Abstract
Transcripts from mitochondrial and chloroplast DNA of land plants often undergo cytidine to uridine conversion-type RNA editing events. RESOPS is a newly built database that specializes in displaying RNA editing sites of land plant organelles on protein three-dimensional (3D) structures to help elucidate the mechanisms of RNA editing for gene expression regulation. RESOPS contains the following information: unedited and edited cDNA sequences with notes for the target nucleotides of RNA editing, conceptual translation from the edited cDNA sequence in pseudo-UniProt format, a list of proteins under the influence of RNA editing, multiple amino acid sequence alignments of edited proteins, the location of amino acid residues coded by codons under the influence of RNA editing in protein 3D structures and the statistics of biased distributions of the edited residues with respect to protein structures. Most of the data processing procedures are automated; hence, it is easy to keep abreast of updated genome and protein 3D structural data. In the RESOPS database, we clarified that the locations of residues switched by RNA editing are significantly biased to protein structural cores. The integration of different types of data in the database also help advance the understanding of RNA editing mechanisms. RESOPS is accessible at http://cib.cf.ocha.ac.jp/RNAEDITING/.
Keywords: Chloroplast, Mitochondrion, Molecular evolution, Organelle genome, Protein 3D structure, RNA editing
Introduction
RNA editing is a process that inserts, deletes and converts nucleotides in RNA after transcription, distinct from RNA splicing (Gray and Covello 1993, Gott and Emeson 2000, Keegan et al. 2001). The conversion type of RNA editing was first discovered in mammalian mRNA for apolipoprotein B (apoB) (Chen et al. 1987, Powell et al. 1987), but most of the known cytidine to uridine conversion-type RNA editing events are mainly found on mRNAs transcribed from mitochondrial and chloroplast DNA of land plants (Covello and Gray 1989, Hoch et al. 1991, Hiesel et al. 1994, Wakasugi et al. 1996,Yoshinaga et al. 1996, Freyer et al. 1997, Giege and Brennicke 1999, Kugita et al. 2003). In hornwort chloroplasts, uridine to cytidine conversion was also found (Kugita et al. 2003). RNA editing is not a rare event. The Anthoceros formosae chloroplast genome has at least 942 RNA editing sites (Kugita et al. 2003), and the Arabidopsis thaliana mitochondrial genome has at least 441 RNA editing sites (Giege and Brennicke 1999). Most of these conversions occur in protein-coding regions, suggesting that RNA editing should impact protein structure and function. The top three patterns of amino acid residue conversions in RNA editing are serine to leucine, proline to leucine and serine to phenylalanine (Bock 2000), all of which are conversions from hydrophilic to hydrophobic residues. This conversion pattern further supports the notion that RNA editing has a substantial impact on protein structure and function. Many experiments have been carried out to demonstrate that the conversion of amino acid residues via RNA editing is crucial for protein function (Covello and Gray 1990, Bock et al. 1994, Bonnard and Grienenberger 1995, Phreaner et al. 1996, Zito et al. 1997, Kozaki et al. 2001, Sasaki et al. 2001); however, it was seldom the case that a converted residue was included in a protein active site (Yura and Go 2008). Hence, the molecular mechanism for function regulation via RNA editing has not been clarified.
Genome sequencing and structural genomics projects have produced massive quantities of data, including RNA editing sites, organelle genome sequences and protein three-dimensional (3D) structures. Based on these data, we reported previously that amino acid residues that are converted by RNA editing (hereafter called edited residues) tend to be located in protein structural cores (Yura and Go 2008). Combinations of genome and protein structure data enabled us to determine that the locations of edited residues were significantly biased toward the structurally important sites of proteins. RNA editing, therefore, seems to regulate protein function through protein folding, because in general when a protein has a hydrophilic mutation in the protein structural core, the protein becomes unstable at best and does not fold at worst (Vos et al. 2001, Loladze et al. 2002).
