor class a domain containing 1 | 1.7 (0.005) | 1.8 (0.003) | |||
| PPL | Periplakin | Keratinization | 1.9 (0.029) | 2.0 (0.020) | ||
| PTAFR | Platelet-activating factor receptor | Chemotaxis/immune response/signal transduction | 2.6 (2E-5) | 2.0 (0.002) | ||
| RAB11FIP4 | rab11 family interacting protein 4 (class ii) | 1.8 (2E-4) | 1.6 (0.004) | |||
| RASSF4 | ras association (ralgds/af-6) domain family 4 | Cell cycle/signal transduction | 1.7 (0.009) | 1.7 (0.004) | ||
| SCNN1A | Sodium channel, nonvoltage-gated 1 α | Sodium ion transport/excretion | 3.4 (4E-4) | 2.5 (0.022) | ||
| SERPINB9 | Serpin peptidase inhibitor, clade b, member 9 | Antiapoptosis/signal transduction | 2.1 (0.001) | 2.1 (0.002) | ||
| SGK | Serum/glucocorticoid regulated kinase 1 | Apoptosis/sodium ion transport | 1.8 (0.020) | 2.0 (0.004) | ||
| SLC16A6 | Solute carrier family 16, member 6 | Monocarboxylic acid transport | 2.5 (0.012) | 2.4 (0.023) | ||
| TNS4 | Tensin 4 | Apoptosis/intracellular signaling cascade | 2.8 (9E-5) | 2.0 (0.020) | ||
| TP53INP2 | Tumor protein p53-inducible nuclear protein 2 | 3.1 (0.001) | 2.9 (0.003) | |||
| USP30 | Ubiquitin-specific peptidase 30 | Ubiquitin cycle | 1.4 (0.016) | 1.4 (0.041) | ||
| SLC2A1 | Solute carrier family 2, member 1 | Glucose transport | 0.7 (0.012) | 0.6 (0.004) | ||
|
TRα1
|
TRβ1
|
NR
|
||||
| TRα1/TRβ1/NR | COQ10A | Coenzyme Q10 homolog A | 3.6 (3E-8) | 3.2 (3E-7) | 2.6 (1E-5) | |
Genes were identified as T3-responsive at a Benjamini-Hochberg (BH) adjusted P value <0.05. NR, No receptor.
T3 response values are presented as fold change, and the BH adjusted (adj.) P values are shown in parentheses.
Genes T3 regulated by both TRα1 and TRβ1 at a BH adjusted P < 0.1.
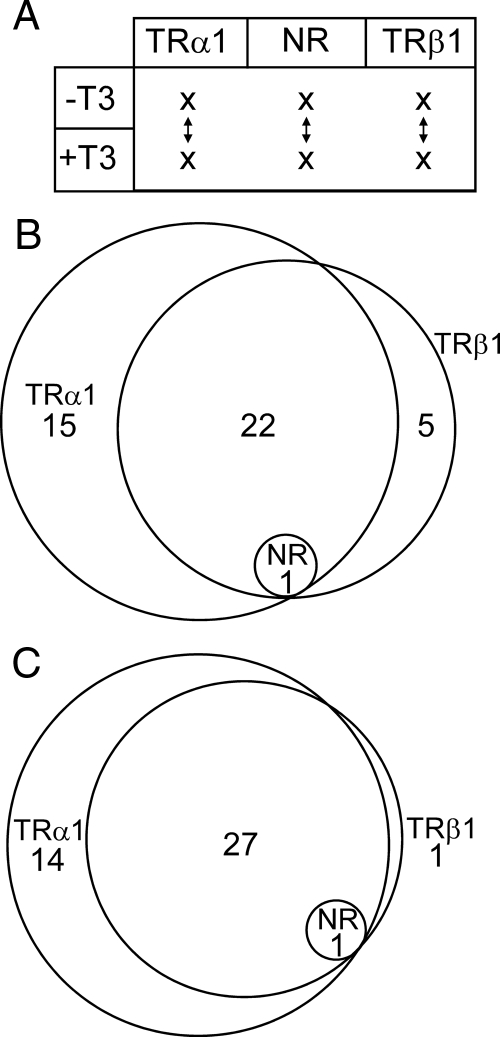
Figure 2.
TRα1 and TRβ1 regulate overlapping sets of T3-responsive genes. A, Schematic of pairwise comparisons employed: HepG2 cells stably transformed by TRα1, by TRβ1, or by the empty plasmid control were treated with vehicle only or with T3 for 6 h. RNA was isolated, and the genes in each transformant were up- or down-regulated by T3 were identified in the following pairwise comparisons: TRα1 transformants minus vs. plus T3, TRβ1 transformants minus vs. plus T3, and empty plasmid (NR) transformants minus vs. plus T3. B, Venn diagram of genes regulated by T3 in TRα1 transformants, by T3 in TRβ1 transformants, or by T3 in the empty plasmid control (NR), identified at a Benjamini-Hochberg (BH) adjusted P value of <0.05. The number of T3-regulated genes shared, or not, among the three different transformants are enumerated and are shown schematically as overlapping circles. C, The same genes identified as T3-up-regulated in panel B are presented again, but at a BH-adjusted P value of <0.1. NR, No receptor.
Under these stringent statistical criteria, only one gene, COQ10A, was flagged as up-regulated by T3 in the empty plasmid control transformants; examination of the microarray intensity values illustrates this more quantitatively (Fig. 3A), and this result was confirmed by RT-PCR assay (Fig. 4A). By the same criteria, 37 additional genes (plus COQ10A) were flagged as up-regulated by T3 in the TRα1 transformants, whereas 27 additional genes (plus COQ10A) were flagged as up-regulated by T3 in the TRβ1 transformants. Of the genes flagged as up-regulated by TRα1, 23 were independently flagged as up-regulated by TRβ1 (Fig. 2B) by the adjusted P < 0.05 criterion. One gene, SLC2A1, was found to be down-regulated by T3 in the TRα1 transformants (Table 1).
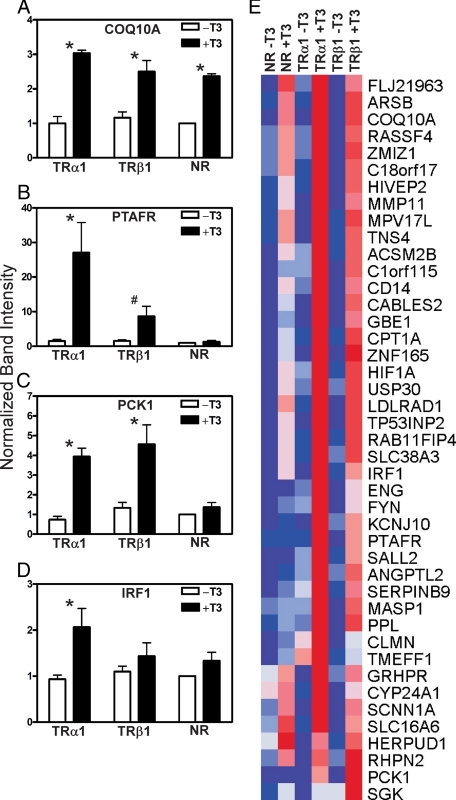
Figure 3.
T3 response of representative target genes. A–J, Microarray intensity signal values of mRNAs, minus and plus T3, for genes flagged as T3-regulated in the TRα1, TRβ1, or empty plasmid transformants (adjusted P < 0.05; mean + sd; n = 3). Asterisk indicates that the difference between minus and plus T3 was significant at a P value ≤0.05. NR, No receptor.
Figure 4.
Validation of microarray data by RT-PCR analysis and heat map of genes regulated by T3 in TRα1 or TRβ1 transformants. A–D, The mRNAs from TRα1, TRβ1, or empty plasmid (NR) transformants, minus or plus 6 h of T3 treatment, were subjected to RT-PCR using the gene-specific primers described in Materials and Methods. The resulting PCR products were stained with SYBR Green, and the gel band intensity values were quantified using an 8900 gel documentation system (Alpha Innotech, San Leandro, CA). The level of expression of each gene product was defined as 1 for the empty-vector control in the absence of T3 (mean + sem; n = 3). E, Heat map clustering of genes up-regulated by T3 in the HepG2 TRα1, TRβ1, or empty plasmid transformants (P <0.05). Dark blue indicates low expression, dark red indicates high expression, with intermediate values represented by lighter shades. *, The difference between minus and plus T3 was significant at a P value ≤0.05; #, the difference between minus and plus T3 was significant at a P value ≤0.1. NR, No receptor.
To examine whether these distinct sets of genes were truly unique, the same gene list in Table 1 was analyzed at a less stringent adjusted P value cutoff of P < 0.1. A Venn diagram at this P value shows that four of the five genes originally identified as being regulated by only TRβ1 at P < 0.05, are moved to the shared group of target genes under this criterion, indicating that they are also likely regulated by TRα1 (Fig. 2C). One gene, originally identified as only TRα1 regulated, also moved to the shared group of target genes under the adjusted P < 0.1 (Fig. 2C). We conclude that a majority of the genes regulated by T3 in these assays were shared targets of both TRα1 and TRβ1. Further individual analysis of these T3-regulated genes, below, elaborated on this concept.
