Abstract
For systematic and genome-wide analyses of rice gene functions, we took advantage of the full-length cDNA overexpresser (FOX) gene-hunting system and generated >12 000 independent FOX-rice lines from >25 000 rice calli treated with the rice-FOX Agrobacterium library. We found two FOX-rice lines generating green calli on a callus-inducing medium containing 2,4-D, on which wild-type rice calli became ivory yellow. In both lines, OsGLK1 cDNA encoding a GARP transcription factor was ectopically overexpressed. Using rice expression-microarray and northern blot analyses, we found that a large number of nucleus-encoded genes involved in chloroplast functions were highly expressed and transcripts of plastid-encoded genes, psaA, psbA and rbcL, increased in the OsGLK1-FOX calli. Transmission electron microscopy showed the existence of differentiated chloroplasts with grana stacks in OsGLK1-FOX calli cells. However, in darkness, OsGLK1-FOX calli did not show a green color or develop grana stacks. Furthermore, we found developed chloroplasts in vascular bundle and bundle sheath cells of coleoptiles and leaves from OsGLK1-FOX seedlings. The OsGLK1-FOX calli exhibited high photosynthetic activity and were able to grow on sucrose-depleted media, indicating that developed chloroplasts in OsGLK1-FOX rice calli are functional and active. We also observed that the endogenous OsGLK1 mRNA level increased synchronously with the greening of wild-type calli after transfer to plantlet regeneration medium. These results strongly suggest that OsGLK1 regulates chloroplast development under the control of light and phytohormones, and that it is a key regulator of chloroplast development.
Keywords: Chloroplast development • FOX hunting system • GARP transcription factor • OsGLK1 • Oryza sativa • Rice
Introduction
Proplastids develop into mature organelles, such as chloroplasts, amyloplasts and chromoplasts, during the life cycle of land plants, and the mature organelles perform important functions in various plant vegetative and reproductive organs (Possingham 1980, Link 1991, Kobayashi 1991, López-Juez and Pyke 2005). Among the plastids, the chloroplast is one of the most essential organelles, since the growth of sessile plants largely depends on the chemical energy produced by photosynthesis in the chloroplast. In darkness, proplastids are converted into etioplasts, while etioplasts in leaves rapidly differentiate into chloroplasts upon exposure to light, and the leaves exhibit photosynthetic activity. This indicates that plastid development is regulated by a light-related factor(s). Wild-type rice calli grown on 2,4-D-containing media are usually ivory yellow, and proplastids do not develop into chloroplasts in these cells, even under illuminated conditions. However, when such calli are transferred to regeneration medium containing a low level of auxin and a high concentration of cytokinin, chloroplast development is induced and the calli gradually turn green. Therefore, in addition to light, chloroplast development may be regulated by an ingredient(s) such as phytohormone.
Such environmental and hormonal signals are thought to activate transcription factors, which positively regulate gene expression required for chloroplast development. Because components in the photosynthetic apparatus of the chloroplast are encoded in the nuclear and plastid genomes, such transcription factors may coordinate gene expression in both nuclei and plastids. Currently, only a few transcription factors have been described that positively regulate photosynthesis-related genes or plastid differentiation, e.g. HY5 (Oyama et al. 1997, Chattopadhyay et al. 1998, McCormac and Terry 2002), the HY5 homolog HYH (Holm et al. 2002) and HFR1/REP1/RSF1 (Fairchild et al. 2000, Soh et al. 2000, Spiegelman et al. 2000) from Arabidopsis. However, we have not yet clarified the overall mechanism(s) underlying chloroplast development induced by environmental and endogenous factors.