The molecular mechanism of the regulation suggested above is based on current advances in data production from omics analysis, and a suggested mechanism should be continuously tested as data are augmented by new results. In addition, combining data related to RNA editing will advance our understanding of the mechanisms and origin of RNA editing in land plant organelles, allowing, for example, the development of RNA editing site prediction methods (Cummings and Myers 2004, Mower 2005, Thompson and Gopal 2006, Du et al. 2007, Yura et al. 2008, Du et al. 2009). So far, there are no databases providing information about the relationship between RNA editing sites and protein 3D structures, multiple sequence alignments of homologous proteins or statistics on RNA editing sites. We therefore launched RESOPS, a database of RNA editing sites of land plant organelles that contains up-to-date RNA editing site raw data, multiple amino acid sequence alignments with editing site information in detail and edited residues in protein 3D structures. The database is freely accessible at http://cib.cf.ocha.ac.jp/RNAEDITING/.
Results
Collection of RNA editing sites from the GenBank and PDB database
In the August 2009 version of RESOPS, based mainly on the GenBank database release 172, there are 710 entries that contain at least one edited residue in an amino acid sequence from plant mitochondria and chloroplasts. A single flat file with 710 entries in pseudo-UniProt format, containing amino acid and cDNA sequences marked with RNA editing sites, can be obtained from the download page. The download page describes the details of the format and the history of manual corrections. A comparison between homologous sequences in the data set is performed via the construction of multiple sequence alignments.
The current data contain 5,754 RNA editing sites, of which 2,059 (35.8%) sites are located on the first letter of a codon, 3,165 (55.0%) are on the second letter and 530 (9.2%) are on the third letter. These figures are dynamically calculated by summing over the alignment data. The distribution of the RNA editing sites on codons is similar to a distribution calculated previously (Bock 2000).
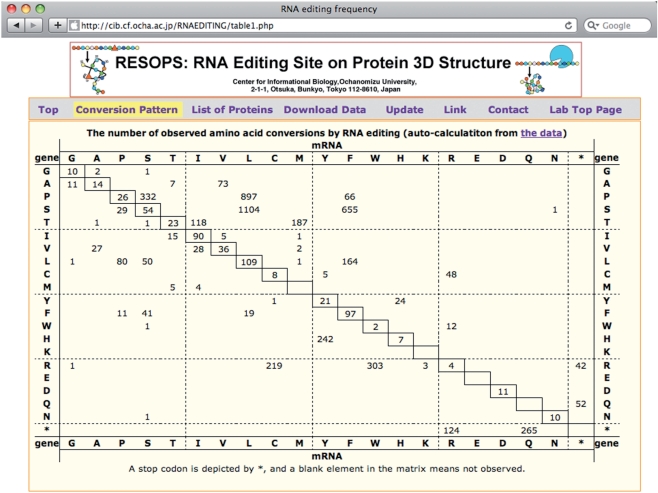
RNA editing events frequently convert coded amino acid residues, because >90% of RNA editing sites are located on either the first or the second letter of codons. The conversion pattern of amino acid residues is automatically tabulated from the flat file as shown in Fig. 1. The most frequent conversion in amino acid residues is from serine to leucine, followed by proline to leucine and serine to phenylalanine. The trend of altering from hydrophilic to hydrophobic residues, mentioned before (Gray and Covello 1993, Bock 2000, Yura and Go 2008), still holds.
Fig. 1.
Conversion patterns of amino acid residues by RNA editing. The vertical axis is the type of amino acid coded in the genome, and the horizontal axis is the type of amino acid in mature mRNAs. The table is dynamically generated from the flat file data each time a user accesses the website. The trend in the pattern is very similar to the trend previously reported.