Despite their ability to recognize a shared set of target genes, TRα1 and TRβ1 differ dramatically in the extent to which they can activate each target gene in response to T3
We next examined specific representatives of the T3-regulated genes characteristic of the three major categories: 1) shared, 2) TRα1 specific, or 3) TRβ1 specific. Two target genes, PTAFR and PPL, flagged as “shared” at an adjusted P < 0.05, were, as anticipated, induced by T3 in both the TRα1 and TRβ1 transformants, but not in the empty plasmid control cells; this was observed by microarray intensity measurement in three biological repeats and, for PTAFR, confirmed by RT-PCR (Fig. 3, B and C, and Fig. 4B). Two representative target genes, PCK1 and MASP1, flagged as “TRβ1-specific” at an adjusted P value <0.05, were induced by T3 in the TRβ1 transformants, as expected, but also were detectably induced by T3 in the TRα1 transformants (Fig. 3, D and E, and Fig. 4C). In fact, MASP1 was among the TRβ1-specific genes that joined the shared category when the stringency of the array analysis was decreased to an adjusted P < 0.1. We conclude that our initial exclusion of MASP1 and PCK1 from the shared group more likely reflected our initial choice of a statistical cutoff in our overall array analysis, rather than a true, TRβ1-specific regulation of these genes.
Similarly, a more detailed analysis of two target genes, KCNJ10 and MPV17L, flagged as “TRα1-specific” at an adjusted P < 0.05, showed that these genes were indeed activated by T3 in the TRα1 transformants; however, it appeared that these genes were also activated, although to a lesser extent, in the TRβ1 transformants (Fig. 3, F and G); indeed MPV17L was one of the TRα1-specific genes that became regrouped as “shared” when the adjusted P value stringency was lowered to P < 0.1 (Fig. 2C). KCNJ10 and MPV17L were not regulated by T3, or were regulated to a lesser degree, in transformants bearing the empty plasmid (Fig. 3, F and G).
Our results indicate that TRα1 and TRβ1 regulate a largely overlapping set of target genes in response to T3; however, what is also clear is that these two different isoforms display very different transcriptional properties on each of these different targets. TRα1 was a stronger activator in response to T3 than is TRβ1 on several genes (as was also observed using the artificial DR-4-tk-luciferase reporter; Fig. 1B). Nonetheless, there was a wide range of relative activities of TRα1 to TRβ1 on the individual target genes, with certain target genes exhibiting equal or slightly greater T3 activation in the TRβ1 transformants than in the TRα1 transformants (e.g. PCK1), other target genes displaying a somewhat lower T3 response in the TRβ1 transformants than in the TRα1 transformants (e.g. PTAFR or KCNJ10), and a few target genes (e.g. IRF1, CLMN, and TMEFF1) displaying a T3 response statistically distinct from the empty plasmid controls in the TRα1 transformants, but not in the TRβ1 transformants (Table 1). Although the final category may truly represent TRα1-specific target genes, individual analysis of these genes (Fig. 3, H–J, and Fig. 4D) suggest they are also potentially, if weakly, TRβ1 responsive. We therefore cannot exclude the possibility that the lack of a clearly enhanced T3 response in the TRβ1 transformants in these cases (compared with empty plasmid controls) simply reflects a much weaker transcriptional response of TRβ1, rather than an absolute isoform specificity.
The range of transcriptional responses for all the genes flagged by the microarray analysis as regulated by T3 is displayed in a heat map in Fig. 4E. Many of these genes displayed little or no detectable regulation by T3 in the absence of an ectopically introduced TR, whereas other genes were regulated by T3 in the empty plasmid control, but were regulated more strongly in one or both TR transformants. Parallel results were observed when a variety of selected genes from the different categories were validated by RT-PCR (data not shown).
To confirm that these patterns of gene regulation were not distorted by our strategy of analyzing pooled transformants, we repeated RT-PCR validations on several target genes using individual clonal populations of our TRα1 and TRβ2 transformants (choosing clones displaying comparable levels of TR expression). Notably, very similar patterns of target gene expression were obtained on these individual clones as were seen for the pooled analysis (e.g. supplemental Figs. S1A and S1B published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). We conclude that TRα1 and TRβ1 recognize a large panel of shared target genes, but that these two isoforms differ in the transcriptional response they can initiate once arrayed on each individual target gene.
TRα1 and TRβ1 display additional regulatory effects on target genes that are observable irrespective of the presence or absence of T3
Our results to this point focused on genes that displayed a statistically significant change in expression in response to T3 in our HepG2 transformants (Fig. 2A). However, this focus on T3 response could potentially ignore genes that are regulated by TRs irrespective of the absence or presence of T3, i.e. in a constitutive fashion. To explore this aspect, the microarray data for our various stable transformants were reanalyzed in four pairwise comparisons (Fig. 5A) to identify: 1) genes that are down-regulated by introducing TRα1 relative to the empty plasmid, regardless of T3 status; 2) genes that are down-regulated by introducing TRβ1 relative to the empty plasmid, regardless of T3 status; 3) genes that are up-regulated by introducing TRα1 relative to the empty plasmid, regardless of T3 status; and 4) genes that are up-regulated by introducing TRβ1 relative to the empty plasmid, regardless of T3 status.
Figure 5.
A subset of genes are down- or up-regulated by TRα1 or TRβ1 irrespective of T3 status. A, Schematic of pairwise comparisons employed: HepG2 cells stably transformed by TRα1, by TRβ1, or by the empty plasmid control were treated with vehicle only or with T3 for 6 h. RNA was isolated, and the genes down- or up-regulated by the presence of the ectopic TRα1 or TRβ1 were identified in the following pairwise comparisons: TRα1 transformants vs. empty plasmid (NR) transformants in the absence of T3, TRβ1 transformants vs. empty plasmid (NR) transformants in the absence of T3, TRα1 transformants vs. empty plasmid (NR) transformants in the presence of T3, TRβ1 transformants vs. empty plasmid (NR) transformants in the presence of T3. B, Venn diagram of genes down-regulated in TRα1 transformants compared with the empty plasmid (NR) control, or down-regulated in TRβ1 transformants compared with the empty plasmid (NR) control regardless of T3 concentration. All were flagged using a Benjamini-Hochberg (BH) adjusted P value of <0.05. The number of down-regulated genes shared, or not, between the TRα1 and TRβ1 transformants is enumerated and is shown schematically as overlapping circles. C, Venn diagram of genes up-regulated in TRα1 transformants compared with the empty plasmid (NR) control, or up-regulated in TRβ1 transformants compared with the empty plasmid (NR) control regardless of T3 concentration. All were flagged using a Benjamini-Hochberg (BH) adjusted P value <0.05. The number of up-regulated genes shared, or unshared, between the TRα1 and TRβ1 transformants is enumerated and is shown schematically as overlapping circles. NR, No receptor.
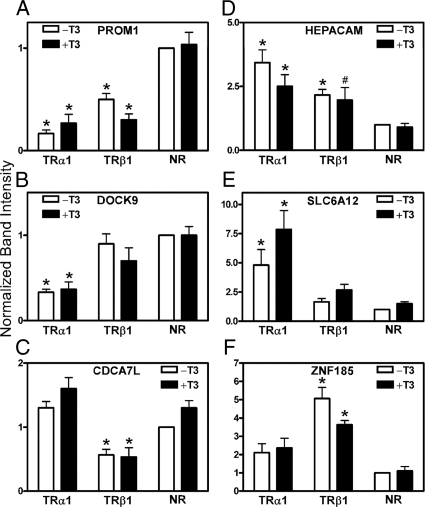
Using an adjusted P value <0.05, relative to the empty plasmid control, the introduction of TRα1 repressed 33 genes, and the introduction of TRβ1 repressed 12 genes in both the absence and presence of T3 (Fig. 5B and Table 2). Nine of these genes were repressed in both the TRα1 and TRβ1 transformants. The PROM1 gene, for example, was repressed by both TR isoforms in either the absence or presence of T3 (Table 2 and confirmed by RT-PCR; Fig. 6A). Several of the repressed genes flagged as TRα1 or TRβ1 specific in the microarray analysis appeared to be truly isoform specific on additional analysis. DOCK9, for example, was significantly repressed only in the TRα1 transformants, whereas CDCA7L was significantly repressed only in the TRβ1 transformants (Table 2 and Fig. 6, B and C). Several of these genes were repressed constitutively and displayed no detectable response to T3 (e.g. DOCK9 in the TRα1 transformant), whereas others were marginally repressed further in the presence of T3 than in its absence (e.g. PROM1 for TRβ1) (Fig. 6); these changes in response to T3 were too modest to be detected as T3 responsive in our initial analysis. Additional specific examples were confirmed by RT-PCR validation (e.g. supplemental Fig. S1C), and a complete list of genes repressed by the introduction of TRα1 or TRβ1 in either the absence or presence of T3 is found in Table 2.
Table 2.