Gain-of-function gene-hunting systems are thought to be powerful tools to explore positively regulating factors involved in chloroplast development. The full-length cDNA overexpresser (FOX) gene-hunting system is a recently developed genome-wide gene screening procedure (Ichikawa et al. 2006, Nakamura et al. 2007, Fujita et al. 2007, Kondou et al. 2009). The Rice Full-Length cDNA Consortium (2003) collected rice full-length cDNAs (FL-cDNAs) and clustered them into 28 469 non-redundant clones. Subsequently, the clones were used to develop the FOX hunting system in rice for systematic functional analysis of rice genes and generated >12 000 FOX-rice lines (Nakamura et al. 2007, Hakata, Nakamura, Okada, Miyao, Kajikawa, Amano, Toki, Pang, Horikawa, Tsuchida-Mayama, Song, Igarashi, Kitamoto, Ichikawa, Matsui, Nagamura, Hirochika and Ichikawa. unpublished). All transgenic rice lines produced ivory-yellow calli, except two independent lines that produced pale-green calli in the presence of 2,4-D. In both exceptions, a FL-cDNA encoding OsGLK1 was integrated and highly expressed (Nakamura et al. 2007).
OsGLK1 is orthologous to maize Golden2-like 1 (ZmGLK1) gene (Rossini et al. 2001). ZmGLK1 is homologous to Golden2 (G2), and the maize g2 mutant shows pale-green leaf blades (Hall et al. 1998). GLK genes encode transcription factors that belong to the plant-specific GARP family (Riechmann et al. 2000, Fitter et al. 2002). Arabidopsis GLKs (AtGLK1/GPRI1 and AtGLK2/GPRI2) interact with proline-rich regions of G-box-binding bZIP factors, and both the N-terminal and the C-terminal regions of AtGLKs can transactivate transcription in Arabidopsis cells (Tamai et al. 2002). Double mutants of AtGLK1 and AtGLK2 are pale green in all photosynthetic tissues and show reduced granal thylakoids in chloroplasts (Fitter et al. 2002). Overexpression of either AtGLK1 or AtGLK2 rescued the pale green phenotype of the double mutant in a cell-autonomous manner (Waters et al. 2008). Yasumura et al. (2005) isolated a pair of GLK genes from a moss, Physcomitrella patens, and showed that they regulate chloroplast development in the moss. These findings indicate that transcription factors encoded by GLK genes positively regulate chloroplast development by a mechanism conserved widely in the plant kingdom.
In this study, overexpression of OsGLK1 promoted transcription of a set of nucleus-encoded genes related to chloroplast functions and plastid-encoded genes. In the green calli of OsGLK1-FOX rice, well-developed thylakoid membranes and grana stacks were observed. The overexpression of OsGLK1 also induced chloroplast development in vascular bundle (VB) and vascular bundle sheath (VBS) cells of young leaves. OsGLK1-FOX calli had much higher chlorophyll (Chl) content and photosynthetic activity than the yellowish-white calli of wild-type rice, and showed photoautotrophic growth. Based on these data, we suggest that GLKs are the main regulators of chloroplast development.
Results
Screening of green calli in FOX-rice lines
In a previous paper (Nakamura et al. 2007), we reported the production of ∼12 000 independent FOX-rice lines. To obtain these FOX-rice lines, primary calli derived from >25 000 rice seeds were treated with Agrobacterium containing the rice-FOX library, and the treated calli were selected on 2,4-D-containing selection (N6D) medium supplemented with hygromycin B (Hyg) at 30 mg l−1. During selection of Agrobacterium-treated calli, we noticed that two lines (AQ190 and BW244) exhibited a pale green color on N6D medium. Sequence analysis of cDNA-containing fragments obtained by genomic PCR demonstrated that an identical rice FL-cDNA (accession No. AK098909) was integrated into both these lines. Searches in the Rice Annotation Project Database (RAP-DB, http://rapdb.dna.affrc.go.jp/; Ohyanagi et al. 2006, Rice Annotation Project 2007) and the Knowledge-based Oryza Molecular biological Encyclopedia (KOME, http://cdna01.dna.affrc.go.jp/cDNA/; Rice Full-Length cDNA Consortium 2003) revealed that AK098909 encoded the OsGLK1 protein. We also obtained two additional lines (AI109 and AX290), in which AK098909 cDNA was integrated, although we could not observe greening of calli in these two lines during the screening of antibiotic-resistant primary calli.