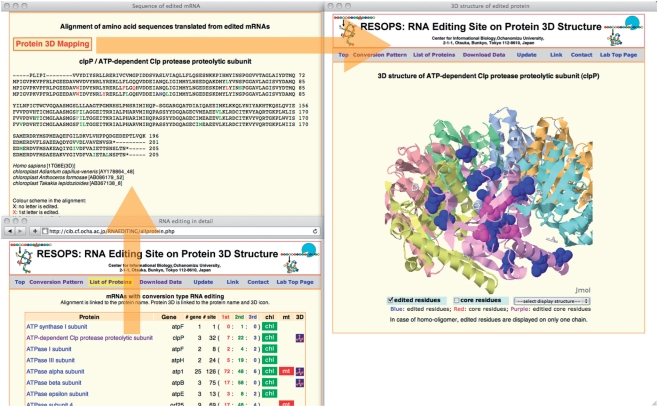
RESOPS stores data for the location of edited residues in both the primary and tertiary structures (Fig. 2). Edited residues are shown in color in the multiple amino acid sequence alignment. If the first letter of the codon is edited then the residue is colored in red, if the second letter then in green and if the third letter then in blue. If more than one letter is edited, then the residue is in the mixed color. In a multiple sequence alignment, the conservation patterns of edited residues amongst species show that RNA editing improves sequence identity among homologous proteins. This evidence further supports the notion that RNA editing is a process of ‘transcript repair’ (Bock 2000). When a group of homologous proteins includes one protein for which the 3D structure has been determined and stored in the Protein Data Bank (PDB) (Berman et al. 2003), the amino acid sequence of the structurally determined protein is shown at the top of the alignment. This alignment forms the basis for mapping edited residues onto protein 3D structures. The 3D structure of a protein is shown as a ribbon model, in which each chain is in a different color, and residues corresponding to the edited residues are marked by space-filling representations using the molecular graphic software, Jmol (http://www.jmol.org/). The edited residues that reside in the protein structural core are shown in purple, and the others are in blue.
Fig. 2.
A typical analysis in RESOPS. A user can find a protein/gene name in the List of Proteins if the protein expression is influenced by RNA editing. The List of Proteins also indicates the number of genes, locations of the edited sites in a codon, the origin (chloroplast and mitochondrion) of the sequence and whether a 3D structure of the protein is known. The multiple sequence alignment of homologous edited proteins is linked to the name of the protein. If the 3D structure of the protein is known, then all edited residues are mapped to the 3D structure. The 3D structure is also directly accessible by an icon link in the 3D column of the List of Proteins.
Bias of RNA editing sites toward a protein structural core
It was shown that the location of edited residues was significantly biased in favor of the protein structural core (Yura and Go 2008). In this database, the statistical test for this biased distribution can be automatically performed. In the August 2009 version of RESOPS, 3D structures of 48 groups of proteins were assigned. In these 48 proteins, 1,985 residues resided in the structural cores (41 residues per protein) and 14,290 residues were categorized as non-core residues. Therefore, about 12% of residues were categorized as residues in the structural cores and 88% were non-core residues. Multiple sequence alignments in RESOPS were able to map edited residues onto a protein 3D structure. It was found that 251 out of 1,277 edited residues resided in protein structural cores, and 1,026 were non-core residues. The expected number of residues in structural cores, based on a random distribution model, is ∼153 (= 1,277 × 0.12), whereas the number of expected non-core residues is ∼1,124 (= 1,277 × 0.88). A χ2 test with one degree of freedom yields 66.3 (P < 3.8 × 10−16). This result indicates that the distribution of edited residues is biased toward protein structural cores in the current data set. The biased distribution of edited residues to the protein structural core might be derived from the fact that edited residues tend to be hydrophobic residues and that hydrophobic residues tend to be buried inside the protein. RESOPS has a function to test automatically the distribution of hydrophobic residues only (phenylalanine and leucine), which eliminates the inherent biased distribution of hydrophobic residues in protein 3D structures from the test. A χ2 test on the adjusted data set still yields 10.3 (P < 1.3 × 10−3), and the distribution of the edited hydrophobic residues is found to be significantly biased toward protein structural cores.