List of genes down-regulated by TRα1 and/or TRβ1 compared with control
| Regulated by | Symbol | Gene | Biological process | Fold change
|
|||
|---|---|---|---|---|---|---|---|
| −T3 | +T3 | ||||||
| TRα1 | AFP | α-Fetoprotein | Transport/immune response | NS | 0.8 | ||
| ATP10A | ATPase, class v, type 10c | Phospholipid transport | NS | 0.6 | |||
| C14orf126 | Chromosome 14 open reading frame 126 | d-Amino acid catabolic process | NS | 0.7 | |||
| CAP2 | Cap, adenylate cyclase-associated protein, 2 | Signal transduction | 0.5 | 0.5 | |||
| CYP7A1 | Cytochrome p450, family 7, subfamily a, polypeptide 1 | Electron transport/lipid metabolic process | NS | 0.5 | |||
| DOCK9 | Dedicator of cytokinesis 9 | 0.4 | 0.4 | ||||
| F13B | Coagulation factor xiii, b polypeptide | Blood coagulation | NS | 0.4 | |||
| FKBP14 | fk506 Binding protein 14, 22 kDa | Protein folding | NS | 0.7 | |||
| FRMD3 | FERM domain containing 3 | NS | 0.6 | ||||
| GDA | Guanine deaminase | Nucleotide and nucleic acid metabolic process | 0.5 | 0.5 | |||
| LYZ | Lysozyme | Inflammatory response/metabolic process | NS | 0.7 | |||
| NR0B2 | Nuclear receptor subfamily 0, group b, member 2 | Transcription/cholesterol metabolic process | 0.6 | NS | |||
| NUBPL | Nucleotide-binding protein-like | NS | 0.8 | ||||
| OXA1 liter | Oxidase (cytochrome c) assembly 1-like | Electron transport | 0.8 | NS | |||
| PAWR | prkc, apoptosis, wt1, regulator | Transcription/apoptosis | NS | 0.7 | |||
| PCSK5 | Proprotein convertase subtilisin/kexin type 5 | Proteolysis/cell-cell signaling | 0.7 | NS | |||
| PELI1 | Pellino homolog 1 | 0.7 | NS | ||||
| SCN1A | Sodium channel, voltage-gated, type i, α | Sodium ion transport | NS | 0.5 | |||
| SGEF | src Homology 3 domain-containing guanine nucleotide exchange factor | Regulation of ρ protein signal transduction | 0.6 | NS | |||
| SLC2A1 | Solute carrier family 2, member 1 | Glucose transport | NS | 0.7 | |||
| SNX6 | Sorting nexin 6 | Intracellular protein transport/cell communication | NS | 0.7 | |||
| TFPI | Tissue factor pathway inhibitor | Blood coagulation | NS | 0.6 | |||
| TTR | Transthyretin | Thyroid hormone generation/transport | 0.7 | 0.6 | |||
| VPS54 | Vacuolar protein sorting 54 | Protein transport | 0.7 | 0.7 | |||
|
TRα1
|
TRβ1
|
||||||
|
−T3
|
+T3
|
−T3
|
+T3
|
||||
| TRα1/TRβ1 | ANKRD1 | Ankyrin repeat domain 1 | Signal transduction | 0.5 | 0.4 | NS | 0.4 |
| BICC1 | Bicaudal c homolog 1 | Visual perception/response to stimulus | 0.4 | 0.3 | 0.4 | 0.4 | |
| DPYD | Dihydropyrimidine dehydrogenase | Multicellular organismal development | 0.6 | 0.6 | 0.6 | 0.6 | |
| ERRFI1 | erbb receptor feedback inhibitor 1 | Electron transport/metabolic process | 0.7 | NS | 0.7 | NS | |
| GPC3 | Glypican 3 | Cell proliferation/growth | 0.6 | 0.7 | 0.7 | 0.7 | |
| HDAC1 | Histone deacetylase 1 | Transcription/chromatin modification | 0.7 | 0.7 | NS | 0.7 | |
| LTB4DH | Leukotriene b4 12-hydroxydehydrogenase | Response to stress | 0.5 | 0.5 | 0.5 | 0.5 | |
| PROM1 | Prominin 1 | Leukotriene metabolic process | 0.3 | 0.2 | 0.5 | 0.4 | |
| SCHIP1 | Schwannomin interacting protein 1 | NS | 0.6 | NS | 0.5 | ||
|
−T3
|
+T3
|
||||||
| TRβ1 | BHMT2 | Betaine-homocysteine methyltransferase 2 | 0.6 | NS | |||
| C3orf57 | Chromosome 3 open reading frame 57 | 0.6 | NS | ||||
| CDCA7 liter | Cell division cycle associated 7-like | Transcription | 0.5 | 0.5 | |||
Genes were identified at a Benjamini-Hochberg adjusted P value <0.05. NS, Not significant (difference from control did not meet a P value cutoff of 0.05).
Figure 6.
Validation of microarray data by RT-PCR analysis: genes down- or up-regulated by TRα1 or TRβ1 regardless of T3 status. A–F, The mRNAs from TRα1, TRβ1, or empty plasmid (NR) transformants, minus or plus 6 h of T3 treatment, were subjected to RT-PCR using the gene-specific primers described in Materials and Methods. The resulting PCR products were stained with SYBR Green, and the gel band intensity values were quantified using an 8900 gel documentation system (Alpha Innotech). The level of expression of each gene product was defined as 1 for the empty-vector control in the absence of T3 (mean + sem; n = 3). *, The difference between vector only and plus the TR isoform was significant at a P value ≤ 0.05; #, the difference was significant at a P value ≤0.1. NR, No receptor.
Finally we flagged the genes that were activated in response to expression of TRα1 or TRβ1 in both the absence and presence of hormone. Compared with the empty plasmid control, introduction of TRα1 up-regulated 105 target genes in both the absence and presence of T3, whereas introduction of TRβ1 up-regulated 38 genes in both the absence and presence of T3; 30 of these up-regulated genes were shared between both isoforms (Fig. 5C and Table 3). HEPACAM, for example, was up-regulated by the ectopic expression of either TR isoform (Fig. 6D). SLC6A12 was strongly up-regulated in the TRα1 transformants both minus or plus T3 (although more activation was detected in the presence of T3), but was only slightly up-regulated in the TRβ1 transformants (Fig. 6E). Conversely, ZNF185 was more strongly up-regulated in the TRβ1 transformants than in the TRα1 transformants (Fig. 6F). Additional specific examples were confirmed by RT-PCR validation (e.g. supplemental Fig. S1D), and a complete list of genes up-regulated by TRα1 or TRβ1 in both the presence and absence of T3 is displayed in Table 3.
Table 3.
List of genes up-regulated by TRα1 and/or TRβ1 compared with control
| Regulated by | Symbol | Gene | Biological process | Fold change
|
|||
|---|---|---|---|---|---|---|---|
| −T3 | +T3 | ||||||
| TRα1 | ALDH8A1 | Aldehyde dehydrogenase 8 family, member a1 | Retinoic acid metabolic process | 2.3 | 1.8 | ||
| ANGPTL2 | Angiopoietin-like 2 | Signal transduction | NS | 1.7 | |||
| APCDD1 | Adenomatosis polyposis coli down-regulated 1 | 2.0 | 1.7 | ||||
| ARSI | Arylsulfatase i | Metabolic process | 2.7 | NS | |||
| BHMT | Betaine-homocysteine methyltransferase | Protein amino acid methylation | 2.0 | NS | |||
| C12orf40 | Chromosome 12 open reading frame 40 | 2.1 | NS | ||||
| C1S | Complement component 1, s subcomponent | Proteolysis/complement activation/immune response | NS | 2.3 | |||
| C1orf115 | Chromosome 1 open reading frame 115 | NS | 1.5 | ||||
| C2orf54 | Hypothetical protein flj22671 | NS | 1.5 | ||||
| CCL16 | Chemokine (c-c motif) ligand 16 | Chemotaxis/immune response/cell-cell signaling | NS | 2.2 | |||
| CD14 | cd14 antigen | Apoptosis/immune response/signal transduction | NS | 1.5 | |||
| CENTD1 | Centaurin, δ 1 | Signal transduction | 1.8 | 1.5 | |||
| CES4 | Carboxylesterase 4-like | Xenobiotic metabolic process | 1.9 | 2.0 | |||
| CLDN6 | Claudin 6 | Calcium-independent cell-cell adhesion | 3.0 | 3.3 | |||
| CLMN | Calmin | NS | 2.1 | ||||
| CORIN | Corin, serine peptidase | Proteolysis/lipid metabolic process | NS | 2.1 | |||
| CYBRD1 | Cytochrome b reductase 1 | Electron transport | 2.8 | 3.1 | |||
| DSC3 | Desmocollin 3 | Cell adhesion | NS | 2.6 | |||
| FGF2 | Fibroblast growth factor 2 (basic) | Angiogenesis/chemotaxis/signal transduction/cell signaling/proliferation | 1.7 | 2.0 | |||
| G6PD | Glucose-6-phosphate dehydrogenase | Glucose metabolic process | 1.3 | 1.4 | |||
| GAL3ST1 | Galactose-3-o-sulfotransferase 1 | Galactosylceramide biosynthetic process/myelination | 1.5 | NS | |||
| H2AFY2 | h2a histone family, member y2 | Nucleosome assembly/dosage compensation/chromatin modification | 1.7 | 1.6 | |||
| HMOX1 | Heme oxygenase (decycling) 1 | Heme oxidation/regulation of I-κB kinase/NF-κB cascade | NS | 2.1 | |||
| HPX | Hemopexin | Cellular iron ion homeostasis/heme transport | 2.6 | NS | |||
| HSPB1 | Heat shock 27 kDa protein 1 | Regulation of translational initiation/antiapoptosis/cell motility | 1.4 | NS | |||
| IL1R1 | IL-1 receptor, type I | Immune response | 1.7 | 1.5 | |||
| ITGB7 | Integrin, β 7 | Cell adhesion/integrin-mediated signaling | NS | 1.4 | |||
| ITIH4 | inter-α (globulin) inhibitor h4 | Hyaluronan metabolic process | 1.5 | NS | |||
| KCNJ10 | Potassium inwardly rectifying channel, subfamily j, member 10 | Potassium ion transport | NS | 2.5 | |||
| KLF12 | Kruppel-like factor 12 | Transcription | NS | 1.9 | |||
| MAGEA3 | Melanoma antigen family A, 3 | 2.8 | 3.2 | ||||
| MASP1 | Mannan-binding lectin serine peptidase 1 | Complement activation/proteolysis | NS | 1.5 | |||
| MCC | Mutated in colorectal cancers | Cell cycle/signal transduction | 1.7 | NS | |||
| MEP1A | Meprin a, α (PABA peptide hydrolase) | Proteolysis | NS | 1.9 | |||
| MKRN3 | Makorin, ring finger protein, 3 | 1.6 | 1.5 | ||||
| MNS1 | Meiosis-specific nuclear structural 1 | Meiosis | 2.0 | 2.0 | |||
| MOGAT1 | Monoacylglycerol o-acyltransferase 1 | Lipid metabolic/biosynthetic process | 1.5 | NS | |||
| MSRB3 | Methionine sulfoxide reductase b3 | Protein repair | NS | 1.6 | |||
| MTMR8 | Myotubularin related protein 8 | Phospholipid dephosphorylation | 1.6 | NS | |||
| MYL9 | Myosin, light polypeptide 9, regulatory | Regulation of muscle contraction | 1.9 | 2.1 | |||
| NOTCH2NL | Notch homolog 2 (Drosophila) N-terminal like | Notch signaling pathway/cell differentiation | 1.4 | NS | |||
| NPNT | Nephronectin | Cell adhesion/cell differentiation | 2.2 | 2.2 | |||
| PAK1 | p21/cdc42/rac1-activated kinase 1 | Apoptosis/ER-nuclear signaling pathway/cytoskeleton organization | 2.2 | 2.0 | |||
| PLAGL1 | Pleiomorphic adenoma gene-like 1 | Transcription/induction of apoptosis/cell cycle arrest | 1.8 | 2.1 | |||
| PLCE1 | Phospholipase c, ε 1 | Lipid metabolic process/signal transduction/cell proliferation | 1.9 | 1.6 | |||
| PLP1 | Proteolipid protein 1 | Synaptic transmission | 1.5 | 1.7 | |||
| PPL | Periplakin | Keratinization | NS | 1.8 | |||
| PRKCE | Protein kinase c, ε | Apoptosis/intracellular signaling cascade | NS | 1.6 | |||
| PRRG4 | Praline-rich gla (g-carboxyglutamic acid) 4 | NS | 1.8 | ||||
| PTPRJ | Protein tyrosine phosphatase, receptor type, j | Cell-cell signaling | 1.7 | 2.0 | |||
| RCN1 | Reticulocalbin 1, ef-hand calcium binding domain | 1.7 | 1.6 | ||||
| (Continued) | |||||||
Table 3A.