Regenerated plants (T0 generation) from calli of the two independent lines (AI109 and AQ190) were grown in a greenhouse and their progeny seeds were obtained. Then, calli induced from the seeds of the two lines and a control line transformed with the empty pRiceFOX vector were induced and selected on N6D medium with 30 mg l−1 Hyg. All Hyg-resistant (HygR) calli derived from the seeds of both AI109 and AQ190 lines were a pale green color, while those from the control line were ivory yellow (Fig. 1A). Furthermore, there was good correlation between the level of greening, Chl content and OsGLK1 expression in these calli (Fig. 1A–C). Since the transcript levels of OsGLK1 were greatly enhanced in the calli from these two lines (Fig. 1C), we designated these lines as OsGLK1-FOX lines.
Fig. 1.
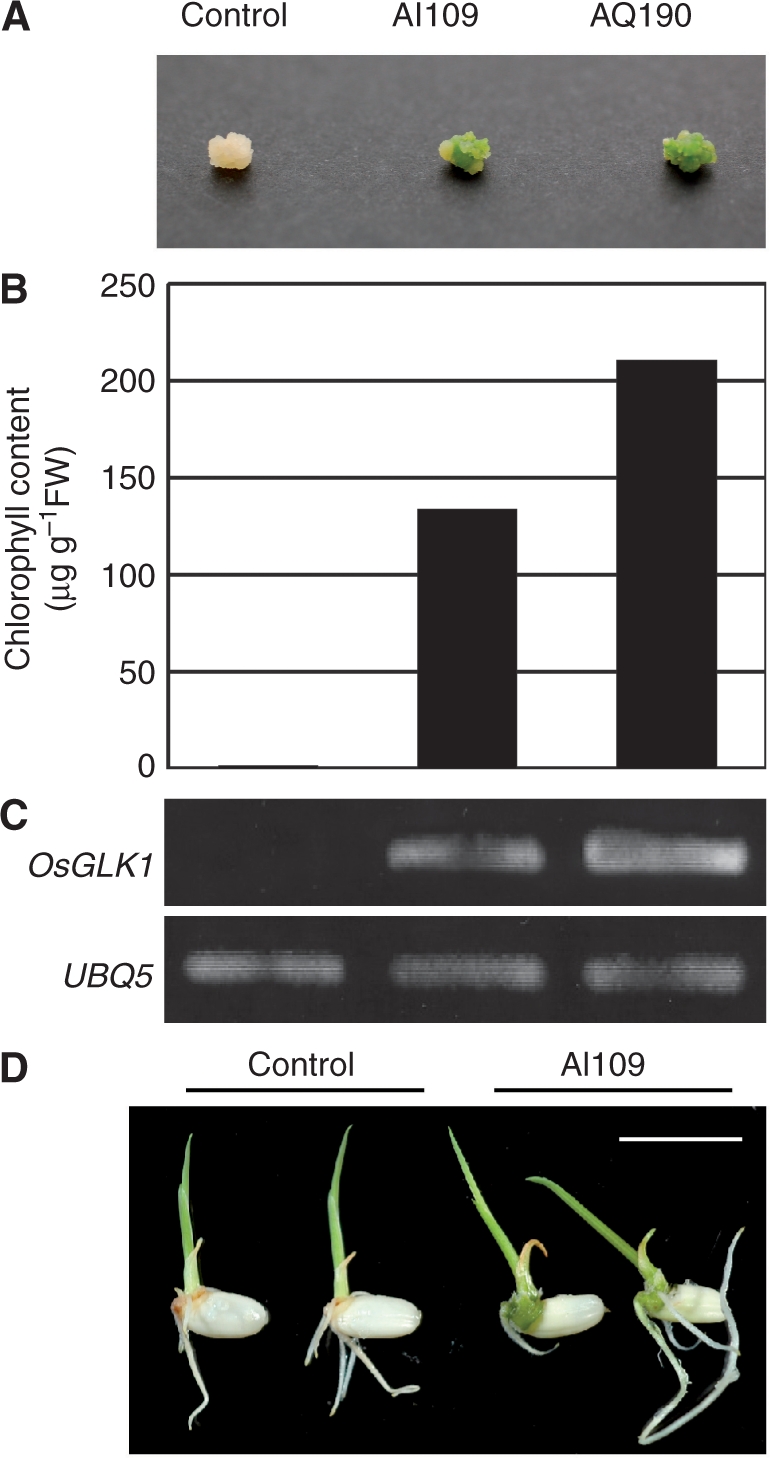
Phenotypes of OsGLK1-FOX rice. (A) Color of calli from a vector control (T1 generation) and OsGLK1-FOX lines, AI109 (T3) and AQ190 (T1). Progeny seeds from the transgenic lines were sown and grown on callus-induction medium (N6D) containing 2,4-D at 2 mg l−1 (Toki et al. 2006) for 6 weeks. (B) Chl content of the control and the OsGLK1-FOX calli shown in (A). (C) Semi-quantitative RT–PCR analysis of calli from the control (T1) and the OsGLK1-FOX lines as in (A). Upper panel shows the transcript levels of OsGLK1 cDNA. Lower panel represents those of UBQ5 used for loading adjustment. (D) Color of shoots from the control and the OsGLK1-FOX (AI109) lines. T1 seeds were sown and grown on hormone-free medium for 4 d. Scale bar = 1 cm.
When T1 seeds of the OsGLK1-FOX lines were grown on hormone-free medium with 30 mg l−1 Hyg for 4 d, imbibed embryos and germinated shoots turned dark green (Fig. 1D). The dark green color, however, did not continue to be significant when compared with the color of control plants as they grew (data not shown).
