Abstract
Mutations and polymorphisms in PPARG have been linked to adiposity and partial lipodystrophy in humans. However, how disturbances in PPARG lead to depot-specific effects on adipose tissue, as shown by the characteristic aberrant fat distribution in patients, remains unclear. By manipulating the 3′-untranslated region of the Pparg gene, we have generated mice with peroxisome proliferator-activated receptor γ (PPARγ) gene expression ranging from 25% to 100% normal. Basal levels of PPARγ transcripts between 50% and approximately 100% had no significant effect on body weight, fat mass, and insulin sensitivity. In contrast, mice with 25% normal PPARγ expression exhibited reduced body weight and total fat mass, insulin resistance, and dyslipidemia. Interestingly, fat mass was selectively reduced in perigonadal depot without significant changes in inguinal and other depots. Expression of adipogenic factor CCAAT enhancer binding protein-α and some other metabolic genes containing peroxisome proliferator response element were reduced in a perigonadal depot-specific fashion. This was further associated with depot-specific reduction in the expression of adipokines, increased expression of TNFα, and increased ectopic lipid deposition in muscles. Together, these results underscore the differential sensitivity of the individual fat depots on PPARγ availability as an underlying mechanism of partial lipodystrophy.
Higher than 30% normal PPARgamma gene expression is necessary to maintain mass and functionality of body fat, and insulin sensitivity in mice.
Nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ) plays critical roles in adipocyte differentiation and fat tissue maturation (1). Alternative splicing and differential promoter utilization generate two protein isoforms, PPARγ1 and PPARγ2. Human PPARγ2 contains 30 extra amino acids (28 in mice) in its N terminus compared with PPARγ1 and is predominant in adipose tissue. PPARγ1 is expressed at lower levels in many adult tissues including liver, intestine, and skeletal muscle (2). PPARγ heterodimerizes with retinoid X receptor (RXR), and, upon binding of specific ligands, activates the transcription of target genes by binding to their peroxisome proliferator response elements (PPREs). The majority of known target genes are involved in triglyceride hydrolysis, fatty acid and glycerol uptake, and fatty acid reesterification and lipid storage. Endogenous PPARγ ligands include various fatty acids and their metabolites (2). Thiazolidinediones (TZDs), which are specific, high-affinity ligands for PPARγ, are used clinically to treat insulin resistance (IR) and hyperglycemia associated with type 2 diabetes mellitus.
Studies in humans and in mice suggest that mild reduction of PPARγ is beneficial. For example, P12A substitution, which leads to a lower affinity for PPRE and decreased transactivation in in vitro studies (3,4), is associated with lower body mass index (BMI) and enhanced insulin sensitivity in some, although not all, population studies (3,5). Concurring to this, mice heterozygous for PPARγ deficiency are protected from diet-induced weight gain and decrease in insulin sensitivity (6). However, a complete lack of PPARγ is embryonically lethal in mice (6,7), and severe generalized PPARγ deficiency, as well as selective PPARγ deficiency in adipose tissues, causes lipodystrophy and insulin resistance in mice (8,9,10). In addition, a series of PPARγ mutations have been found in humans with regional as opposed to systemic loss of fat. These include missense mutations that are located in DNA-binding domain (11,12,13) or ligand-binding domain (14,15,16), chain termination mutations (11,17), and a base substitution in the promoter region affecting PPARγ1 expression (18). Some of these mutations have been assessed in vitro to exert a dominant-negative effect, whereas others showed reduced activity of the mutant PPARγ, suggesting haploinsufficiency of PPARγ as the basis of partial lipodystrophy among affected heterozygotes rather than the specific mutations. However, how disturbances in PPARG lead to depot-specific effects on adipose tissue, as shown by the characteristic aberrant fat distribution in patients, remains unclear.
Our present study was initiated with a hypothesis that body fat distribution and insulin sensitivity can be influenced by the allelic variations of Pparg conferring defined gene expression levels without disturbances in normal transcriptional regulation and protein sequence. Taking advantage of message destabilization by the AU-rich elements (ARE) in the 3′-untranslated region (3′-UTR) of transcripts (19), we have inserted the ARE of c-fos gene into the 3′-UTR of mouse Pparg gene (20,21). With this allele, we have generated a series of mice with basal PPARγ mRNA levels ranging from 25% to 100% normal. Here we report that basal expression between 25% and approximately 50% normal level of PPARγ is critical in mice to maintain their perigonadal fat mass and function. Insulin resistance was accompanied by reduced expressions of genes responsible for lipid metabolism selectively in perigonadal fat depot and by ectopic lipid accumulation in muscles.
Results
Mice with reduced PPARγ expression
We previously described the production of mice carrying the Pparg gene with c-fos ARE sequence inserted into its 3′-UTR region (Pparg-C allele) (21). The transcripts of Pparg-C encode normal protein but are less stable than wild-type transcripts. Consequently the steady state expression of PPARγ of PpargC/C homozygotes in adipose tissue, the primary site of PPARγ function, was reduced to 52% of Pparg+/+ mice (Fig. 1A). The levels of PPARγ mRNA in the kidneys, livers, and hearts of PpargC/C mice were similarly decreased to 52%, 30%, and 48% of Pparg+/+ mice, respectively. Expression in PpargC/+ heterozygotes was about 75% normal in all tissues (supplemental Fig. 1A published as supplemental data on The Endocrine Society’s Online Journals web site at http://mend.endojournals.org).
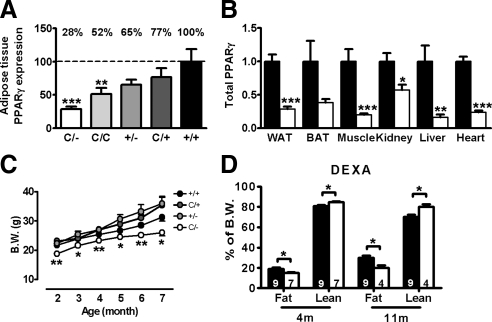
Figure 1.
Expression and quantitative effects of PPARγ in mutant mice. A, Titration of PPARγ mRNA levels in perigonadal adipose tissue of mutant mice. Relative PPARγ expression to Pparg+/+ (100%) are indicated above. B, PPARγ mRNA levels in PpargC/− (□, n = 5) tissues relative to those of Pparg+/+ tissues (▪, n = 5). C, Slower growth of female PpargC/− mice compared with their littermates (n = 6∼14 in each group) D, Body composition analyses of 4- and 11-month-old female mice by DEXA. Numbers of mice are inside bars. ▪, Pparg+/+; □, PpargC/−. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 against Pparg+/+. B.W., Body weight.
To generate mice with further reduction in the PPARγ expression, we bred PpargC/+ mice with Pparg+/− mice. Pparg+/+, PpargC/+, and Pparg+/− pups were produced at an expected Mendelian ratio (56:65:64), but the number of PpargC/− pups (37) was significantly reduced (P = 0.028 by Chi-square test). As expected, expression of PPARγ in PpargC/− mice was decreased to 28%, 38%, 20%, 57%, 16%, and 23% of Pparg+/+ mice in white adipose tissue (WAT), brown adipose tissue (BAT), skeletal muscle, kidney, liver, and heart, respectively (Fig. 1B). Together, we generated mice with a wide spectrum of generalized reduction in the Pparg gene expression down to 25% normal (Fig. 1A).