Cont.
| Regulated by | Symbol | Gene | Biological process | Fold change
|
|||
|---|---|---|---|---|---|---|---|
| −T3 | +T3 | ||||||
| RNF157 | Ring finger protein 157 | NS | 1.7 | ||||
| SALL2 | sal-like 2 | Transcription | NS | 2.0 | |||
| SERPIND1 | Serpin peptidase inhibitor, clade d, member 1 | Chemotaxis/blood coagulation | 1.3 | 1.3 | |||
| SERPINF2 | Serpin peptidase inhibitor, clade f, member 2 | Acute-phase response | 1.4 | 1.4 | |||
| SERPING1 | Serpin peptidase inhibitor, clade g, member 1 | Complement activation | 2.2 | 2.3 | |||
| SIRPA | Protein tyrosine phosphatase, nonreceptor type substrate 1 | Cell adhesion | NS | 2.0 | |||
| SLC22A7 | Solute carrier family 22, member 7 | Ion transport | 2.2 | 2.1 | |||
| SLC6A12 | Solute carrier family 6, member 12 | Neurotransmitter transport | 1.9 | 2.4 | |||
| SLCO2B1 | Solute carrier organic anion transporter family, member 2b1 | Ion transport | NS | 2.4 | |||
| SPARC | Secreted protein, acidic, cysteine-rich | Transmembrane receptor protein tyrosine kinase signaling pathway | 2.4 | 2.3 | |||
| TEAD2 | TEA domain family member 2 | Transcription | 1.5 | 1.5 | |||
| TGM4 | Transglutaminase 4 | Protein amino acid polyamination | 1.5 | NS | |||
| THRA | TRα | Transcription | NS | 1.9 | |||
| TIMP3 | TIMP metallopeptidase inhibitor 3 | Visual perception/apoptosis | NS | 1.6 | |||
| TMEFF1 | Transmembrane protein with EGF-like and two FSH-like domains 1 | Multicellular organismal development | 1.9 | 2.1 | |||
| TNS1 | Tensin 1 | Intracellular signaling cascade | NS | 1.9 | |||
| TRIM2 | Tripartite motif-containing 2 | 1.8 | 1.7 | ||||
| TRIM73 | Tripartite motif-containing 50b | 2.6 | NS | ||||
| TRIM74 | Tripartite motif-containing 50c | NS | 1.4 | ||||
| TSPAN7 | Tetraspanin 7 | Protein amino acid N-linked glycosylation | 1.7 | 2.1 | |||
| TTC7B | Tetratricopeptide repeat domain 7b | NS | 1.8 | ||||
| WNT11 | Wingless-type MMTV integration site family, member 11 | Signal transduction/Wnt receptor signaling | NS | 2.1 | |||
| WT1 | Wilms tumor 1 | Transcription/cell cycle | 1.6 | 1.7 | |||
| ZAK | Sterile α motif and leucine zipper containing kinase azk | Cell cycle/DNA damage checkpoint/cell death/proliferation/differentiation | 1.6 | 1.5 | |||
|
TRα1
|
TRβ1
|
||||||
|
−T3
|
+T3
|
−T3
|
+T3
|
||||
| TRα1/TR1 | ARHGEF9 | cdc42 guanine nucleotide exchange factor (gef) 9 | Regulation of ρ protein signal transduction | 2.1 | 1.7 | 1.7 | NS |
| AYTL1 | Acyltransferase like 1 | Phospholipid biosynthetic process | 1.4 | 1.5 | 1.8 | 1.8 | |
| CES1 | Carboxylesterase 1 | Metabolic process/response to toxin | 1.9 | 2.2 | NS | 1.6 | |
| CHN1 | Chimerin 1 | Intracellular signaling cascade | 3.9 | 4.8 | 2.2 | 2.2 | |
| CNNM1 | Cyclin m1 | ion transport | 2.0 | 1.9 | 1.5 | 1.5 | |
| ENG | Endoglin | Cell adhesion/regulation of transcription | NS | 3.3 | NS | 1.9 | |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | Cell motility/chemotaxis/lipid catabolic process | 3.6 | 5.0 | 2.0 | 2.3 | |
| FLJ21986 | 2.2 | 2.1 | 2.0 | 1.9 | |||
| FYN | fyn Oncogene related to src, fgr, yes | T cell receptor signaling pathway | 1.8 | 4.0 | NS | 1.8 | |
| HEPACAM | Hepatocyte cell adhesion molecule | Cell adhesion/cell cycle/cell growth | 2.0 | 1.7 | 2.1 | 1.8 | |
| HIST1H2AH | Histone cluster 1, H2ah | Nucleosome assembly | 3.5 | 2.8 | 2.2 | 2.1 | |
| HIST1H2BK | Histone 1, h2bk | Nucleosome assembly | 6.6 | 9.7 | NS | 6.8 | |
| HTR1D | 5-Hydroxytryptamine (serotonin) receptor 1d | Signal transduction | 1.9 | 2.3 | 1.9 | 2.1 | |
| INS | Insulin | Glucose metabolic process/cell-cell signaling/cell death | NS | 1.4 | NS | 1.4 | |
| KLHL13 | Kelch-like 13 | 2.0 | 2.1 | 2.3 | 2.0 | ||
| MUC15 | Mucin 15 | 1.9 | 2.0 | 1.9 | 1.9 | ||
| MX1 | Myxovirus resistance 1, interferon-inducible protein p78 | Immune response/signal transduction | 1.4 | 1.5 | 1.5 | NS | |
| MX2 | Myxovirus resistance 2 | Immune response | 1.6 | 1.6 | 1.6 | 1.6 | |
| NAT8 liter | Hypothetical protein loc339983 | Metabolic process | 1.8 | 2.1 | 1.6 | 1.8 | |
| OLFML1 | Olfactomedin-like 1 | 3.3 | 2.6 | 2.5 | NS | ||
| PRKCH | Protein kinase c, η | Intracellular signaling cascade | 2.4 | 2.2 | 2.1 | 1.7 | |
| PTAFR | Platelet-activating factor receptor | Chemotaxis/immune response/signal transduction | NS | 2.7 | NS | 1.8 | |
| RNASE2 | Ribonuclease, rnase a family, 2 | RNA catabolic process/chemotaxis | NS | 1.5 | NS | 1.6 | |
| (Continued) | |||||||
Table 3B.
Cont.
| Regulated by | Symbol | Gene | Biological process | Fold change
|
|||
|---|---|---|---|---|---|---|---|
| −T3 | +T3 | ||||||
| SERPINB9 | Serpin peptidase inhibitor, clade b, member 9 | Antiapoptosis/signal transduction | NS | 2.5 | NS | 2.0 | |
| SLAIN1 | Hypothetical protein flj30046 | 2.7 | 2.7 | 2.0 | NS | ||
| SLC37A2 | Solute carrier family 37, member 2 | Transport | NS | 3.1 | NS | 2.1 | |
| SULT1C2 | Sulfotransferase family, cytosolic, 1c, member 1 | Amine metabolic process | 3.2 | 2.9 | 2.3 | NS | |
| TRIB2 | Tribbles homolog 2 | Regulation of MAPK activity | 2.2 | 2.9 | 1.9 | 2.3 | |
| UGT2B4 | UDP glucuronosyltransferase 2 family, polypeptide b4 | Estrogen/xenobiotic metabolic process | 3.6 | 4.1 | 2.5 | 2.9 | |
| VCAN | Chondroitin sulfate proteoglycan 2 | Cell adhesion/cell recognition | 5.5 | 5.3 | 3.6 | 2.9 | |
|
−T3
|
+T3
|
||||||
| TRβ1 | BBS5 | Bardet-Biedl syndrome 5 | Visual perception/response to stimulus | 1.6 | NS | ||
| ENPP4 | Ectonucleotide pyrophosphatase/phosphodiesterase 4 | Nucleotide metabolic process | NS | 1.6 | |||
| HERC5 | HECT domain and rld 5 | Regulation of cyclin-dependent protein kinase activity/ubiquitin cycle | 1.7 | NS | |||
| NEGR1 | Neuronal growth regulator 1 | Cell adhesion | NS | 1.5 | |||
| NRCAM | Neuronal cell adhesion molecule | Central nervous system development/cell-cell adhesion | 1.8 | 1.8 | |||
| PCK1 | Phosphoenolpyruvate carboxykinase 1 | Gluconeogenesis/lipid metabolic process | NS | 3.0 | |||
| SGK | Serum/glucocorticoid-regulated kinase 1 | Apoptosis/sodium ion transport | NS | 2.0 | |||
| ZNF185 | Zinc finger protein 185 | 1.8 | 1.7 | ||||
Genes were identified at a Benjamini-Hochberg adjusted P value <0.05. NS, Not significant (difference from control did not meet a P value cutoff of < 0.05).