To confirm whether the greening phenotype of the four lines could be ascribed to overexpression of the GLK1 gene, an expression vector pRiceFOX-OsGLK1 was newly constructed by inserting OsGLK1 FL-cDNA into the pRiceFOX vector and then introduced into rice cells. Subsequently, calli treated with Agrobacterium were grown and selected on N6D medium with 30 mg l−1 Hyg. As expected, some portions of calli (31 out of 70 HygR blocks) showed a pale green color when treated with pRiceFOX-OsGLK1, whereas none of the 70 blocks of HygR calli treated with pRiceFOX exhibited a pale green color. These results confirmed that the green phenotype of the calli was the result of ectopic OsGLK1 overexpression. Transgenic plants harboring T-DNA from pRiceFOX-OsGLK1 were regenerated and designated as OsGLK1-FOX-RE.
Seeds from T1 progenies of OsGLK1-FOX-RE #1, #5, and #7 were sown and grown on N6D medium without Hyg and produced both green and ivory-yellow calli. Green calli were distinguishable from ivory-yellow calli within 5 weeks of sowing. Subsequently, both green and ivory-yellow calli were transferred to N6D medium with 30 mg l−1 Hyg. All green calli were resistant to Hyg, whereas ivory-yellow calli were not.
Expression of endogenous OsGLK1 is synchronized with greening of regenerating calli
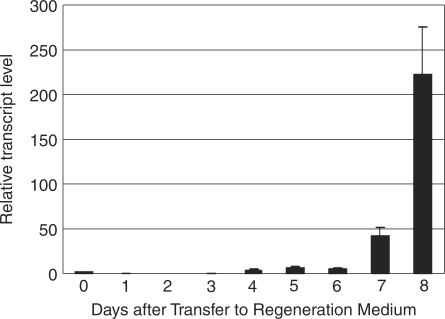
Wild-type rice calli normally show a green color 7–10 d after transfer to regeneration medium. To test whether the expression level of endogenous OsGLK1 changes during this period, we performed quantitative real-time RT–PCR analysis using OsGLK1-specific primers. The wild-type calli grown on N6D medium for 14 d were transferred to the regeneration medium, and total RNA was periodically extracted from them. Actin1 transcript was employed as an internal normalization standard. The OsGLK1 transcript level began to increase markedly 7 d after transfer to the regeneration medium (Fig. 2). Concurrent with the increase in OsGLK1 mRNA level, the calli began to exhibit a green color 7–8 d after transfer to the regeneration medium (data not shown).