Discussion
Impact of RNA editing on protein structure and function
The role of RNA editing in plant organelles was suggested to be a means of regulating organellar protein expression. A number of experiments were performed to test the function of unedited proteins, most of which turned out to be less functional than the edited proteins (Covello and Gray 1990, Bock et al. 1994, Bonnard and Grienenberger 1995, Phreaner et al. 1996, Zito et al. 1997, Kozaki et al. 2001, Sasaki et al. 2001). However, the molecular mechanism for regulation has yet to be uncovered, because only a few of the edited sites comprise the active sites of proteins (Yura and Go 2008). Based on the biased distribution of edited residues toward protein structural cores, we suggest that the expression of function should be regulated via protein folding, because unedited proteins tend to contain more hydrophilic residues in the parts that are supposed to be protein structural cores. Mutation to hydrophilic residues in the protein structural core destabilizes the protein, because the hydrophobic core is required to build a functional protein 3D structure (Vos et al. 2001, Loladze et al. 2002). An unedited protein has, on average, two to three hydrophilic mutations in its protein structural core, and a single hydrophilic mutation in a protein structural core destabilizes proteins by ∼5 kcal mol−1, which is comparable in magnitude with the reduction of free energy in protein folding, ∼10–15 kcal mol−1 (Creighton 1990).
Kotera et al. (2005) identified a nuclear protein CRR4 involved in RNA editing in A. thaliana. It was shown that CRR4 protein was specifically involved in RNA editing on the initiation codon of ndhD, because mutation of CRR4 changed the extent of the RNA editing. Following this study, many nuclear proteins involved in RNA editing of chloroplasts and mitochondria were identified (Chateigner-Boutin et al. 2008, Cai et al. 2009, Kim et al. 2009, Robbins et al. 2009, Yu et al. 2009, Zehrmann et al. 2009, Zhou et al. 2009). These studies identified the target sites of the nuclear proteins for RNA editing, and the functional effect of mutation on the nuclear proteins, mainly the impact of suppressing RNA editing of the target sites. We found that many effects in these cases could be qualitatively explained based on protein 3D structures in RESOPS. The result is summarized in Table 1 and the details are described below.
Table 1.
Correspondence between functional/structural effects by suppressing RNA editing and the suppressed RNA editing sites on protein 3D structures
| Gene | Positiona | Effect | 3Db | Reference |
|---|---|---|---|---|
| rpoA | C200 | May prevent PEP assembly | Core | Chateigner-Boutin et al. (2008) |
| rpoB | C338 | Partial loss of activity | Non-core | Zhou et al. (2009) |
| clpP | C559 | Unclear | Surface | Chateigner-Boutin et al. (2008) |
| cox2 | C167 | Malfunction in electron transport chain | Surface | Kim et al. (2009) |
| cox3 | C572 | Malfunction in electron transport chain | Core | Kim et al. (2009) |
| rps4 | C954 | No effect | ND | Zehrmann et al. (2009) |
| accD | C794 | Low solubility | Core | Sasaki et al. (2001) |
| Albino phenotype | Yu et al. (2009) | |||
| No effect | Robbins et al. (2009) |
aThe position number is taken from the original paper and it is the nucleotide number of the edited nucleotide in mRNA.
bND indicates that the corresponding residue is not included in the 3D structure determined. Each 3D structure is given in Supplementary data 1–6.