These data suggest that TRα1 and TRβ1 can not only regulate genes in a T3-responsive manner but also in a hormone-independent fashion. This hormone-independent regulation occurs upon introduction of the TR irrespective of the presence of T3; in certain cases this regulation by receptor is truly neutral to hormone, whereas in other cases the effect of adding the receptor in the absence of T3 is further modified by the addition of T3. Intriguingly, somewhat greater isoform-specific regulation was observed for these hormone-independent target genes (Fig. 5, B and C, and Fig. 6) than for the target genes identified through their T3 responsiveness (Figs. 2, 3, and 6).
Discussion
TRα1 and TRβ1 regulate a shared panel of target genes
TRα1 and TRβ1 represent the products of an ancestral gene duplication and divergence that occurred early in the vertebrate lineage approximately 550 million years ago (38,39); their maintenance as separate loci in all extant gnathostomes (including all cartilaginous and bony fish, amphibians, reptiles, and mammals examined) powerfully suggests that they play nonredundant functions in evolutionary terms. In fact, there is much stronger sequence conservation between the same isoform compared among different species than between the TRα1 and TRβ1 isoforms compared within the same species (1,3,11,33,34,35). Their patterns of expression are also indicative of distinct biological functions: TRα1 is expressed very early in embryonic development and is found in virtually all tissues, whereas TRβ1 expression arises later in fetal development, cotemporal with the appearance of T3 hormone, and exhibits a more organ-specific pattern and distinct diurnal cycle than does TRα1 (1,3,11,33,34,35,55). Parsing these characteristics together with those of other nuclear receptors suggests that TRα1 maps to a receptor clade primarily involved in regulation of central nervous system, circadian and basal metabolic functions, whereas TRβ1 maps to a distinct clade responsible for control of lipid metabolism and energy utilization (56,57).
Our goal in the current study was to determine whether TRα1 and TRβ1 play their different biological roles primarily by regulating different panels of target genes. We explored this question by using a hepatocyte-derived cell line, HepG2, that is innately T3 responsive, yet which, through overexpression of one or the other isoform, could be converted into either a TRα1-dominant or TRβ1-dominant background. We chose these cells, rather than a cell line entirely lacking an endogenous T3 response, to ensure that all the components required for appropriate TR-mediated regulation were present. Confirming the validity of this approach, our HepG2 transformants behaved as expected, expressing the ectopic TR isoform at levels 4 to 5 times that of the endogenous receptor, regulated an artificial reporter gene in an appropriate manner, and displayed a target gene response readily distinguishable from that observed in the absence of an ectopic receptor.
We first identified genes that demonstrated a statistically relevant response, up or down, to T3, intentionally choosing a relatively stringent, adjusted P value cutoff of <0.05. Notably, even at this first pass, the majority of T3-regulated genes were flagged by the array analysis as regulated by both TRα1 and TRβ1 (23 of 43). Detailed further analysis indicated that 23 was, in fact, an underestimate and that most of the genes initially identified as specific for a given isoform were actually regulated by both TRα1 and TRβ1, but to different extents. These results were confirmed for representative target gene candidates using RT-PCR.
It is interesting that most of the genes identified as T3 responsive in our array experiments were up-regulated by this hormone; SLC2A1 was the only gene flagged by our approach as down-regulated (using the Benjamini-Hochberg adjusted P-value <0.05) by T3. However, use of a less stringent statistical approach flagged additional candidate genes as down-regulated by T3 in our TRα1 or TRβ1 transformants; the absence of these genes from our original analysis appears to reflect a lesser dynamic range for down-regulation compared with up-regulation. It should be noted that we purposely chose conditions to minimize false positives and to focus on genes that were specifically regulated as a consequence of introduction of the ectopic TR isoforms; therefore, the list of T3-regulated genes presented in this study is not meant to be comprehensive either for up- or down-regulated genes.
A significant number of the TR target genes identified in our analysis study have been previously found to be T3 regulated in prior microarray or gene-specific studies, although many are newly identified by our current characterization, and some previously identified were not flagged in our own study (5,58,59,60,61,62,63,64,65,66,67,68). Differences between our array analysis and previously published array studies on T3-responsive genes are likely to be due to differences in the time course chosen (we employed 6 h of T3 treatment to focus on primary response genes, but at the risk of missing additional target genes only detectable at later time points), differences in the statistical analysis (we used a highly stringent approach), and/or differences in the specific target cells examined (e.g. primary liver vs. cultured HepG2 cells).
Despite regulating a largely overlapping set of target genes, TRα1 and TRβ1 mediate isoform-specific, individually tailored, levels of transcriptional activity once recruited to a given target gene
Although regulating a mostly shared set of target genes, it was nonetheless apparent that the ability of TRα1 and TRβ1 to activate any given target gene in response to T3 did display isoform specificity. TRα1 was a stronger transcriptional activator in the HepG2 cells than was TRβ1 on a consensus DR4 reporter and on several endogenous target genes. However, the TRα1 ≫ TRβ1 response clearly was not universally the case, and the relative ability of TRβ1 to activate a given target gene in response to T3 ranged from virtually undetectable to slightly greater than that of TRα1. Although modest differences in the expression level of TRα1 and TRβ1 at the protein level might alter the precise ratio of the response at each target gene, it is clear that each isoform displays a unique ability to regulate each target gene. We suggest that it is this gene-specific, isoform-specific amplitude of transcriptional regulation that is the likely basis for the appearance and maintenance of TRα1 and TRβ1 over evolutionary time. In essence, TRα1 and TRβ1 can adjust the magnitude of the transcriptional response at different genes to different levels in much the same way as an audiographic equalizer permits the volume to be individually adjusted at different frequencies. By altering the ratio of TRα1 to TRβ1 in different tissues or at different developmental times, the magnitude of the T3 response can be individually tailored for different target genes, customizing the hormone response under different conditions to different physiological needs.
This proposal is consistent with observations that although the DNA-binding domain is strongly conserved between TRα1 and TRβ1, the transcriptional regulatory domains (denoted AF-1 and AF-2 (activator proteins 1 and 2) located in the receptor N- and C-terminal regions, respectively] are much more highly divergent between the two isoforms (4,6,7,8,9,10,11). It is likely that by recruiting distinct sets of coregulators to TRα1 and β1, and by working combinatorially with other transcription factors arrayed on each promoter, these divergent AF-1 and AF-2 domains confer the target gene-specific regulatory functions observed in our experiments. As a result, the magnitude of each transcriptional response by TRα1 and TRβ1 may also vary in different cell types that express different repertoires of coregulator proteins.
It is worth noting that our analysis of TRα1 and TRβ1 revealed less isoform specificity in their target gene repertoire than has been found in otherwise similar studies of several steroid receptors. Estrogen receptor (ER) α and β, for example, up-regulated 34 shared genes, 33 ERα-unique genes, and 87 ERβ-unique genes in transfected U2OS cells (69). Of 94 genes regulated in response to progesterone in transfected T47D cells, 25 were jointly regulated by both progesterone receptor (PR) A and B, four were uniquely regulated by PR-A, and 65 were uniquely regulated by PR-B (70). The basis for this difference between the TRs and the reproductive steroid receptors is not clear, but it may be relevant that ERs and PRs have larger, more complex A/B domains than do TRs, providing more versatile platforms for target- and isoform-specific coregulator recruitment (and regulation) by the former. ERs and PRs also are known to participate in an extensive network of protein-protein interactions that contribute to their target gene specificity (e.g. Ref. 71); isoform-specific differences in these protein-protein interaction sites may result in a more strict isoform-specific access to or regulation of genes by ERs or PRs compared with TRs.
TRα1 and TRβ1 can also repress or activate a subset of target genes regardless of hormone status
The TRs have traditionally been viewed as repressing transcription below basal levels in the absence of T3 and activating above basal levels in the presence of T3. However, this bipolar mode of regulation is not the only one possible (e.g. Refs. 72,73,74). By parsing our analysis to compare genes regulated by the introduction of the ectopic receptors independent of hormone status, we were able to further identify target genes that were repressed in both the absence and presence of hormone. We also were able to identify other target genes that were activated in both the absence and presence of hormone. The regulation of certain of these genes appeared to be truly hormone independent; for others, T3 further modified the transcriptional response but did not change its fundamental direction relative to that seen in the absence of ectopic receptor. Presumably these distinct transcriptional regulatory responses reflect the differing contributions of the TRα1 and TRβ1 AF-1 (hormone-independent) and AF-2 (hormone-dependent) domains when recruited to these different target genes, and the interactions of these TR activation domains with coregulators, with other transcription factors, and with epigenetic marks present at each target gene location.