Fig. 2.
Induction of OsGLK1 expression during regeneration of rice calli. Wild-type rice seeds were sown and cultured on N6D medium for 2 weeks and calli were transferred to regeneration medium. Total RNA was extracted on the indicated day. Quantitative real-time PCR was used to assess OsGLK1 mRNA levels. Data are the mean ± SD of three independent measurements. OsGLK1 mRNA levels were calculated relative to the abundance of Actin1 mRNA, with the OsGLK1 mRNA level of calli at day 0 set to 1.
Overexpression of OsGLK1 induces expression of genes related to chloroplast function
To confirm whether overexpression of OsGLK1 induces expression of chloroplast-related genes, we conducted an expression microarray analysis of ∼44 000 rice genes. Total RNA was extracted from calli derived from T1 seeds of the AI109 and control lines grown on N6D medium with 30 mg l−1 Hyg for 13 d at 30°C, and subjected to 44k oligo-DNA microarray analysis with four biological replicates.
Expression levels of 294 genes were upregulated >5-fold in OsGLK1-FOX rice calli compared with the control (Supplementary Table S1). Rice genes homologous to the previously reported Arabidopsis genes related to chloroplast function (Leister and Schneider 2003, Eckhardt et al. 2004, Lysenko 2007) were selected by BLAST search. Among them, genes with transcript levels that were markedly increased by overexpression of OsGLK1 are listed in Table 1. The transcript levels for a large number of genes involved in photosystem I (PSI), photosystem II (PSII), intersystemic electron transport, CO2 assimilation, Chl biosynthesis and the transcriptional machinery required for plastid-encoded gene expression were found to be markedly elevated in AI109 OsGLK1-FOX rice calli.
Table 1.
Chloroplast-related genes upregulated in green calli from OsGLK1-FOX rice compared with calli from control rice transformed with the empty pRiceFOX vector
| Accession No. | Gene product | Mean fold change (OsGLK1-FOX/control) |
|---|---|---|
| AK098909 | OsGLK1 | 193.26 |
| (i) PSI subunits | ||
| AK120372 | PSI reaction center subunit II (PSI-D) | 38.34 |
| AK120598 | PSI reaction center subunit IV (PSI-E) | 35.96 |
| AK060493 | PSI reaction center subunit III (PSI-F) | 16.52 |
| AK098847 | PSI reaction center subunit V (PSI-G) | 27.23 |
| AK060254 | PSI reaction center subunit VI (PSI-H) | 18.46 |
| AK058788 | PSI reaction center subunit X (PSI-K) | 122.64 |
| AK058207 | PSI reaction center subunit XI (PSI-L) | 38.56 |
| AK059037 | PSI reaction center subunit N (PSI-N) | 97.43 |
| Os05g0242400 | PSI reaction center subunit N (PSI-N) | 10.49 |
| AK058848 | Conserved hypothetical protein (PSI-O) | 36.00 |
| AK060904 | Chl a/b-binding protein family protein (Lhca1) | 80.21 |
| AK104283 | Lhca2 protein (Lhca2) | 18.31 |
| AK106085 | Chl a/b-binding protein type III (Lhca3) | 169.12 |
| AK060222 | Lhca4 protein (Lhca4) | 147.84 |
| AK066291 | Lhca5 protein (Lhca5) | 12.00 |
| AK067780 | Chl a/b-binding protein family protein (Lhca6) | 21.74 |
| (ii) PSII subunits | ||
| AK103937 | 33 kDa subunit of oxygen evolving system of PSII (PsbO) | 130.98 |