Chateigner-Boutin et al. (2008) speculated that abolishing RNA editing on amino acid residue 67 of RpoA in Arabidopsis chloroplast mutants may prevent assembly of plastid-encoded RNA polymerase (PEP). The speculation implies that RpoA becomes unstable. In RESOPS, we find that the residue forms a protein structural core of RpoA, and alteration of the residue to a small hydrophilic amino acid probably destabilizes the protein, and hence affects interactions with other subunits of the polymerase (Supplementary data 1). Zhou et al, (2009) showed that ys1 mutants had a defect in RNA editing of rpoB in Arabidopsis chloroplast and that the defect possibly caused a partial loss of RpoB activity. In RESOPS, we find that the residue is buried, but not in a structural core (Supplementary data 2) and hence the alteration of the residue probably has a partial impact on protein stability. Chateigner-Boutin et al. (2008) found that their clb19 mutants abolished one of the RNA editing events on clpP. The impact of abolishing the RNA editing event on clpP was not clear in their work. In RESOPS, the edited residue is found on the surface of the protein, even though it is a hydrophobic residue (Supplementary data 3). We speculate that ClpP is stable and functional in the mutant. Kim et al (2009) demonstrated that rice ogr1 mutants had defects in RNA editing of cox2 and cox3 and speculated that the defects caused malfunction in the mitochondrial electron transport chain. The edited residue in Cox2 is found on the surface of a transmembrane helix, which suggests that the residue is in contact with membrane lipids (Supplementary data 4). The edited residue in Cox3 is found in the protein structural core, in the internal interfaces of the helix bundle (Supplementary data 5). The structural data, therefore, suggest that the mutation on Cox3 should have a more significant impact on protein function than that on Cox2. Robbins et al. (2009) showed that rare1 mutants abolished RNA editing at C794 of accD. The mutants were unexpectedly robust and they suggested that RNA editing at C794 of accD was not essential for acetyl-CoA carboxylase activity, or that other carboxylases should compensate for the loss of accD function. In RESOPS, we find that the edited residue is included in a protein structural core, and mutation of the residue evidently has an impact on protein stability (Supplementary data 6). Our analysis is consistent with previous works by Sasaki et al. (2001) and Yu et al. (2009), and we suggest that the second suggestion by Robbins et al. (2009) is much more likely than the first one.
Possible origin of RNA editing in land plant organelles
Multiple sequence alignment of the edited proteins suggests a multiple origin of RNA editing in organelles. Most of the sites with RNA editing are not unanimously edited in homologous proteins. When the type of amino acid is compared at each site, amino acids of non-edited sequences are almost always the same as the residues of the edited sequence, but not the unedited sequence. This suggests that RNA editing was not introduced into the non-edited sequences at the site. If RNA editing had been introduced in the common ancestor of the genes, and if the current non-edited sequence had lost its RNA editing mechanism, then the type of amino acid residue should be the same as the type of unedited amino acid. Hence, this observation suggests that RNA editing should be introduced at a site in the most recent common ancestor of the genes that share RNA editing sites at the same position, which also suggests that the introduction of an RNA editing site should have occurred many times in many genes. It is well known that RNA editing in plant organelles has only been found in land plants (Gray and Covello 1993, Bock 2000). This suggests that RNA editing was introduced at the time land plants came into being (Yoshinaga et al. 1996). Because RNA editing is introduced later than the time that plants acquired two organelles, it should be rare to find RNA editing events in homologous sites of proteins in mitochondria and chloroplasts. By checking through multiple sequence alignments in RESOPS, however, we found 12 such events in five genes, as shown in Table 2. The amino acid sequence alignment of ndhC/nad3 products is shown in Fig. 3. Other alignments are given in Supplementary data 7. These correspondences could reflect preferred sites for introduction of RNA editing events.
Table 2.
RNA editing events in homologous sites in genes from chloroplasts and mitochondria
| Gene name | Position in alignmenta | Chloroplast | Mitochondrion |
|---|---|---|---|
| rps7 | 195 (2) | S→L | S→L |
| rps19 | 16 (2) | S→L | S→L |
| ndhB/nad2 | 186 (2) | T→M | S→L |
| 242 (2) | S→F | S→F | |
| 285 (2) | S→L | S→F | |
| ndhC/nad3 | 130 (2) | S→F | S→F |
| 144 (2) | S→L | S→F | |
| 153 (2) | S→L | S→L | |
| petB/cob | 64 (2) | T→M | S→L |
| 68 (1) | H→Y | H→Y | |
| 116 (2) | S→L | S→F/F→S | |
| 209 (2) | T→M | W→S |
aDetail of the amino acid sequence alignment is given in Supplementary data 7. The number in parentheses is the position of the edited nucleotide in the codon.
Fig. 3.