Many of the genes that were regulated by TRs independent of hormone displayed a somewhat stronger isoform specificity than did the genes flagged by virtue of their T3 response. For several of the target genes repressed by receptor regardless of T3 status, this isoform specificity was near absolute (e.g. CDCA7L). Repression by nuclear receptors is often mediated by inhibitory interactions with other transcription factors that are conferred by protein-protein interactions, rather than by direct DNA binding. This protein-protein mode of inhibition is likely to be particularly dependent on, and therefore to reflect the sequence differences between, the AF-1 and AF-2 domains of TRα1 and TRβ1.
The isoform specificity observed in the TR transcriptional response, rather than in the TR target gene repertoire per se, helps explain the partial functional redundancy observed for TRα1 and TRβ1
Targeted disruptions of murine TRα vs. TRβ clearly produce different phenotypes: TRα knockouts display defects in linear growth, bone deposition, and cardiac function, whereas TRβ knockouts present with a distinct phenotype of elevated circulating T3/T4 (due to loss of a negative feedback regulation in the hypothalamus/pituitary/thyroid gland axis), defects in inner ear and cone cell differentiation, and altered hepatic metabolism (1,33,35,37,42,43,44,45). Despite this clear division of biological function between TRα and TRβ, it is notable that neither knockout is lethal, and that many of the physiological and developmental roles attributed to a given TR isoform remain detectable, if impaired, in the knockout of the corresponding isoform. Similarly, naturally occurring dominant-negative mutations in TRα or TRβ are found in several human diseases (75); these mutant receptors inhibit in trans not only their own, cognate isoform, but also the corresponding paralog, and the resulting neoplastic and endocrine disorders present clinically with defects that are a blend of both isoform-specific and nonspecific characteristics.
This pattern of partial redundancy is not readily explained if TRα1 and TRβ1 regulate rigidly separate sets of target genes, but is fully compatible with the differentially programmed graphic equalizers concept we suggest here. In this model, knocking out either TRα or TRβ would not halt the overall concert of gene regulation involved in differentiation and physiology: most genes regulated by one isoform would still be recognized and regulated by the other. Nonetheless, the resulting music will be distorted by being filtered through only one, instead of both, TRα and TRβ equalizers, and the resulting organism, although viable, will display a range of developmental and regulatory pathological impairments.
Our results also complement and may reconcile two prior elegant studies examining the effects of TR isoform-specific knockouts on T3-regulated liver gene expression. In Yen et al. (74), the authors reported an overall reduction in the T3 transcriptional response in TRβ knockout mice vs. wild-type or TRα knockout animals, but observed little or no target gene-specific effects resulting from disruption of either isoform. In contrast, Flores-Morales et al. (60) reported that a specific subset of genes regulated by T3 in wild-type mice were significantly impaired in their regulation in TRβ knockout animals. These results foreshadow our own conclusions that TRα1 and TRβ1 regulate largely overlapping sets of target genes, but to a significantly different extent. It is likely that both prior studies were observing the same fundamental phenomenon but obtained incompletely congruent results due to technical differences in their approaches. We note also that compensatory changes known to occur in genetically manipulated animals may have attenuated in the knockout mice the isoform-specific differences that we observe in the transcriptional activities of TRα1 and TRβ1 in cultured cells.
Although we focused this study on a comparison of target genes regulation by TRα1 and TRβ1 (the most abundant and widely expressed TR isoform), the TRα and TRβ loci generate additional isoforms through alternative mRNA splicing. TRα2, for example, contains the TRα1 N-terminal and DNA-binding domains, but possesses a unique C-terminal domain that cannot bind T3. TRα2 appears to operate in most studies as a general inhibitor of TR function (e.g. Ref. 76). However, our observations that TRα1 and TRβ1 can exert hormone-independent, target gene-specific transcriptional properties raise the possibility that TRα2 may similarly display hormone-independent activating functions on a subset of target genes (perhaps mediated by the A/B domain that is shared by both TRα1 and TRα2). Conversely, the alternative splice product of the TRβ locus, TRβ2, contains the same DNA-binding and hormone-binding domains of TRβ1 but possesses a distinct N-terminal A/B domain. We have already shown that TRβ2 displays very different transcriptional properties compared with either TRα1 or TRβ1 (77,78,79); our current study suggests that the TRβ2 transcriptional program is likely to also be gene specific, a question we are currently exploring.
Materials and Methods
Molecular clones
Molecular clones of human TRα1 and TRβ1, described elsewhere (80), were first subjected to standard PCR using oligonucleotide primers designed to incorporate the restriction sites NotI and SalI at 5′- and 3′-ends, respectively. The amplified products were subsequently subcloned into the pBI-L vector within the NotI and SalI multiple cloning site plasmid (CLONTECH Laboratories, Inc., Mountain View, CA). The TRs were then subcloned from pBI-L into the pCI-neo vector using NheI (5′) and SalI (3′) restriction sites (Promega Corp., Madison WI). The thymidine kinase promoter (TK)-luciferase reporter bearing a direct-repeat 4 TRE (DR4-tk-luciferase) was previously described (80). Molecular clones of the human TRα1 and TRβ1 were also subjected to PCR to incorporate the restriction sites SacII (5′) and EcoRI (3′) and subcloned into the pUHD10–3 vector (81) for use as qRT-PCR standards. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-pOTB7 (IMAGE clone 2989348) was obtained from American Type Culture Collection (Manassas, VA).
Stable transformants
HepG2 cells were maintained at 37 C in DMEM supplemented with 5% fetal bovine serum using bicarbonate buffer and a 5% CO2 atmosphere. HepG2 transformants expressing ectopic TRα1, ectopic TRβ1, or an empty plasmid control were generated as previously described using a G418 coselection methodology (80). Cell clones arising from individual transformation events were propagated and screened for integration and expression of the expression plasmid by PCR/RT-PCR. To minimize any clone-to-clone variation, six, 12, and 11 independent cell clones, scored as positive for the presence/expression of the introduced construct and (excluding the vector-only transformants) for TR expression, were pooled for the TRα1, TRβ1, and empty plasmid control cell populations, respectively, and used in the expression analysis. Analysis of individual clones revealed some clone-to-clone variation in the overall strength of the T3 responsive, but confirmed that the pooled populations accurately recapitulated the patterns of specific target gene regulation observed in individual clones.
Transient transfection assays
The pooled HepG2 stable transformant populations were subsequently analyzed for T3 responsiveness by introduction of a luciferase reporter using an Effectene transient transfection protocol (80). Briefly, HepG2 cells expressing TRα1, TRβ1, or the empty plasmid control were switched to DMEM containing 10% serum-stripped fetal bovine serum. Following the manufacturer’s recommendations (QIAGEN, Valencia CA), 50 ng of the DR4-tk-luciferase reporter, 50 ng of pCH110-lacZ (as an internal transfection control), and sufficient pUC18 to bring the total DNA to 250 ng were mixed with 2 μl of Enhancer and 2.5 μl Effectene and were added to 7 × 104 HepG2 cells. The transfection medium was replaced 24 h later with fresh medium containing either T3 or ethanol carrier alone, as indicated. Luciferase and β-galactosidase activities were determined 24 h later, and relative luciferase activity was calculated.
Microarrays
The HepG2 transformant pools expressing ectopic TRα1, ectopic TRβ1, or the empty plasmid control were treated with 100 nm T3 or with ethanol carrier alone for 6 h in DMEM containing 10% hormone-stripped fetal bovine serum. Separate biological replicates were performed on different days to generate independent samples for analysis. The cells were harvested and the RNA was isolated using an RNeasy kit (QIAGEN). The purified RNA was provided to the University of California, Davis, Cancer Center Gene Expression Resource, for subsequent cDNA probe generation and hybridization to Affymetrix GeneChip Human Gene 1.0 ST microarrays (Affymetrix, Inc., Santa Clara, CA). After scanning, the raw microarray data were normalized by the Robust Multichip Array method, with P value adjustments performed by the Benjamini-Hochberg method using R software and the affylmGUI package (82,83). Heat maps were generated with GenePattern (84). Three independent biological repeats were analyzed for each of the three transformant pools (empty plasmid control, TRα1, and TRβ1).