Quantitative effects of PPARγ on body and fat weights
The growth curves and major organ weights of both female and male PpargC/C and PpargC/+ mice were not different from those of Pparg+/+ mice (supplemental Fig. 1B and supplemental Table 1). These demonstrate that the expression of PPARγ at 50% normal levels is sufficient to maintain normal body composition. However, as PPARγ expression reduced further to 25% normal, the difference in body weight became evident. PpargC/− mice, both male and female, were significantly smaller than their littermates (Fig. 1C and supplemental Fig. 2A), becoming apparent as early as postnatal d 15 (supplemental Fig. 2B). Body lengths of PpargC/− mice were shorter than Pparg+/+ mice by about 3% (supplemental Fig. 2C). Body composition analysis by dual energy x-ray absorptiometry (DEXA) scan identified that the reduction of body weight of PpargC/− mice is mainly the result of smaller fat mass, which is accompanied by a significant increase in the lean mass (Fig. 1D). PpargC/− mice are leaner than Pparg+/+ mice at both young (4 months) and old (11 months) ages (Fig. 1D). Spleen and liver weights relative to their body weights in PpargC/− mice were larger than those in Pparg+/+ mice, but the difference was statistically significant only in females (supplemental Fig. 2D).
Failure of PpargC/− mice to deposit fat in perigonadal fat pad
To further analyze the individual fat weight, we dissected perigonadal, retroperitoneal, and mesenteric fat to represent intraabdominal WAT, and inguinal and subscapular fat to represent sc WAT. The weights of these WATs and interscapular BATs were not distinguishable in Pparg+/−, PpargC/C, PpargC/+, and Pparg+/+ mice, the PPARγ expression levels of which were between 50% and 100% (Fig. 2A and supplemental Table 1). No genotype effect was observed even when these mice were on a high-fat diet or aged up to 18 months (supplemental Tables 1 and 2).
Figure 2.
Partial lipodystrophy of PpargC/− mice. A, Perigonadal and inguinal fat mass of female PPARγ quantitative mutants relative to those of Pparg+/+ mice. Numbers of mice are inside bars. B, Adipose tissue mass from 4-month-old female Pparg+/+ (▪) and PpargC/− (□) mice expressed as percent body weight. Per, Ret, Mes, Ing, Sub represent perigonadal, retroperitoneal, mesenteric, inguinal, and subscapular WAT, respectively. BAT indicates interscapular brown adipose tissue. C, Fat mass in abdominal and thoracic regions and extremities of 4- and 11-month-old female Pparg+/+ (▪) and PpargC/− (□) mice by DEXA. D, Morphology of perigonadal (top panels) and inguinal (bottom panels) WAT from 4-month-old mice. E, Distribution of cell size in perigonadal (n = 3 and 4) and inguinal (n = 5 each) WAT of Pparg+/+ (•) and PpargC/− (○) mice. F, Age effects on the deposition of fat in individual depots. (•, Pparg+/+, n = 3∼4; ○, PpargC/−, n = 3∼5). *, P < 0.05; and **, P < 0.01 against Pparg+/+. B.W., Body weight.
In a marked contrast, perigonadal WAT mass of PpargC/− mice was significantly reduced to 20% normal (Fig. 2B). However, retroperitoneal, mesenteric, inguinal, and subscapular WAT mass was normal (Fig. 2B). The perigonadal depot-specific reduction in WAT mass of PpargC/− mice was consistent in both females and males (supplemental Fig. 2, E and F). DEXA analyses confirmed a significant loss of fat from the abdominal region but remained normal in the thoracic region and extremities (Fig. 2C). Thus 25% normal expression of Pparg causes alterations in the overall distribution of body fat in mice.
Histologically, adipocytes in perigonadal WAT of PpargC/− mice were smaller than Pparg+/+ cells (median area 760 μm2 in PpargC/− vs. 1073 μm2 in Pparg+/+), whereas such differences were not detectable in inguinal WAT of Pparg C/− mice (median area 626 μm2 in PpargC/− vs. 634 μm2 in Pparg+/+) (Fig. 2, D and E). Thus, the reduction of PPARγ brought a shift of adipocyte size-distribution toward smaller in perigonadal WAT in a depot-specific manner. Based on the calculation by Di Girolamo et al. (22), we estimate that the number of adipocytes in perigonadal WAT of PpargC/− mice (3.5 × 106) was markedly less than that of Pparg+/+ mice (12.1 × 106), but the numbers in the inguinal WAT did not differ (16.9 × 106 in PpargC/− vs. 18.8 ×106 in Pparg+/+). A 30% reduction in cell volume together with 70% reduction in cell number thus accounts for almost 80% reduction in fat mass of perigonadal depot of PpargC/− mice. As Pparg+/+ mice aged, the proportion of total fat to lean mass increased significantly, whereas the ratio remained the same in PpargC/− mice (Fig. 1D). When individual fat tissues are compared, tissue mass of the retroperitoneal, mesenteric, inguinal, and subscapular WAT, as well as interscapular BAT, in PpargC/− mice was similar with those of Pparg+/+ mice throughout their life (Fig. 2F). Notably, whereas the proportion of perigonadal fat mass to body weight doubled in Pparg+/+ mice from 4 months to 16 months, the proportion in PpargC/− mice fell significantly behind at 4 months and failed to increase throughout their life. Thus, whereas Pparg+/+ mice store excess energy preferentially in perigonadal fat and some in inguinal fat as they age, PpargC/− mice appear to be defected in the fuel storage in perigonadal fat.
Energy homeostasis and insulin resistance in PpargC/− mice
PpargC/− mice had similar daily energy intake and feeding efficiency with those of Pparg+/+ littermates (supplemental Fig. 3A). There was no evidence for fat malabsorption, because fecal output and fecal lipid content were similar between Pparg+/+ and PpargC/− mice (supplemental Fig. 3B). In addition, body temperature was similar (supplemental Fig. 3C), and gene expression for uncoupling protein (UCP)1, which plays a crucial role in thermogenesis, in BAT and WAT was not altered in PpargC/− mice (supplemental Fig. 3D). Telemetry recording showed that the average activity of PpargC/− mice was slightly higher than Pparg+/+ mice, but the difference was not statistically significant (supplemental Fig. 3E). Thus, energy dissipation appeared normal in PpargC/− mice.