Amino acid sequence alignment of ndhC/nad3 products. An edited residue is differently colored based on the edited letter in the codon. When the first letter is edited then the residue is colored in red, the second letter in green and the third letter in blue. When the first and the second letters are edited, then the residue is colored in orange. The top three sequences are derived from chloroplast genomes and the others are from mitochondrion genomes. Three positions with a triangle have edited residues in sequences from both chloroplasts and mitochondria. The number on the triangle is the alignment position given in the RESOPS database.
Future development of the database
RESOPS will be updated regularly following the major update of GenBank every 2 months. The procedure for updating is automatic, except for the initial process of adding data from the literature and of checking the consistency of the GenBank database (see Materials and Methods). We hope that the inconsistencies in the public database may be resolved by the original depositors in the near future. In the last 2 years, we have seen some corrections introduced into the annotations of RNA editing events listed in the GenBank database. To promote corrections, we continue to contact the original depositors when we find ambiguous annotations. RESOPS will also be upgraded as a tool for mapping RNA editing sites on protein 3D structures in the future.
Materials and Methods
Collection of nucleotide sequences with conversion-type RNA editing in plant organelles
GenBank/EMBL/DDBJ (Cochrane et al. 2008, Benson et al. 2009, Sugawara et al. 2009) stores the nucleotide position numbers for RNA editing sites without a standardized description and, therefore, interpretation of the collection of RNA editing sites in nucleotide and amino acid sequences is not straightforward. Manual inspection, with the aid of in-house C programs, was performed to decipher the GenBank/EMBL/DDBJ database descriptions, specifically for plant organelle conversion-type RNA editing site descriptions. A whole character search was performed to find a string of characters that matched ‘RNA’ and ‘editing’ in the ‘/note’ field of ‘misc_feature’ lines for plant entries in the GenBank database release 172. The C program then extracted protein-coding regions with RNA editing sites, generated both edited and unedited cDNA sequences, and translated edited mRNAs into amino acid sequences.
Some of the entries contained an error in the nucleotide position number of the RNA editing site, or a discrepancy between the types of nucleotide described in the misc_ feature line and the corresponding nucleotide in the deposited nucleotide sequence. In these cases, manual correction was done based either on the literature or by communication with the depositors. In the GenBank database release 172, corrections were needed to AJ006146, BA000029, DQ645537, DQ984517, X07566, X69720, X80170, X92735, X96536, Y14434, Y14435 and Y17812. We could not correct all errors encountered, because we could not make contact with all depositors. The entries with errors were discarded. We started the error correction procedure about 3 years ago, and the depositors of AB254134, AY521591 and AY820131 have evidently made contact with GenBank to rectify the annotations; the annotations of these three entries are corrected in the latest version of GenBank.
Manual checks also included the curation of RNA editing information from the literature and a check for duplicated data. For RNA editing on rbcL transcripts from a number of different species, we copied the RNA editing site described in the table from the literature (Freyer et al. 1997) into the following entries: D14882, D43696, L11055, L11056 and L13485. Occasionally, the same gene and cDNA were sequenced by different groups and independently deposited with different IDs. These entries were stored as they were, because the editing sites may differ, even if the sequences were the same.
Multiple amino acid sequence alignments and protein 3D structures
Amino acid sequences were clustered based on sequence identity. When a cluster contained more than one sequence, a sequence in the cluster had at least one different sequence with identity no less than 25%. Representative sequences of each cluster were then used as a query to find homologous proteins in the PDB (Berman et al. 2003) with BLAST (Altschul et al. 1997). When the amino acid sequences with identity no less than 30% were found, we selected the largest structure in the PDB with the highest sequence identity for assigning structural properties to the amino acid sequences in the cluster. Multiple sequence alignments, including amino acid sequences of proteins from the PDB, were then performed for each cluster, and edited residues were located both in the alignment and in the protein 3D structure.