RT-PCR
RNA was isolated from cells using either TRIzol (Invitrogen, Carlsbad, CA) or RNeasy kit (QIAGEN) following the manufacturer’s protocols. cDNA was synthesized from 1 μg of each RNA preparation using a QuantiTect kit (QIAGEN) and quantitative real-time PCR was performed using 0.5 μl of the resulting cDNA and the ABsolute QPCR SYBR Green Mix kit (ABgene/Thermo Scientific, Epsom Kent, UK) with the following primers. TRα: sense, 5′-CAAGGTGGAGTGTGGGTCA-3′; antisense, 5′-CCGTTCTTTCTTTTTCGCTTT-3′; TRβ1: sense, 5′-ATGACTCCCAACAGTATGACAG-3′; antisense, 5′-TCCAGTCGTGTTCTCGGTCT-3′; and GAPDH: sense, 5′-GAAGGTGAAGGTCGGAGTC-3′; antisense, 5′-GAAGATGGTGATGGGATTTC-3′. Serial dilutions of TRα1 and TRβ1 plasmid clones served as copy number standards. Semiquantitative PCR was performed using 0.5 μl cDNA, GoTaq DNA Polymerase and run for 25 cycles with the following primers. COQ10A: sense, 5′-TGTGCCCTGGTGTAAGAAG-3′; antisense, 5′-TCAGTACAAACAGCCTTGACC-3′; PTAFR: sense, 5′-CAGAGACACACGGTCACTGC-3′; antisense, 5′-GTCCATGTGGGAGGAGTCAT-3′; IRF1: sense, 5′-GACCCTGGCTAGAGATGCAG-3′; antisense, 5′-AGGCATCCTTGTTGATGTCC-3′; PROM1: sense, 5′-ACTCCCATAAAGCTGGACCC-3′; antisense, 5′-TTTGGATTCATATGCCTTCTGT-3′; PCK1: sense, 5′-GAGAAAGCGTTCAATGCCAG-3′; antisense, 5′-ATGCCGATCTTTGACAGAGG-3′; SLC6A12: sense, 5′-CAATGGACCAACAAGATGGA-3′; antisense: 5′-GCTCCACCTCCGTTTTTGTA-3′; DOCK9: sense, 5′-GGAGACTCGGAAGTTCACCC-3′; antisense, 5′-GTGGCTCAATTAGCTTTGGC-3′; CDCA7L: sense, 5′-TAGGAAGAATGGAGTTGGCG-3′; antisense, 5′-TCTGACGAGAGGGTTTCCAT-3′; HEPACAM: sense, 5′-CGCCTTGCTCCTTTTGTCTA-3′; antisense, 5′-ACTGCACAGAAAGCAGAGCC-3′; ZNF185: sense, 5′-AGCGGATCTGAGCAACTTGT-3′; antisense, 5′-GTACAGCTGGGTCTCCTTGG-3′. PCR products were subsequently analyzed on 8% Tris-acetate-EDTA polyacrylamide gels and visualized with SYBR Green. Bands were quantified with a Flurochem900 Imager (Alpha Innotech, San Leandro, CA). Very good matches were observed between the microarray and RT-PCR methods, with the former often, in fact, the more precise of the two approaches.
Supplementary Material
Acknowledgments
We thank Liming Liu for technical support and Michael L. Goodson and Elsie L. Campbell (Department of Microbiology, University of California at Davis) for valuable advice and assistance.
Footnotes
This work was supported by Public Health Service/National Cancer Institute award R37-CA53394 and by the University of California Davis Cancer Center Gene Expression Resource (NCI P30 CA93373). I.H.C. was supported by PHS Pre-Doctoral Training Award T32-GM007377 from the National Institute of General Medical Sciences.
Disclosure Summary: The authors have nothing to disclose.
First Published Online July 23, 2009
Abbreviations: AF-1, Activation function 1; ER, estrogen receptor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PR, progesterone receptor; qRT-PCR, quantitative RT-PCR; TR, thyroid hormone receptor; TRE, thyroid hormone response element.
References
- Brent GA 2000 Tissue-specific actions of thyroid hormone: insights from animal models. Rev Endocr Metab Disord 1:27–33 [DOI] [PubMed] [Google Scholar]
- Buchholz DR, Paul BD, Fu L, Shi YB 2006 Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol 145:1–19 [DOI] [PubMed] [Google Scholar]
- Flamant F, Baxter JD, Forrest D, Refetoff S, Samuels H, Scanlan TS, Vennström B, Samarut J 2006 International Union of Pharmacology. LIX. The pharmacology and classification of the nuclear receptor superfamily: thyroid hormone receptors. Pharmacol Rev 58:705–711 [DOI] [PubMed] [Google Scholar]
- Larsen PR, Davies TF, Schlumberger M-J, Hay ID 2003 Thyroid physiology and diagnostic evaluation of patients with thyroid disorders. In: Larsen PR, Kronenberg HM, Melmed S, Polonsky KS, eds. Williams textbook of endocrinology. Philadelphia: Saunders; 331–373 [Google Scholar]
- Viguerie N, Langin D 2003 Effect of thyroid hormone on gene expression. Curr Opin Clin Nutr Metab Care 6:377–381 [DOI] [PubMed] [Google Scholar]
- Apriletti JW, Ribeiro RC, Wagner RL, Feng W, Webb P, Kushner PJ, West BL, Nilsson S, Scanlan TS, Fletterick RJ, Baxter JD 1998 Molecular and structural biology of thyroid hormone receptors. Clin Exp Pharmacol Physiol (Suppl) 25:S2–S11 [DOI] [PubMed] [Google Scholar]
- Bassett JH, Harvey CB, Williams GR 2003 Mechanisms of thyroid hormone receptor-specific nuclear and extra nuclear actions. Mol Cell Endocrinol 213:1–11 [DOI] [PubMed] [Google Scholar]
- Claessens F, Gewirth DT 2004 DNA recognition by nuclear receptors. Essays Biochem 40:59–72 [DOI] [PubMed] [Google Scholar]
- Glass CK 1996 Some new twists in the regulation of gene expression by thyroid hormone and retinoic acid receptors. J Endocrinol 150:349–357 [DOI] [PubMed] [Google Scholar]
- Yen PM 2001 Physiological and molecular basis of thyroid hormone action. Physiol Rev 81:1097–1142 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lazar MA 2000 The mechanism of action of thyroid hormones. Annu Rev Physiol 62:439–466 [DOI] [PubMed] [Google Scholar]
- Brent GA, Williams GR, Harney JW, Forman BM, Samuels HH, Moore DD, Larsen PR 1992 Capacity for cooperative binding of thyroid hormone (T3) receptor dimers defines wild type T3 response elements. Mol Endocrinol 6:502–514 [DOI] [PubMed] [Google Scholar]
- Forman BM, Casanova J, Raaka BM, Ghysdael J, Samuels HH 1992 Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol 6:429–442 [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Yu VC, Näär A, Kyakumoto S, Han Z, Silverman S, Rosenfeld MG, Glass CK 1993 Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Gene Dev 7:1423–1435 [DOI] [PubMed] [Google Scholar]
- Lazar MA, Berrodin TJ, Harding HP 1991 Differential DNA binding by monomeric, homodimeric, and potentially heteromeric forms of the thyroid hormone receptor. Mol Cell Biol 11:5005–5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DD, Brent GA 1991 Thyroid hormone—half-sites and insights. New Biol 3:835–844 [PubMed] [Google Scholar]
- Näär AM, Boutin JM, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG 1991 The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell 65:1267–1279 [DOI] [PubMed] [Google Scholar]
- Perlmann T, Rangarajan PN, Umesono K, Evans RM 1993 Determinants for selective RAR and TR recognition of direct repeat HREs. Gene Dev 7:1411–1422 [DOI] [PubMed] [Google Scholar]
- Rastinejad F, Perlmann T, Evans RM, Sigler PB 1995 Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 375:203–211 [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM 1991 Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65:1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GR, Zavacki AM, Harney JW, Brent GA 1994 Thyroid hormone receptor binds with unique properties to response elements that contain hexamer domains in an inverted palindrome arrangement. Endocrinology 134:1888–1896 [DOI] [PubMed] [Google Scholar]
- Davis PJ, Tillmann HC, Davis FB, Wehling M 2002 Comparison of the mechanisms of nongenomic actions of thyroid hormone and steroid hormones. J Endocrinol Invest 25:377–388 [DOI] [PubMed] [Google Scholar]
- Furuya F, Ying H, Zhao L, Cheng SY 2007 Novel functions of thyroid hormone receptor mutants: beyond nucleus-initiated transcription. Steroids 72:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatcioglu F, Claret FX, Karin M 1994 Negative transcriptional regulation by nuclear receptors. Semin Cancer Biol 5:347–359 [PubMed] [Google Scholar]
- Cheng SY 2000 Multiple mechanisms for regulation of the transcriptional activity of thyroid hormone receptors. Rev Endocr Metab Disord 1:9–18 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG 2000 The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14:121–141 [PubMed] [Google Scholar]
- Koenig RJ 1998 Thyroid hormone receptor coactivators and corepressors. Thyroid 8:703–713 [DOI] [PubMed] [Google Scholar]
- Lee H, Yen PM 1999 Recent advances in understanding thyroid hormone receptor coregulators. J Biomed Sci 6:71–78 [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG 2004 A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511–526 [DOI] [PubMed] [Google Scholar]
- Perissi V, Scafoglio C, Zhang J, Ohgi KA, Rose DW, Glass CK, Rosenfeld MG 2008 TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol Cell 29:755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Wolffe AP, Wahli W 2000 Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol Endocrinol 14:329–347 [DOI] [PubMed] [Google Scholar]
- Tsai CC, Fondell JD 2004 Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam Horm 68:93–122 [DOI] [PubMed] [Google Scholar]
- Cheng SY 2005 Isoform-dependent actions of thyroid hormone nuclear receptors: lessons from knockin mutant mice. Steroids 70:450–454 [DOI] [PubMed] [Google Scholar]
- Murata Y 1998 Multiple isoforms of thyroid hormone receptor: an analysis of their relative contribution in mediating thyroid hormone action. Nagoya J Med Sci 61:103–115 [PubMed] [Google Scholar]
- Wondisford FE 2003 Thyroid hormone action: insight from transgenic mouse models. J Investig Med 51:215–220 [DOI] [PubMed] [Google Scholar]
- Forrest D, Vennström B 2000 Functions of thyroid hormone receptors in mice. Thyroid 10:41–52 [DOI] [PubMed] [Google Scholar]
- Abel ED, Boers ME, Pazos-Moura C, Moura E, Kaulbach H, Zakaria M, Lowell B, Radovick S, Liberman MC, Wondisford F 1999 Divergent roles for thyroid hormone receptor β isoforms in the endocrine axis and auditory system. J Clin Invest 104:291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S, Brunet FG, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M 2004 Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol 21:1923–1937 [DOI] [PubMed] [Google Scholar]