| AK065248 | 23 kDa polypeptide of PSII (PsbP) | 37.98 |
| CI426428 | PSII oxygen evolving complex protein (PsbQ) | 5.70 |
| AK121083 | PSII 10 kDa polypeptide (PsbR) | 31.68 |
| AK058284 | PSII subunit PsbS (PsbS) | 59.45 |
| AK119161 | PSII reaction center W protein (PsbW2) | 20.93 |
| AK105813 | PSII protein PsbX family protein (PsbX) | 105.94 |
| AK060602 | PSII core complex proteins psbY (PsbY) | 6.04 |
| AK061619 | Chl a/b-binding protein 2 (Cab2/Lhcb1) | 126.82 |
| AK060851 | Chl a/b-binding protein 1 (Cab1/Lhcb1) | 53.08 |
| AK058289 | Chl a/b-binding protein 1 (Cab1/Lhcb1) | 18.61 |
| AK066762 | PSII type II Chl a/b-binding protein (Lhcb2) | 255.52 |
| AK109399 | Type III Chl a/b-binding protein (Lhcb3) | 47.46 |
| AK119534 | Chl a/b-binding protein CP29 precursor (Lhcb4) | 63.38 |
| AK098872 | Chl a/b-binding protein family protein (Lhcb5) | 22.57 |
| AK066070 | Chl a/b-binding protein CP24 (Lhcb6) | 73.99 |
| (iii) Intersystemic electron transport | ||
| AK071634 | Rieske iron–sulfur protein | 6.30 |
| CI439180 | Rieske [2Fe–2S] region domain containing protein | 4.33 |
| AK120704 | Rieske iron–sulfur protein | 4.15 |
| AK067025 | Rieske iron–sulfur protein | 3.84 |
| AF093636 | Plastocyanin | 164.42 |
| AK120393 | Ferredoxin I | 108.76 |
| AK120232 | Ferredoxin I | 2.20 |
| AK059896 | Ferredoxin | 2.58 |
| D17790 | Ferredoxin-NADP reductase | 9.93 |
| AK106213 | Ferredoxin-NADP reductase | 126.56 |
| (iv) ATPase | ||
| AK072104 | ATP synthase γ-subunit | 5.12 |
| Os02g0750100 | H+-transporting ATP synthase δ-subunit | 2.11 |
| AK066019 | H+-transporting ATP synthase chain 9-like protein | 4.23 |
| (v) CO2 assimilation | ||
| AK068266 | Ribulose 1,5-bisphosphate carboxylase small subunit | 217.61 |
| AK121444 | Ribulose 1,5-bisphosphate carboxylase small subunit | 16.13 |
| AK068555 | Ribulose 1,5-bisphosphate carboxylase small subunit | 22.68 |
| AK099574 | Ribulose 1,5-bisphosphate carboxylase small subunit | 14.01 |
| Os12g0291200 | Ribulose 1,5-bisphosphate carboxylase small subunit | 10.58 |
| AK104332 | Ribulose bisphosphate carboxylase activase | 62.26 |
| AK066594 | 3-phosphoglycerate kinase | 2.85 |
| AK067755 | GAPDH | 168.48 |
| AK071685 | GAPDH | 140.74 |
| AK102013 | CP12 protein-like protein | 94.00 |
| AK103722 | CP12 | 50.81 |
| AK073758 | Fructose 1,6-bisphosphate aldolase | 77.34 |
| AK070516 | Fructose 1,6-bisphosphatase | 31.79 |
| AK065201 | Fructose 1,6-bisphosphatase | 16.53 |
| AK119209 | Sedoheptulose 1,7-bisphosphatase | 33.94 |
| AK066306 | Ribulose-phosphate 3-epimerase | 2.23 |
| AF529237 | Phosphoribulokinase | 107.57 |
| (vi) Plastid genome expression | ||
| AK065997 | Sigma factor SIG1 | 3.92 |
| AK067693 | Sigma factor SIG2B | 2.11 |
| AK105697 | Sigma factor SIG5 | 4.00 |
| AK068874 | Sigma factor SIG6 | 50.17 |
A data set for all the upregulated genes by overexpression of OsGLK1 cDNA with mean fold change values of >5.0 can be found in Supplementary Table SI on line. A complete set of microarray data was deposited to the Gene Expression Omnibus (GEO) repository under accession number GSE11451.