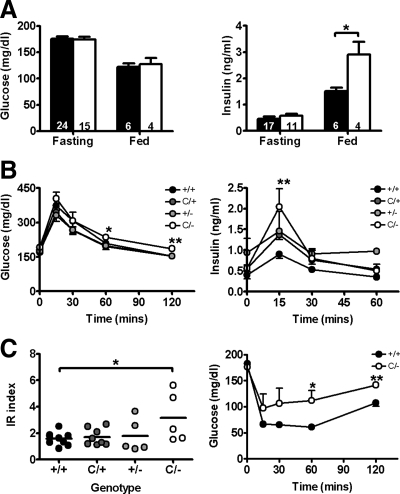
A reduction of PPARγ level to 50% normal in PpargC/C mice did not elicit significant changes in glucose metabolism and insulin sensitivity (supplemental Fig. 4). Although both age and high-fat feeding reduced insulin sensitivity, no effect of 50% reduction in the PPARγ gene expression was observed (supplemental Table 2 and supplemental Fig. 4). In contrast, PpargC/− mice with 25% normal expression of PPARγ showed significantly impaired insulin sensitivity. PpargC/− mice have normal fasting glucose and insulin levels, but their postprandial insulin levels were significantly higher than those in Pparg+/+ mice (Fig. 3A). Furthermore, clearance of glucose after a glucose load in PpargC/− mice was slower than in Pparg+/+ mice (Fig. 3B, left panel). The insulin levels 15 min after the glucose load were significantly higher in PpargC/− mice than in Pparg+/+ mice (Fig. 3B, right panel). The IR index of PpargC/− mice was significantly elevated (Fig. 3C, left panel). Consistently, insulin tolerance tests showed that glucose-lowering effects of insulin were impaired in mice with 25% normal level of PPARγ (Fig. 3C, right panel). These results demonstrate that the dysregulation of glucose metabolism and insulin sensitivity become evident when the basal level of PPARγ expression falls to 25%.
Figure 3.
IR in PpargC/− mice. A, Serum glucose and insulin levels during 5-h fasting or 2-h postfeeding of Pparg+/+ (▪) and PpargC/− (□) mice. Numbers of mice are inside bars. B, Serum glucose and insulin levels during glucose tolerance tests in approximately 3- to 4-month-old female littermates (n = 5∼9). C, Impaired insulin sensitivity revealed by IR index (left panel) and insulin tolerance tests (right panel) of approximately 3- to 4-month-old female PpargC/− mice (n = 6) compared with Pparg+/+ mice (n = 8). *, P < 0.05; and **, P < 0.01 against Pparg+/+.
Reduced expression of CCAAT enhancer binding protein-α (C/EBPα) and adipogenesis in perigonadal fat of PpargC/− mice
Both perigonadal and inguinal depots from PpargC/− mice showed a comparable decrease in the reduction of total PPARγ messages (Fig. 4A). The messages for both PPARγ1 and PPARγ2 protein isoforms were reduced in both depots, although the reduction of PPARγ2 was slightly more in perigonadal depot (85%) than in inguinal depot (65%). In parallel with the reduction in mRNA expression, we found reduced PPARγ protein levels in both perigonadal and inguinal depots of PpargC/− mice (Fig. 4B). The ratios of PPARγ2/PPARγ1 protein isoforms were about 2.4 in perigonadal depot and 1.7 in inguinal depot of Pparg+/+ mice, and the ratios in PpargC/− mice appeared to be similar (Fig. 4B). No compensatory alteration in expression of RXRα, PGC-1α, PPARα, and PPARβ/δ was observed (supplemental Fig. 5).
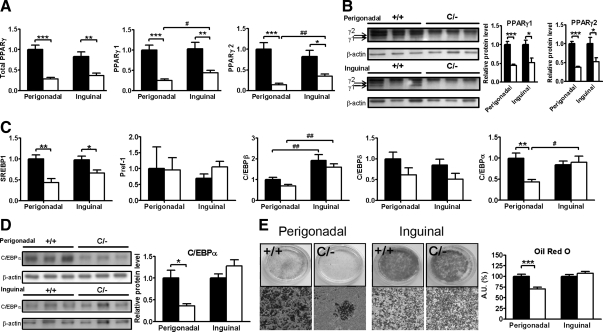
Figure 4.
Gene expression and protein analysis in PpargC/− WAT. Expression of genes for (A) PPARγ isoforms and (C) adipogenic transcription factors in perigonadal and inguinal WAT of Pparg+/+ (▪, n = 8) and PpargC/− (□, n = 5) mice. mRNA amount is expressed relative to the average expression in perigonadal WAT of Pparg+/+ mice. *, P < 0.05, **, P < 0.01, and ***, P < 0.001 between two genotypes and #, P < 0.05, ##, P < 0.01, and ###, P < 0.001 between two depots by t test. Protein Western blot analysis (left panels) and densitometer quantification (right panels) for (B) PPARγ and (D) C/EBPα in perigonadal and inguinal WAT of Pparg+/+ (▪) and PpargC/− (□) mice. E, Differentiation and quantification (right panel) of preadipocytes from perigonadal and inguinal WAT of Pparg+/+ (▪) and PpargC/− (□) mice. Oil Red O staining of plates (left upper panels) and microscopic examination (left lower panels) after 10 d of differentiation. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 against Pparg+/+. AU, Arbitrary units.
Adipogenesis involves complex regulatory pathways controlled by the coordinated expressions of specific genes and several other transcription factors. We found that sterol-regulatory element-binding protein-1 (SREBP1) was expressed at significantly lower levels in both perigonadal and inguinal depots of PpargC/− mice (Fig. 4C). The expression of the preadipocyte factor-1 (Pref1), a negative regulator of adipogenesis, was similar in both depots and did not differ by the genotype. Although there was a trend toward reduction in the expression of C/EBPβ and C/EBPδ genes in both perigonadal and inguinal depots of PpargC/− mice, the reductions were not statistically significant. In a marked contrast, the expression of C/EBPα in perigonadal fat of PpargC/− mice was significantly lower than Pparg+/+ mice, whereas the expression in inguinal fat remained unaltered. Consistently, C/EBPα protein levels were significantly reduced in perigonadal depot but not in inguinal depot of PpargC/− mice (Fig. 4D). In vitro differentiation of stromovascular preadipocytes isolated from these two depots of PpargC/− and Pparg+/+ mice showed that adipogenic ability judged by formation of Oil Red O-positive cells after hormone stimulation was preserved in inguinal fat of PpargC/− mice, whereas the induction of adipogenesis was severely impaired in perigonadal fat of PpargC/− mice (Fig. 4E). Taken together, the failure to enlarge perigonadal fat mass is a cell-autonomous effect of PPARγ in this tissue and likely involves the depot-specific reduction of the adipogenic factor C/EBPα.
Dysregulated lipid metabolism in perigonadal fat of PpargC/− mice
To dissect the molecular mechanism by which a reduced production of PPARγ limits the accumulation of fat in a depot-specific fashion, we examined the differences in the gene expression of key proteins important for lipid flux and synthesis in perigonadal and inguinal depots of Pparg+/+ and PpargC/− mice (Fig. 5). All of these genes contain well-characterized or putative PPRE in their promoter regions. The expressions of CD36 and fatty acid transport protein-1, which are involved in free fatty acid (FFA) uptake and transport, were similar in these two depots and not significantly affected by the genotype. Similarly, adipose triglyceride lipase, an important determinant of basal lipolysis in WAT, was equally expressed in both depots and did not differ significantly by the genotype. However, lipoprotein lipase (LPL), hormone-sensitive lipase (HSL), and fatty acid binding protein aP2 were expressed 2- to 3-fold higher in perigonadal fat than in inguinal fat of Pparg+/+ mice. LPL expression in inguinal but not in perigonadal depot of PpargC/− mice was significantly increased, whereas expressions of HSL and aP2 were markedly reduced only in perigonadal fat of PpargC/− mice to the level similar to that in inguinal fat. Together, these data suggest that lipolysis and mobilization of FFA are specifically reduced in perigonadal fat of PpargC/− mice.