Identification of protein structural cores
A structural core was determined by identifying clusters of buried residues and peripheral residues as described previously (Yura and Go 2008). The solvent-accessible surface area of each residue was calculated (Shrake and Rupley 1973) and solvent-inaccessible residues were identified first. When carbon atoms from two different solvent-inaccessible residues were in contact (≤4.0 Å), then the pair of residues was defined as a cluster. In the next step, every carbon atom in residues with accessibilities to solvent molecules (Go and Miyazawa 1980) between 0 and 0.05 was selected, and if the atom was in contact (≤4.0 Å) with one of the carbon atoms in the cluster residues, then the residue not in the cluster was defined as peripheral. Both cluster and peripheral residues were defined as structural core residues.
Supplementary data
Supplementary data are available at PCP online.
Funding
Japan Society for the Promotion of Science KAKENHI [Grant-in-Aid for Scientific Research (B) No. 18370061 to M.G.]; University Education Internationalization Promotion Program of the Ministry of Education, Culture, Sports, Science, and Technology-Japan (to Ochanomizu University).
Acknowledgments
K.Y. and M.G. thank the late Professor Hans Kössel for introducing us to his RNA editing work in RuBisCO large subunit transcripts when he visited Nagoya University. K.Y. also thanks Mr. Kazuhiro Kobayashi, Ms. Kazuko Kaji and Ms. Atsuko Doi for gathering RNA editing data from the literature.
Supplementary Material
Glossary
Abbreviations
- 3D
three-dimensional
- PDB
Protein Data Bank.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nature Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Bock R, Kössel H, Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie. 2000;82:549–557. doi: 10.1016/s0300-9084(00)00610-6. [DOI] [PubMed] [Google Scholar]
- Bonnard G, Grienenberger JM. A gene proposed to encode a transmembrane domain of an ABC transporter is expressed in wheat mitochondria. Mol. Gen. Genet. 1995;246:91–99. doi: 10.1007/BF00290137. [DOI] [PubMed] [Google Scholar]
- Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, et al. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 2009;150:1260–1271. doi: 10.1104/pp.109.136812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A-L, Ramos-Vega M, Guevara-García A, Andrés C, Gutiérrez-Nava ML, Cantero A, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008;56:590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Cochrane G, Akhtar R, Aldebert P, Althorpe N, Baldwin A, Bates K, et al. Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2008;36:D5–D12. doi: 10.1093/nar/gkm1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello PS, Gray MW. RNA editing in plant mitochondria. Nature. 1989;341:662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. RNA sequence and the nature of the CuA-binding site in cytochrome c oxidase. FEBS Lett. 1990;268:5–7. doi: 10.1016/0014-5793(90)80958-l. [DOI] [PubMed] [Google Scholar]
- Creighton TE. Protein folding. Biochem. J. 1990;270:1–16. doi: 10.1042/bj2700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MP, Myers DS. Simple statistical models predict C-to-U edited sites in plant mitochondrial RNA. BMC Bioinformatics. 2004;5:132. doi: 10.1186/1471-2105-5-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, He T, Li Y. Prediction of C-to-U RNA editing sites in higher plant mitochondria using only nucleotide sequence features. Biochem. Biophys. Res. Commun. 2007;358:336–341. doi: 10.1016/j.bbrc.2007.04.130. [DOI] [PubMed] [Google Scholar]
- Du P, Jia L, Li, Y CURE-Chloroplast: a chloroplast C-to-U RNA editing predictor for seed plants. BMC Bioinformatics. 2009;10:135. doi: 10.1186/1471-2105-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer R, Kiefer-Meyer M-C, Kössel H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl Acad. Sci. USA. 1997;94:6285–6290. doi: 10.1073/pnas.94.12.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go M, Miyazawa S. Relationship between mutability, polarity and exteriority of amino acid residues in protein evolution. Int. J. Pept. Protein Res. 1980;15:211–224. doi: 10.1111/j.1399-3011.1980.tb02570.x. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Gray MW, Covello PS. RNA editing in plant mitochondria and chloroplasts. FASEB J. 1993;7:64–71. doi: 10.1096/fasebj.7.1.8422976. [DOI] [PubMed] [Google Scholar]