Interestingly, genes involved in the late steps of Chl biosynthesis pathway, CHLH, CHLM, CHL27, PORA, PORB and CHLP (Eckhardt et al. 2004), were highly expressed in the OsGLK1-FOX calli, while increases in the expression levels of genes involved in the early steps, which lead to protoporphyrin synthesis, were not obvious in the OsGLK1-FOX line (Table 2).
Table 2 .
Changes in transcript level of rice genes involved in Chl biosynthesis in the microarray analysis
| Accession No. | Gene product | Mean fold change | P value |
|---|---|---|---|
| AK099931 | Glutamyl-tRNA synthetase (GltX) | 1.07 | 0.452 |
| AK099393 | Glutamyl-tRNA reductase (HemA) | 1.13 | 0.451 |
| AK064826 | Glutamate-1-semialdehyde aminotransferase (HemL) | 1.60 | <0.001 |
| AK101836 | Delta-aminolevylinic acid dehydratase (Alad) | 1.44 | <0.001 |
| AK060914 | Porphobilinogen deaminase | 1.08 | 0.793 |
| AK107127 | Uroporphyrinogen III (HemD) | 1.28 | 0.003 |
| AK070859 | Uroporphyrinogen decarboxylase (HemE) | 1.88 | <0.001 |
| AK106203 | Uroporphyrinogen decarboxylase (HemE) | 1.15 | 0.298 |
| AK070391 | Coproporphyrinogen III oxidase (Lin2) | 1.31 | 0.084 |
| AK108365 | Protoporphyrinogen (PpxI) | 1.19 | 0.406 |
| AK060389 | Magnesium-chelatase subunit chlI (ChlI) | 1.62 | 0.007 |
| AK072463 | Magnesium-chelatase subunit chlD (ChlD) | 1.51 | 0.003 |
| AK067323 | Magnesium-chelatase subunit chlH (ChlH/Gun5) | 7.18 | <0.001 |
| AK059151 | Magnesium-protoporphyrin O-methyltransferase (ChlM) | 5.42 | <0.001 |
| AK069333 | Magnesium-protoporphyrin IX monomethyl ester aerobic oxidative cyclase (Chl27) | 26.81 | <0.001 |
| AK103940 | Protochlorophyllide reductase (PorA) | 219.73 | <0.001 |
| AK068143 | NADPH:protochlorophyllide oxidoreductase (PorB) | 24.88 | <0.001 |
| AK061968 | Geranylgeranyl reductase (ChlP) | 5.10 | <0.001 |
| AK068855 | Chl synthase (ChlG) | 1.12 | 0.107 |
| AF284781 | Chlorophyllide a oxygenase 1 (Cao1) | 1.92 | 0.002 |
| AK063367 | Chlorophyllide a oxygenase 2 (Cao2) | 0.38 | 0.024 |
Results of differential expression of individual genes for Chl biosynthesis between the OsGLK1-FOX callus and the control callus are shown with mean fold changes and P values (t-test).
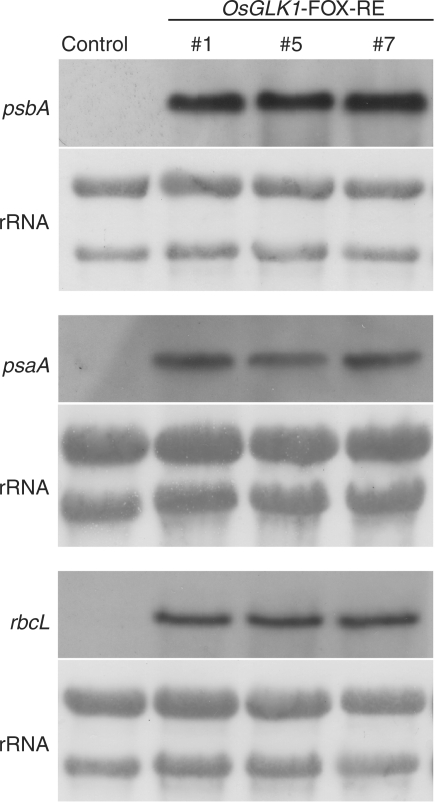
The transcript levels of sigma factors, OsSIG1, OsSIG2B, OsSIG5, and OsSIG6, were also enhanced in the OsGLK1-FOX calli. In various plant species, it is known that the photosystem genes encoded by the plastid genome are transcribed by plastid-encoded plastid RNA polymerase (PEP), which is composed of core enzyme subunits encoded by the plastid genome and a sigma factor encoded by the nuclear