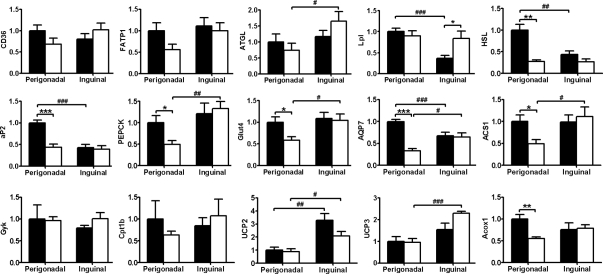
Figure 5.
Gene expression in PpargC/− WAT. Expression of genes for PPARγ target genes in perigonadal and inguinal WAT of Pparg+/+ (▪, n = 8) and PpargC/− (□, n = 5) mice. mRNA amount is expressed relative to the average expression in perigonadal WAT of Pparg+/+ mice. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 between two genotypes; #, P < 0.05; ##, P < 0.01; and ###, P < 0.001 between two depots by t test. FATP1, Fatty acid transport protein-1; ATGL, adipose triglyceride lipase; AQP7, aquaporin-7; Gyk, glycerol kinase; Cpt1b, carnitine palmitoyltransferase 1b; Acox1, acyl-coenzyme A oxidase.
A perigonadal depot-specific reduction of de novo lipogenesis from carbohydrates in PpargC/− mice was evidenced by significant decreases in glucose transporter-4 (Glut4) and phosphoenolpyruvate carboxykinase (PEPCK), which transport glucose and provide the cells with glycerol backbone for triglyceride synthesis, respectively. Similarly, mRNA levels of aquaporin-7, which is involved in adipose glycerol transport, and acyl-coenzyme A synthetase-1 (ACS1), which facilitates acylation of FFA in adipocyte, were dramatically decreased in perigonadal fat. Expression of these genes in inguinal depots remained unchanged. Expression of glycerol kinase, which enables the synthesis for glycerol-3-phosphate from glycerol, was not affected in either depot. Among genes related to fatty acid oxidation, expression of carnitine palmitoyltransferase 1b, UCP2, and UCP3 were not affected. Expression of acyl-coenzyme A oxidase, however, was selectively decreased in perigonadal depot of PpargC/− mice, implicating that peroxisomal fatty acid β-oxidation, but not mitochondrial fatty acid oxidation, is affected in perigonadal depot of PpargC/− mice. Taken together, the gene expression pattern indicates that the decrease in lipid uptake and synthesis is associated with the reduction in total fat mass of perigonadal depot of PpargC/− mice.
Adipose-derived molecules and ectopic fat deposition in PpargC/− mice
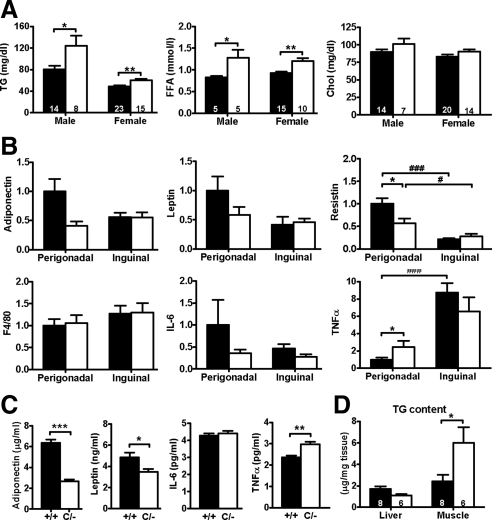
Adipose tissues actively secrete signaling molecules, including FFA, adipokines, and inflammatory molecules, into the circulation and communicate with other organs to regulate insulin sensitivity (23). The dysregulated expression of genes responsible for lipid metabolism in perigonadal fat tissues was reflected in higher fasting serum triglyceride and FFA levels in PpargC/− than in Pparg+/+ mice (Fig. 6A). Serum cholesterol level was not affected. In Pparg+/+ mice, expression of adiponectin, leptin and resistin genes was approximately 2- to 4-fold higher in perigonadal fat compared with inguinal fat (Fig. 6B). In PpargC/− mice, expression of these adipokine genes was reduced in perigonadal fat but not in inguinal fat. The decreased gene expression together with significantly reduced perigonadal fat mass resulted in the significant reductions of circulating adiponectin and leptin proteins in PpargC/− serum (Fig. 6C).
Figure 6.
Adipose-derived molecules in PpargC/− mice. A, Fasting serum triglyceride (TG, left panel), FFA (middle panel), and cholesterol (Chol, right panel). Numbers of mice are inside bars. B, Expression of genes for adipokines, macrophage marker, and inflammatory cytokines in perigonadal and inguinal WAT of Pparg+/+ (▪, n = 8) and PpargC/− (□, n = 5) mice relative to the average mRNA in perigonadal WAT from Pparg+/+ mice. C, Serum levels of adipokines and cytokines of Pparg+/+ (n = 8∼10) and PpargC/− (n = 6) mice. D, Triglyceride content in liver and muscle. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 between two genotypes; #, P < 0.05; ##, P < 0.01; and ###, P < 0.001 between two depots by t test.
Although increased macrophage infiltration into adipose tissues leading to chronic inflammatory conditions has been recognized as a cause of IR, expression of macrophage makers, including F4/80 (Fig. 6B) and CD68 (data not shown), was normal. Expression of proinflammatory cytokine, IL-6, showed a trend toward decrease in both perigonadal and inguinal depots of PpargC/− mice (Fig. 6B), but serum IL-6 level was not different (Fig. 6C). In contrast, expression of TNFα, known as a potent inducer for fat apoptosis and lipolysis, was about 2.5-fold elevated in perigonadal depot but remained unaltered in inguinal depot of PpargC/− mice (Fig. 6B). This is associated with a significant increase of circulating serum TNFα in PpargC/− mice (Fig. 6C). Thus, although chronic inflammation in adipose tissue is not likely to be the primary cause of IR of PpargC/− mice, the increased expression of TNFα specifically in perigonadal WAT may, at least in part, account for the inability of adipocyte to store excess fat leading to increased circulating lipids in PpargC/− mice.
Lipid may be stored in the nonadipose tissues. Although triglyceride content in liver was not altered, muscle triglyceride content was approximately 2.5-fold higher in PpargC/− mice than in Pparg+/+ mice (P < 0.05; Fig. 6D). Thus, these data suggest that inability to store extra fat into perigonadal fat depots in PpargC/− mice is causing an increase in the serum lipids and ectopic fat deposition in nonadipose tissues.