- Hiesel R, Combettes B, Brennicke A. Evidence for RNA editing in mitochondria of all major groups of land plants except the Bryophyta. Proc. Natl Acad. Sci. USA. 1994;91:629–633. doi: 10.1073/pnas.91.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch B, Maier RM, Appel K, Igloi GL, Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991;353:178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nature Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- Kim S-R, Yang J-I, Moon S, Ryu C-H, An K, Kim, K-M, et al. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria. Plant J. 2009;59:738–749. doi: 10.1111/j.1365-313X.2009.03909.x. [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- Kozaki A, Mayumi K, Sasaki Y. Thiol–disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase. J. Biol. Chem. 2001;276:39919–39925. doi: 10.1074/jbc.M103525200. [DOI] [PubMed] [Google Scholar]
- Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003;31:2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loladze VV, Ermolenko DN, Makhatadze GI. Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. J. Mol. Biol. 2002;320:343–357. doi: 10.1016/S0022-2836(02)00465-5. [DOI] [PubMed] [Google Scholar]
- Mower JP. PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phreaner CG, Williams MA, Mulligan RM. Incomplete editing of rps12 transcripts results in the synthesis of polymorphic polypeptides in plant mitochondria. Plant Cell. 1996;8:107–117. doi: 10.1105/tpc.8.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Robbins JC, Heller WP, Hanson MR. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA. 2009;15:1142–1153. doi: 10.1261/rna.1533909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Ohmori A, Iguchi H, Nagano Y. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem. 2001;276:3937–3940. doi: 10.1074/jbc.M008166200. [DOI] [PubMed] [Google Scholar]
- Shrake A, Rupley JA. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- Sugawara H, Ikeo K, Fukuchi S, Gojobori T, Tateno Y. DDBJ dealing with mass data produced by the second generation sequencer. Nucleic Acids Res. 2009;37:D16–D18. doi: 10.1093/nar/gkn724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Gopal S. Genetic algorithm learning as a robust approach to RNA editing site prediction. BMC Bioinformatics. 2006;7:145. doi: 10.1186/1471-2105-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SD, Backmann J, Prevost M, Steyaert J, Loris R. Hydrophobic core manipulations in ribonuclease T1. Biochemistry. 2001;40:10140–10149. doi: 10.1021/bi010565n. [DOI] [PubMed] [Google Scholar]
- Wakasugi T, Hirose T, Horihata M, Tsudzuki T, Kössel H, Sugiura M. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl Acad. Sci. USA. 1996;93:8766–8770. doi: 10.1073/pnas.93.16.8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K, Iinuma H, Masuzawa T, Ueda K. Extensive RNA editing of U to C in addition to C to U substitution in the rbcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants. Nucleic Acids Res. 1996;24:1008–1014. doi: 10.1093/nar/24.6.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q-B, Jiang Y, Chong K, Yang Z-N. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 2009;59:1011–1023. doi: 10.1111/j.1365-313X.2009.03930.x. [DOI] [PubMed] [Google Scholar]
- Yura K, Go M. Correlation between amino acid residues converted by RNA editing and functional residues in protein three-dimensional structures in plant organelles. BMC Plant Biol. 2008;8:79. doi: 10.1186/1471-2229-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura K, Miyata Y, Arikawa T, Higuchi M, Sugita M. Characteristics and prediction of RNA editing sites in transcripts of the moss Takakia lepidozioides chloroplast. DNA Res. 2008;15:309–321. doi: 10.1093/dnares/dsn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takanaka M. A DYW domain containing pentatricopeptide repeat protein is required for RNA editing at multiple sits in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–567. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin A-L, Delannoy E, Hammani K, et al. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 2009;58:82–96. doi: 10.1111/j.1365-313X.2008.03766.x. [DOI] [PubMed] [Google Scholar]
- Zito F, Kuras R, Choquet Y, Kössel H, Wollman FA. Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 1997;33:79–86. doi: 10.1023/a:1005734809834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.