- Escriva H, Manzon L, Youson J, Laudet V 2002 Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol 19:1440–1450 [DOI] [PubMed] [Google Scholar]
- Escriva H, Bertrand S, Laudet V 2004 The evolution of the nuclear receptor superfamily. Essays Biochem 40:11–26 [DOI] [PubMed] [Google Scholar]
- Flamant F, Samarut J 2003 Thyroid hormone receptors: lessons from knockout and knock-in mutant mice. Trends Endocrinol Metab 14:85–90 [DOI] [PubMed] [Google Scholar]
- Forrest D, Hanebuth E, Smeyne RJ, Everds N, Stewart CL, Wehner JM, Curran T 1996 Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor β: evidence for tissue-specific modulation of receptor function. EMBO J 15:3006–3015 [PMC free article] [PubMed] [Google Scholar]
- Gauthier K, Chassande O, Plateroti M, Roux JP, Legrand C, Pain B, Rousset B, Weiss R, Trouillas J, Samarut J 1999 Different functions for the thyroid hormone receptors TRα and TRβ in the control of thyroid hormone production and post-natal development. EMBO J 18:623–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier K, Plateroti M, Harvey CB, Williams GR, Weiss RE, Refetoff S, Willott JF, Sundin V, Roux JP, Malaval L, Hara M, Samarut J, Chassande O 2001 Genetic analysis reveals different functions for the products of the thyroid hormone receptor α locus. Mol Cell Biol 21:4748–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchia PE, Takeuchi Y, Kawai T, Cua K, Gauthier K, Chassande O, Seo H, Hayashi Y, Samarut J, Murata Y, Weiss RE, Refetoff S 2001 Increased sensitivity to thyroid hormone in mice with complete deficiency of thyroid hormone receptor α. Proc Natl Acad Sci USA 98:349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göthe S, Wang Z, Ng L, Kindblom JM, Barros AC, Ohlsson C, Vennström B, Forrest D 1999 Mice devoid of all known thyroid hormone receptors are viable but exhibit disorders of the pituitary-thyroid axis, growth, and bone maturation. Genes Dev 13:1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro MO 2008 Effects of thyroid hormone analogs on lipid metabolism and thermogenesis. Thyroid 18:197–203 [DOI] [PubMed] [Google Scholar]
- Chang C, Lin Y, O-Lee TW, Chou CK, Lee TS, Liu TJ, P’eng FK, Chen TY, Hu CP 1983 Induction of plasma protein secretion in a newly established human hepatoma cell line. Mol Cell Biol 3:1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby IA, Bouhnik J, Coezy ED, Corvol P 1991 Thyroid hormone receptors and stimulation of angiotensinogen production in HepG2 cells. In Vitro Cell Dev Biol 27:21–24 [DOI] [PubMed] [Google Scholar]
- Ma Y, Freitag P, Zhou J, Brüne B, Frede S, Fandrey J 2004 Thyroid hormone induces erythropoietin gene expression through augmented accumulation of hypoxia-inducible factor-1. Am J Physiol Regul Integr Comp Physiol 287:R600–R607 [DOI] [PubMed] [Google Scholar]
- Nozaki S, Shimomura I, Funahashi T, Menju M, Kubo M, Matsuzawa Y 1992 Stimulation of the activity and mRNA level of hepatic triacylglycerol lipase by triiodothyronine in HepG2 cells. Biochim Biophys Acta 1127:298–302 [DOI] [PubMed] [Google Scholar]
- Theriault A, Ogbonna G, Adeli K 1992 Thyroid hormone modulates apolipoprotein B gene expression in HepG2 cells. Biochem Biophys Res Commun 186:617–623 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamaguchi S, Pohlenz J, Murata Y, Refetoff S, Seo H 1997 Modification of thyroid hormone and 9-cis retinoic acid signaling by overexpression of their cognate receptors using adenoviral vector. Mol Cell Endocrinol 131:59–66 [DOI] [PubMed] [Google Scholar]
- Chamba A, Neuberger J, Strain A, Hopkins J, Sheppard MC, Franklyn JA 1996 Expression and function of thyroid hormone receptor variants in normal and chronically diseased human liver. J Clin Endocrinol Metab 81:360–367 [DOI] [PubMed] [Google Scholar]
- Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escrivà H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V 2007 Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet 3:e188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ 2006 Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM 2006 Nuclear receptor expression links the circadian clock to metabolism. Cell 126:801–810 [DOI] [PubMed] [Google Scholar]
- Clément K, Viguerie N, Diehn M, Alizadeh A, Barbe P, Thalamas C, Storey JD, Brown PO, Barsh GS, Langin D 2002 In vivo regulation of human skeletal muscle gene expression by thyroid hormone. Genome Res 12:281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Jiang Y, Meltzer P, Yen PM 2000 Thyroid hormone regulation of hepatic genes in vivo detected by complementary DNA microarray. Mol Endocrinol 14:947–955 [DOI] [PubMed] [Google Scholar]
- Flores-Morales A, Gullberg H, Fernandez L, Ståhlberg N, Lee NH, Vennström B, Norstedt G 2002 Patterns of liver gene expression governed by TRβ. Mol Endocrinol 16:1257–1268 [DOI] [PubMed] [Google Scholar]
- Huang YH, Tsai MM, Lin KH 2008 Thyroid hormone dependent regulation of target genes and their physiological significance. Chang Gung Med J 31:325–334 [PubMed] [Google Scholar]
- Lin KH, Lee HY, Shih CH, Yen CC, Chen SL, Yang RC, Wang CS 2003 Plasma protein regulation by thyroid hormone. J Endocrinol 179:367–377 [DOI] [PubMed] [Google Scholar]
- Miller LD, Park KS, Guo QM, Alkharouf NW, Malek RL, Lee NH, Liu ET, Cheng SY 2001 Silencing of Wnt signaling and activation of multiple metabolic pathways in response to thyroid hormone-stimulated cell proliferation. Mol Cell Biol 21:6626–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CH, Chen SL, Yen CC, Huang YH, Chen CD, Lee YS, Lin KH 2004 Thyroid hormone receptor-dependent transcriptional regulation of fibrinogen and coagulation proteins. Endocrinology 145:2804–2814 [DOI] [PubMed] [Google Scholar]
- Ventura-Holman T, Mamoon A, Maher JF, Subauste JS 2007 Thyroid hormone responsive genes in the murine hepatocyte cell line AML 12. Gene 396:332–337 [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Hamann S, Jauk M, Lacey M, Filbry A, Radtke C, Iwen KA, Kutz S, Harneit A, Lizardi PM, Seitz HJ 2003 Hepatic gene expression patterns in thyroid hormone-treated hypothyroid rats. J Mol Endocrinol 31:291–303 [DOI] [PubMed] [Google Scholar]
- Weitzel JM, Radtke C, Seitz HJ 2001 Two thyroid hormone-mediated gene expression patterns in vivo identified by cDNA expression arrays in rat. Nucleic Acids Res 29:5148–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WM, Sarapura VD, Dowding JM, Woodmansee WW, Haakinson DJ, Gordon DF, Ridgway EC 2002 Early gene expression changes preceding thyroid hormone-induced involution of a thyrotrope tumor. Endocrinology 143:347–359 [DOI] [PubMed] [Google Scholar]
- Kian Tee M, Rogatsky I, Tzagarakis-Foster C, Cvoro A, An J, Christy RJ, Yamamoto KR, Leitman DC 2004 Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors α and β. Mol Biol Cell 15:1262–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB 2002 Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem 277:5209–5218 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M 2006 Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- Helmer EB, Raaka BM, Samuels HH 1996 Hormone-dependent and -independent transcriptional activation by thyroid hormone receptors are mediated by different mechanisms. Endocrinology 137:390–399 [DOI] [PubMed] [Google Scholar]
- Nagl SB, Nelson CC, Romaniuk PJ, Allison LA 1995 Constitutive transactivation by the thyroid hormone receptor and a novel pattern of activity of its oncogenic homolog v-ErbA in Xenopus oocytes. Mol Endocrinol 9:1522–1532 [DOI] [PubMed] [Google Scholar]
- Yen PM, Feng X, Flamant F, Chen Y, Walker RL, Weiss RE, Chassande O, Samarut J, Refetoff S, Meltzer PS 2003 Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Rep 4:581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SY 2005 Thyroid hormone receptor mutations and disease: beyond thyroid hormone resistance. Trends Endocrinol Metab 16:176–182 [DOI] [PubMed] [Google Scholar]
- Katz D, Lazar MA 1993 Dominant negative activity of an endogenous thyroid hormone receptor variant (α2) is due to competition for binding sites on target genes. J Biol Chem 268:20904–20910 [PubMed] [Google Scholar]
- Yang Z, Hong SH, Privalsky ML 1999 Transcriptional anti-repression. Thyroid hormone receptor β-2 recruits SMRT corepressor but interferes with subsequent assembly of a functional corepressor complex. J Biol Chem 274:37131–37138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Privalsky ML 2001 Isoform-specific transcriptional regulation by thyroid hormone receptors: hormone-independent activation operates through a steroid receptor mode of co-activator interaction. Mol Endocrinol 15:1170–1185 [DOI] [PubMed] [Google Scholar]
- Wan W, Farboud B, Privalsky ML 2005 Pituitary resistance to thyroid hormone syndrome is associated with T3 receptor mutants that selectively impair β2 isoform function. Mol Endocrinol 19:1529–1542 [DOI] [PubMed] [Google Scholar]
- Chan IH, Privalsky ML 2006 Thyroid hormone receptors mutated in liver cancer function as distorted antimorphs. Oncogene 25:3576–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bonin AL, Freundlieb S, Bujard H 1994 Inducible gene expression systems for higher eukaryotic cells. Curr Opin Biotechnol 5:516–520 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003 Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- Smyth GK 2004 Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3 [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP 2006 GenePattern 2.0. Nat Genet 38:500–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.