Responses to high-fat diet and TZD in PpargC/− mice
To investigate the effects of PPARγ hypomorph on metabolic regulation of high-fat feeding and PPARγ activation, mice were placed on a high-fat diet (HFD) with or without TZD. After high-fat feeding, PpargC/− mice gained significantly less weight (Fig. 7A), and their residual perigonadal fat remained lipoatrophic (Fig. 7B). In response to the HFD, both cell size (median area 1521 μm2 in PpargC/− and 1928 μm2 in Pparg+/+) and cell number (6.7 × 106 in PpargC/− and 19.6 × 106 in Pparg+/+) in perigonadal fat of PpargC/− and Pparg+/+ mice doubled compared with those in mice fed regular chow (see above). The difference in the perigonadal fat mass between the two genotypes was nevertheless increased. The size of adipocytes also enlarged equally by 2.5-fold in inguinal fat of both Pparg C/− and Pparg+/+ mice in response to high-fat feeding (median area 1635 μm2 in PpargC/− and 1627 μm2 in Pparg+/+). The numbers of adipocytes in inguinal fat of both genotypes were reduced by high-fat feeding (11.4 × 106 in PpargC/− and 11.6 × 106 in Pparg+/+). Thus the relative responses in adiposity to high-fat feeding are the same in PpargC/− and Pparg+/+ mice, but high-fat feeding exaggerates the difference in the absolute amounts of intraabdominal fat.
Figure 7.
Responses to HFD and TZD in PpargC/− mice. A, Representative growth curve of male mice (n = 8∼9 in each group) fed HFD and TZD-containing HFD. B, Adipose tissue mass from HFD and TZD-treated male Pparg+/+ (▪) and PpargC/− (□) mice expressed as percent body weight. Numbers of mice are inside bars. C, Serum glucose and insulin levels during 5-h fasting or 2-h postfeeding of male Pparg+/+ (▪) and PpargC/− (□) mice fed HFD and HFD-TZD. D, Serum glucose and insulin levels and (E) their representative IR indices during glucose tolerance tests in male littermates (n = 8∼9) fed HFD or HFD-TZD for 4 wk. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 against Pparg+/+. HF, High fat.
Pparg+/+ mice fed HFD containing TZD had 30% smaller perigonadal fat mass compared with those fed HFD only (Fig. 7B). In contrast, a reduction of perigonadal fat mass in TZD-treated PpargC/− mice was small compared with that in HFD-fed PpargC/− mice. Therefore, the difference in perigonadal fat mass between the two genotypes was less in the HFD-TZD group than in the HFD group. When fed a HFD, PpargC/− mice remained insulin resistant, evidenced by significantly higher postprandial insulin levels and higher IR index than those of Pparg+/+ mice (Fig. 7, C and D). TZD treatment ameliorated the HFD-induced increase in the serum insulin levels and the intolerance to a glucose challenge in Pparg+/+ mice. Of interest, postprandial hyperinsulinemia of PpargC/− mice was completely abolished, and the impaired insulin sensitivity of PpargC/− mice, as judged by IR index, was normalized by the treatment with TZD (Fig. 7, C and E). These results suggest that the residual PPARγ is not sufficient to rescue perigonadal fat lipoatrophy in response to the exogenous PPARγ agonist but is sufficient to normalize the impaired insulin sensitivity.
Discussion
Our work has shown that a reduction in PPARγ expression down to 50% has no significant impact on body and fat weights, fat distribution, and insulin sensitivity in mice. In contrast, 25% normal PPARγ expression induces a selective reduction in mass and functionality of perigonadal depot with little or no effect on other depots. This was associated with a concurrent decrease, restricted to perigonadal depot only, in mRNA for proteins promoting lipid uptake and retention. Impaired perigonadal WAT function, combined with ectopic lipid deposition in muscles and disturbances in circulating lipid and adipokines, causes metabolic dysfunction and IR in PpargC/− mice.
In humans, variations in body fat distribution have gained considerable interest because fat cells in individual depot have different properties and are differentially linked to the development of metabolic disorders (24). However, little is known about the control of relative amounts and functional heterogeneity among white fat cells in these depots. Our results demonstrating that 25% normal PPARγ expression causes a selective reduction of perigonadal WAT highlight that adipogenesis in the perigonadal WAT in mice is more dependent on the PPARγ-mediated signaling than others. The transition of preadipocytes to adipocytes involves a transcriptional cascade beginning with C/EBPβ and C/EBPδ, followed by C/EBPα and PPARγ (25). C/EBPα and PPARγ synergistically trigger the terminal differentiation program and reciprocally activate transcription of one another (26). Previous studies demonstrated that cells lacking PPARγ express greatly reduced levels of C/EBPα (6,27). Notably, reduced PPARγ expression affected C/EBPα but not C/EBPβ and C/EBPδ in PpargC/− mice. Furthermore, C/EBPα was reduced only in perigonadal WAT, and the depot-specific reduction in C/EBPα was associated with selective decrease in in vitro adipogenic capability. These suggest that the expression of C/EBPα in perigonadal fat is more sensitive to the PPARγ level than that in inguinal fat, and the reduced PPARγ and C/EBPα together limit adipocyte differentiation and maturation, accounting for a reduction of fat cell number and size in this depot. These results further support the notion that the observed reduction in perigonadal fat mass of PpargC/− mice is likely to be a cell-autonomous effect of PPARγ in this depot rather than the secondary effect of PPARγ reduction in other organs. We notice, however, that impacts of PPARγ deficiency in other tissues on the normal functions of perigonadal depot cannot be completely excluded in our study.
Fat storage represents the net balance between energy input (uptake and synthesis) and expenditure (lipolysis, oxidation, and thermogenesis). Gene expression analysis identified that majority of PPARγ downstream genes involved in the regulation of classical lipid metabolism are unchanged in inguinal WAT of PpargC/− mice, with an exception of up-regulated LPL. In contrast, several genes associated with fuel uptake and fatty acid mobilization, most conspicuously aquaporin-7, Glut4, and aP2, are down-regulated only in perigonadal depots of PpargC/− mice. Perigonadal WAT of PpargC/− mice also has reduced triglyceride synthesis from glycerol and fatty acids judged by the decreased expression of ACS1 and PEPCK. The limited amount of intracellular fuel may account for the reduced ability to export, which is reflected on the reduced HSL in perigonadal WAT. Together, these dysregulations in fat metabolism result in ineffectual storage of excess energy selectively in perigonadal fat depot. Examination of gene expression related to lipid metabolism identified a selective reduction of PPARγ target genes in perigonadal fat of PpargC/− mice. Importantly, C/EBPα has also been reported to directly induce some of the PPARγ-regulated genes, including aP2, Glut4, PEPCK, and ACS1 (28,29). Thus the selective reduction in the expression of genes in perigonadal WAT may also be attributed to the selective reduction in C/EBPα. In addition, expression of PPARγ-downstream genes is highly regulated by the recruitment of an array of coactivators and corepressors with widely diverging metabolic consequences. Although expressions of RXRα, PGC-1α, and other PPARs were not affected, differential recruitments of such cofactors in perigonadal and inguinal fat may also explain the depot-specific regulation of PPARγ function. Alternatively, depot-specific environmental factors, such as ligand availability, may be influencing the regulation of downstream genes through PPARγ, as well as the autoregulation of PPARγ gene expression.
Various in vitro studies have suggested that haploinsufficiency underlies lipodystrophy (12,13,15,30), implicating that PPARγ gene dosage is critical for the disease progression. Lipodystrophic subjects heterozygous for some mutations resulting in truncated PPARγ due to the premature stop (11,17,31) or reduced expression due to the altered promoter function (18) certainly represent quantitative deficiency for PPARγ. Other mutations act in a dominant-negative fashion, resulting in a greater reduction in net receptor availability (11,32). Our results unequivocally point out that a critical amount of PPARγ for maintaining normal function of perigonadal fat tissues in mice is between 25 to 50%. However, haploinsufficency may not be the only explanation for partial lipodystrophy seen in dominant-negative PPARG mutations. Thus there are distinctive phenotypic differences between PpargC/− mice with reduced expression of normal PPARγ and PpargP465L/+ mice that carry a P465L dominant-negative mutation (33) (supplemental Table 3). For example, although both mutants exhibit altered body fat distribution with preferential reduction in the perigonadal fat mass, PpargC/− mice have normal inguinal fat whereas the inguinal fat mass is significantly increased to 120% normal in PpargP465L/+ mice (33). Most strikingly, perigonadal fat cells in PpargC/− mice are 70% normal in volume, but they are larger than normal in PpargP465L/+ mice, suggesting that the storage capacity of WAT is not compromised in PpargP465L/+ mice as in PpargC/− mice. A mutant protein can behave quite differently from the deficiency of the normal protein. Reduced promoter turnover and increased repressor binding of mutant proteins have been proposed to contribute to the dominant-negative activity (34). Alternatively, a possibility that the P465L mutation in the ligand-binding domain may have differential selectivity to different ligands cannot be excluded.
Several other mouse models with systemic reduction of PPARγ exhibit severe loss or a near absence of fat tissues systemically (supplemental Table 3). For instance, global deletion of PPARγ1 and PPARγ2 resulted in less than 5% of residual PPARγ expression and generalized fat loss in MORE-PGKO mice (9). Severe loss of WAT in both visceral and sc depots is also demonstrated in PPARγ hypomorphic mice (Pparghyp/hyp) with significantly reduced expression of PPARγ2 (10). Pparghyp/hyp mice also lack expression of PPARγ1 in WAT, but a compensatory increase of PPARγ1 in BAT and normal expression of PPARγ1 in other tissues are likely to confound the complex phenotypes of these mice. Phenotypes such as reduced WAT mass and survival to the adulthood of MORE-PGKO and Pparghyp/hyp mice are more severe than PpargC/− mice, suggesting a direct correlation between the PPARγ expression levels and these phenotypes. However, no lethality in mice selectively lacking PPARγ2 has been reported, although both perigonadal and inguinal WAT are reduced significantly (35,36). This leaves the possibility open that the PPARγ1 expression in fat may be critical for the survival of preweaning pups. In addition, targeted deletion of PPARγ selectively in adipose tissue using aP2 promoter-driven Cre recombinase results in progressive loss of fat in all depots (8,37). We emphasize that, compared with the models in which PPARγ expression is altered in complex manners in different tissues, the PPARγ expression in mice carrying the Pparg-C allele is reduced in all tissues throughout their life, representing genetic variations that likely occur in human populations.
The core clinical phenotype of human partial lipodystrophy is fat loss with subsequent development of secondary metabolic disturbances, such as IR, dyslipidemia, and diabetes (38). The normal lipid ingestion and excretion in PpargC/− mice indicate that lipid is steadily imported into the body. Impaired lipid storage and FFA uptake in perigoandal fat substantially increase serum triglyceride and FFA and likely increase the lipid burden and lipotoxicity in nonadipose tissues, which is known to impair insulin sensitivity in these tissues. The increased lipid content in the muscle of PpargC/− mice reflects this phenomenon, although liver lipid content and hepatic steatosis were surprisingly unremarkable. In addition, the marked reduction of circulating adiponectin, which is capable of potentiating insulin’s actions in muscle and liver, likely contributes to the observed metabolic impairment in PpargC/− mice. Finally, the contribution of significant reduction of PPARγ expression in skeletal muscle and liver on metabolic impairment of PpargC/− mice cannot be excluded.
In conclusion, our titrated reduction of Pparg gene expression in mice demonstrates that the level of expression required to maintain functionality of perigonadal WAT is 25% to 50%, but other WATs can spare 70% of the normal expression. The sensitivity to PPARγ deficiency is therefore depot dependent, as reflected in depot-specific changes in gene expressions for multiple pathways of adipose lipid metabolism and mobilization. Clearly, more studies are needed to gain further insights on how PPARγ dosage affects depot-specific fat biology. However, the resulted phenotypic outcome of PpargC/− mice is extremely rare and requires the rise of public awareness. Our PPARγ quantitative variants also provide an excellent tool for unveiling the underlying mechanisms for partial lipodystrophy in humans with PPARG mutations.
Materials and Methods
Mice
The Pparg-C allele was made by gene targeting in embryonic stem cells by inserting a 69-bp DNA fragment containing ARE of c-fos gene (39) into the 3′-UTR immediately downstream of the stop codon in the mouse Pparg gene. The generation of mice with the modified Pparg locus has been described (21), and the mutant line that does not carry a linked mutation in the Pmca2 gene was used in the present study. PpargC/C, PpargC/+, and Pparg+/+ mice were littermates of F2 generation from a cross between 129S6 and C57BL/6J F1 heterozygotes. The Pparg-C allele was subsequently placed on a C57BL/6J background by backcrossing for eight generations. PpargC/−, Pparg+/−, PpargC/+, and Pparg+/+ mice were F1 littermates from mating of PpargC/+ mice on a C57BL/6J background with Pparg+/− mice on a 129S6 background (kindly provided by Dr. Ronald Evans at the Salk Institute) (7). Mice were fed with either a regular chow (LabDiet 5P76; PMI Nutrition International, Richmond, IN) or a high-fat diet (59% of calories as fat; 58R2; TestDiet, Richmond, IN). For TZD treatment, high-fat diet (58R2) was blended with rosiglitazone and fed to mice at approximately 10 mg/kg·d. Animals were housed in a specific pathogen-free barrier facility and handled following procedures approved by the IACUC of University of North Carolina at Chapel Hill and National Cheng Kung University.
Tissue collection and RNA analysis
Tissues were collected and stored in RNAlater (Ambion, Austin, TX), and RNA was extracted using the TRIzol Reagent (Invitrogen, Carlsbad, CA). mRNA were analyzed with real-time quantitative RT-PCR (Applied Biosystems, Foster City, CA), with β-actin or Gapdh as reference gene in each reaction. Sequences of the primers used for RT-PCR assays are shown in supplemental Table 4.
Protein analysis
Total protein (10 μg) was separated by SDS-PAGE, transferred to PVDF membranes (Pall Gelman Laboratory, Ann Arbor, MI), and probed with antibodies recognizing PPARγ (sc-7196; Santa Cruz Biotechnology, Santa Cruz, CA), C/EBPα (sc-61; Santa Cruz Biotechnology), and β-actin (A5441; Sigma, St. Louis, MO). Immunoreactive proteins were detected using an enhanced chemiluminescence Western blotting detection system (GE Healthcare, Buckinghamshire, UK).
Body composition
Body composition was analyzed in anesthetized animals using a DEXA Scanner (PIXImus2; Lunar Corp., Madison, WI). The boundary between abdominal and thoracic regions was defined by the position of diaphragm. The fat content in extremities was determined by subtracting fat in the trunk from the whole-body fat.
Morphological analysis
Paraffin sections (10 μm) of adipose tissues isolated from female mice (n = 3∼5) at 16 wk of age were stained with hematoxylin and eosin. Adipocyte size was measured in 500 cells per mouse in several parts of the perigonadal and inguinal fat pads using Nikon NIS Elements AR 2.30 software. Mean adipocyte number was determined by dividing the mean fat mass by the mean fat cell size as described previously (22).
Metabolic analysis
Daily food intake was examined using metabolic cages (Solo Mouse Metabolic Cage; Tecniplast, Italy). Daily energy intake was calculated from daily food consumed per mouse multiplied by the gross energy for regular chow 4.10 kcal/g. Feeding efficiency was calculated by dividing the weight increase by their feed consumption over 3 months of the study. Daily fecal output and lipid content were determined (40). Rectal temperature was recorded by thermometer (TH-8; Physitemp Instruments, Clifton, NJ). Activity was measured by implanting a telemetric device (PA-C20; Data Sciences, St. Paul, MN) on 4-month-old male mice. Continuous activities were recorded every 5 min for 6 d beginning 8 d after surgery to allow mice to regain normal diurnal rhythms.
ip glucose and insulin tolerance tests
Mice were fasted for 5 h and injected ip with glucose (1 g/kg body weight) or human regular insulin (0.5 or 0.75 U/kg body weight; Humulin; Eli Lilly, Indianapolis, IN). Blood samples were collected before and at indicated times after injections. Serum glucose concentration was determined by a glucose colorimetric test (Autokit Glucose; Wako Chemicals USA, Richmond, VA). Insulin was measured using mouse insulin ELISA (Ultrasensitive Mouse Insulin ELISA; Mercodia, Sweden). The IR index was calculated as the product of the areas under glucose and insulin curves in glucose tolerance tests as previously described (41).
Differentiation of preadipocyte
Fat tissues from perigonadal and inguinal depots were isolated and digested with 1 mg/ml collagenase (Sigma) in DMEM. The cell suspension was filtered through a 100-μm pore mesh and centrifuged. The stromovascular cells from the resuspended cell pellet were cultured in DMEM supplemented with 10% fetal bovine serum, 33 μm biotin, 100 μm ascorbic acid, 4 nm insulin, and 8.3 mm l-glutamine. Preadipocyte differentiation was induced by adding the induction cocktail containing insulin (1 μg/ml), dexamethasone (1 μm), 1-methyl-3isobutyl-xanthine (0.5 mm), and rosiglitazone (1 μm) to the medium. After 48 h, cells were changed to the medium containing DMEM with 10% fetal bovine serum, insulin (1 μg/ml), and rosiglitazone (1 μm). Lipid content was assessed by Oil Red O staining at the end of differentiation.
Lipid and cytokine assays
Serum total cholesterol (Cholesterol E; Wako, Osaka, Japan), FFA (NEFA C; Wako), and triglyceride (Stanbio Laboratory, San Antonio, TX) were measured following each protocol. Organ lipid content was determined after extraction of lipids (40). Serum levels of leptin and adiponectin were determined by ELISA (R&D Systems, Minneapolis, MN). Serum levels of IL-6 and TNFα were determined using the Luminex multiplex assay system (Linco Research, St. Charles, MO).
Data analysis
Values are reported as mean ± sem. Statistical analysis was conducted using JMP software (SAS Institute) by multifactorial ANOVA with genotype, gender, and age as factors. Student’s t test was used for comparisons between mutant and wild-type mice within each group, and differences were considered to be statistically significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank Drs. C. H. Lee, R. Coleman, J. Harp, N. Takahashi, and O. Smithies for discussions; J. Hagaman, University of North Carolina Animal Metabolism and Phenotyping Core (K. Hua), and National Laboratory Animal Center Pathology Core (C. T. Liang) for technical assistance; and Dr. R. Evans for kindly providing us with Pparg+/− mice.
Footnotes
This work was supported by National Institutes of Health Grants HL42630, HL077145, and DK067320 (to N.M.) and National Science Council Taiwan Grants 96-23-2-B-006-001 and National Cheng Kung University Landmark Project (to Y.-S.T.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online September 11, 2009
Abbreviations: ACS1, acyl-coenzyme A synthetase-1; ARE, AU-rich element; BAT, brown adipose tissue; C/EBPα, CCAAT enhancer binding protein-α; DEXA, dual energy x-ray absorptiometry; FFA, free fatty acid; Glut4, glucose transporter-4; HFD, high-fat diet; HSL, hormone-sensitive lipase; IR, insulin resistance; LPL, lipoprotein lipase; PEPCK, phosphoenolpyruvate carboxykinase; PPAR, peroxisome proliferator-activated receptor; PPRE, peroxisome proliferator response element; RXR, retinoic X receptor; TZD, thiazolidinedione; UCP, uncoupling protein; UTR, untranslated region; WAT, white adipose tissue.
References
- Evans RM, Barish GD, Wang YX 2004 PPARs and the complex journey to obesity. Nat Med 10:355–361 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM 2008 Fat and beyond: the diverse biology of PPARγ. Annu Rev Biochem 77:289–312 [DOI] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J 1998 A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet 20:284–287 [DOI] [PubMed] [Google Scholar]
- Masugi J, Tamori Y, Mori H, Koike T, Kasuga M 2000 Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-γ 2 on thiazolidinedione-induced adipogenesis. Biochem Biophys Res Commun 268:178–182 [DOI] [PubMed] [Google Scholar]
- Doney A, Fischer B, Frew D, Cumming A, Flavell DM, World M, Montgomery HE, Boyle D, Morris A, Palmer CN 2002 Haplotype analysis of the PPARγ Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genet 3:21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, Eto K, Tsubamoto Y, Okuno A, Murakami K, Sekihara H, Hasegawa G, Naito M, Toyoshima Y, Tanaka S, Shiota K, Kitamura T, Fujita T, Ezaki O, Aizawa S, Kadowaki T, et al. 1999 PPAR γ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell 4:597–609 [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM 1999 PPAR γ is required for placental, cardiac, and adipose tissue development. Mol Cell 4:585–595 [DOI] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM 2003 Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA 100:15712–15717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan SZ, Ivashchenko CY, Whitesall SE, D’Alecy LG, Duquaine DC, Brosius III FC, Gonzalez FJ, Vinson C, Pierre MA, Milstone DS, Mortensen RM 2007 Hypotension, lipodystrophy, and insulin resistance in generalized PPARγ-deficient mice rescued from embryonic lethality. J Clin Invest 117:812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutnikova H, Cock TA, Watanabe M, Houten SM, Champy MF, Dierich A, Auwerx J 2003 Compensation by the muscle limits the metabolic consequences of lipodystrophy in PPAR γ hypomorphic mice. Proc Natl Acad Sci USA 100:14457–14462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Schoenmakers E, Mitchell C, Szatmari I, Savage D, Smith A, Rajanayagam O, Semple R, Luan J, Bath L, Zalin A, Labib M, Kumar S, Simpson H, Blom D, Marais D, Schwabe J, Barroso I, Trembath R, Wareham N, Nagy L, Gurnell M, O’Rahilly S, Chatterjee K 2006 Non-DNA binding, dominant-negative, human PPARγ mutations cause lipodystrophic insulin resistance. Cell Metab 4:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüdtke A, Buettner J, Wu W, Muchir A, Schroeter A, Zinn-Justin S, Spuler S, Schmidt HH, Worman HJ 2007 Peroxisome proliferator-activated receptor-γ C190S mutation causes partial lipodystrophy. J Clin Endocrinol Metab 92:2248–2255 [DOI] [PubMed] [Google Scholar]
- Monajemi H, Zhang L, Li G, Jeninga EH, Cao H, Maas M, Brouwer CB, Kalkhoven E, Stroes E, Hegele RA, Leff T 2007 Familial partial lipodystrophy phenotype resulting from a single-base mutation in deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-γ. J Clin Endocrinol Metab 92:1606–1612 [DOI] [PubMed] [Google Scholar]
- Savage DB, Tan GD, Acerini CL, Jebb SA, Agostini M, Gurnell M, Williams RL, Umpleby AM, Thomas EL, Bell JD, Dixon AK, Dunne F, Boiani R, Cinti S, Vidal-Puig A, Karpe F, Chatterjee VK, O’Rahilly S 2003 Human metabolic syndrome resulting from dominant-negative mutations in the nuclear receptor peroxisome proliferator-activated receptor-γ. Diabetes 52:910–917 [DOI] [PubMed] [Google Scholar]
- Hegele RA, Cao H, Frankowski C, Mathews ST, Leff T 2002 PPARG F388L, a transactivation-deficient mutant, in familial partial lipodystrophy. Diabetes 51:3586–3590 [DOI] [PubMed] [Google Scholar]
- Agarwal AK, Garg A 2002 A novel heterozygous mutation in peroxisome proliferator-activated receptor-γ gene in a patient with familial partial lipodystrophy. J Clin Endocrinol Metab 87:408–411 [DOI] [PubMed] [Google Scholar]
- Hegele RA, Ur E, Ransom TP, Cao H 2006 A frameshift mutation in peroxisome-proliferator-activated receptor-γ in familial partial lipodystrophy subtype 3 (FPLD3; MIM 604367). Clin Genet 70:360–362 [DOI] [PubMed] [Google Scholar]
- Al-Shali K, Cao H, Knoers N, Hermus AR, Tack CJ, Hegele RA 2004 A single-base mutation in the peroxisome proliferator-activated receptor γ4 promoter associated with altered in vitro expression and partial lipodystrophy. J Clin Endocrinol Metab 89:5655–5660 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 1995 AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20:465–470 [DOI] [PubMed] [Google Scholar]
- Kakoki M, Tsai YS, Kim HS, Hatada S, Ciavatta DJ, Takahashi N, Arnold LW, Maeda N, Smithies O 2004 Altering the expression in mice of genes by modifying their 3′ regions. Dev Cell 6:597–606 [DOI] [PubMed] [Google Scholar]
- Tsai YS, Pendse A, Moy SS, Mohri I, Perez A, Crawley JN, Suzuki K, Maeda N 2006 A de novo deafwaddler mutation of Pmca2 arising in ES cells and hitchhiking with a targeted modification of the Pparg gene. Mamm Genome 17:716–722 [DOI] [PubMed] [Google Scholar]
- Di Girolamo M, Mendlinger S, Fertig JW 1971 A simple method to determine fat cell size and number in four mammalian species. Am J Physiol 221:850–858 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM 2006 Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR 2007 Developmental origin of fat: tracking obesity to its source. Cell 131:242–256 [DOI] [PubMed] [Google Scholar]
- Lowell BB 1999 PPARγ: an essential regulator of adipogenesis and modulator of fat cell function. Cell 99:239–242 [DOI] [PubMed] [Google Scholar]
- Lazar MA 2002 Becoming fat. Genes Dev 16:1–5 [DOI] [PubMed] [Google Scholar]
- Ren D, Collingwood TN, Rebar EJ, Wolffe AP, Camp HS 2002 PPARγ knockdown by engineered transcription factors: exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes Dev 16:27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougald OA, Lane MD 1995 Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem 64:345–373 [DOI] [PubMed] [Google Scholar]
- Olofsson LE, Orho-Melander M, William-Olsson L, Sjöholm K, Sjöström L, Groop L, Carlsson B, Carlsson LM, Olsson B 2008 CCAAT/enhancer binding protein α (C/EBPα) in adipose tissue regulates genes in lipid and glucose metabolism and a genetic variation in C/EBPα is associated with serum levels of triglycerides. J Clin Endocrinol Metab 93:4880–4886 [DOI] [PubMed] [Google Scholar]
- Jeninga EH, van Beekum O, van Dijk AD, Hamers N, Hendriks-Stegeman BI, Bonvin AM, Berger R, Kalkhoven E 2007 Impaired peroxisome proliferator-activated receptor γ function through mutation of a conserved salt bridge (R425C) in familial partial lipodystrophy. Mol Endocrinol 21:1049–1065 [DOI] [PubMed] [Google Scholar]
- Francis GA, Li G, Casey R, Wang J, Cao H, Leff T, Hegele RA 2006 Peroxisomal proliferator activated receptor-γ deficiency in a Canadian kindred with familial partial lipodystrophy type 3 (FPLD3). BMC Med Genet 7:3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O’Rahilly S 1999 Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880–883 [DOI] [PubMed] [Google Scholar]
- Tsai YS, Kim HJ, Takahashi N, Kim HS, Hagaman JR, Kim JK, Maeda N 2004 Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARγ. J Clin Invest 114:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Leff T 2007 Altered promoter recycling rates contribute to dominant-negative activity of human peroxisome proliferator-activated receptor-γ mutations associated with diabetes. Mol Endocrinol 21:857–864 [DOI] [PubMed] [Google Scholar]
- Zhang J, Fu M, Cui T, Xiong C, Xu K, Zhong W, Xiao Y, Floyd D, Liang J, Li E, Song Q, Chen YE 2004 Selective disruption of PPARγ 2 impairs the development of adipose tissue and insulin sensitivity. Proc Natl Acad Sci USA 101:10703–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez G, Virtue S, Lelliott C, Boiani R, Campbell M, Christodoulides C, Perrin C, Jimenez-Linan M, Blount M, Dixon J, Zahn D, Thresher RR, Aparicio S, Carlton M, Colledge WH, Kettunen MI, Seppänen-Laakso T, Sethi JK, O’Rahilly S, Brindle K, Cinti S, Oresic M, Burcelin R, Vidal-Puig A 2005 The link between nutritional status and insulin sensitivity is dependent on the adipocyte-specific peroxisome proliferator-activated receptor-γ2 isoform. Diabetes 54:1706–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Barrick C, Kim KA, Lindner J, Blondeau B, Fujimoto Y, Shiota M, Kesterson RA, Kahn BB, Magnuson MA 2005 Deletion of PPARγ in adipose tissues of mice protects against high fat diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA 102:6207–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YS, Maeda N 2005 PPARγ: a critical determinant of body fat distribution in humans and mice. Trends Cardiovasc Med 15:81–85 [DOI] [PubMed] [Google Scholar]
- Chen CY, Chen TM, Shyu AB 1994 Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol 14:416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH 1957 A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497–509 [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, Ezaki O, Akanuma Y, Gavrilova O, Vinson C, Reitman ML, Kagechika H, Shudo K, Yoda M, Nakano Y, Tobe K, Nagai R, Kimura S, Tomita M, Froguel P, Kadowaki T 2001 The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med 7:941–946 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.