genome. Sigma factors play a role in donating specific binding of −35 to −10 promoter motifs to the core PEP complex (Weihe and Börner 1999, Allison 2000). In addition, it has been reported that OsSIG1 and OsSIG5 recognize the psaA operon and psbA promoter, respectively, in rice (Tozawa et al. 2007, Kubota et al. 2007). In Arabidopsis, transcription of psbA and psaA is known to be regulated by Sig5 (Tsunoyama et al. 2004), while that of psbA and rbcL is regulated by Sig6 (Ishizaki et al. 2005, Loschelder et al. 2006). Based on these findings, it was expected that an increase in the transcript levels of sigma factors would lead to an upregulation of the expression of plastid genome-encoded photosystem genes in the OsGLK1-FOX calli. Therefore, we analyzed the expression levels of three chloroplast-encoded genes (psbA, psaA and rbcL) by Northern blot hybridization. Predictably, the transcript levels of all three genes were elevated in calli of the OsGLK1-FOX lines (Fig. 3). In contrast to the increased expression levels of sigma factor genes, transcript levels for the core subunits of PEP (rpoA, rpoB, rpoC1 and rpoC2) in the OsGLK1-FOX calli were almost the same as those in wild-type calli (Supplementary Fig. S1).
Fig. 3.
Expression of three plastid-genome-encoded genes, psbA, psaA and rbcL, in the calli of OsGLK1-FOX and control lines. Total RNA was isolated from calli of the OsGLK1-FOX (RE #1, #5 and #7) and control lines grown on N6D medium with 30 mg l−1 hygromycin B (Hyg) for 7 d after sowing. Northern blot hybridization was performed as described in Materials and Methods.
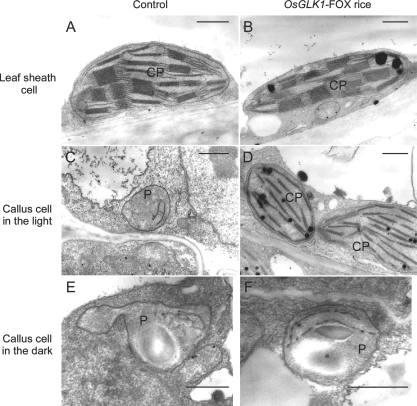
Ectopic development of chloroplasts in OsGLK1-FOX lines
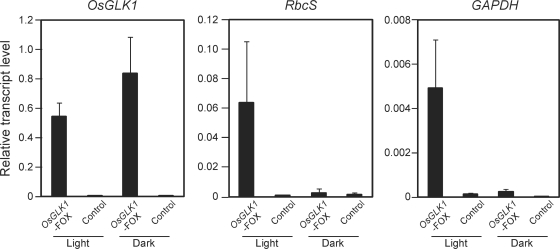
To determine whether chloroplasts developed in the green calli of the OsGLK1-FOX lines or not, intracellular structures of calli from OsGLK1-FOX lines were observed using a transmission electron microscope. In cells of OsGLK1-FOX calli grown under white light, well-developed thylakoid membranes and grana structures were observed in the plastids (Fig. 4D), which can be observed in chloroplasts of leaves (Fig. 4A, B), but not in the cells of the control callus (Fig. 4C). Furthermore, such structures were not observed in cells of OsGLK1-FOX calli when grown in the dark (Fig. 4E, F). Transcript levels of RbcS (accession No. AK068266) and a photosynthetic glyceraldehyde-3-phosphate dehydrogenase (GAPDH: AK067755) genes, which were markedly increased in light-grown OsGLK1-FOX calli (Supplementary Table S1), were not enhanced in dark-grown OsGLK1-FOX calli (Fig. 5). These observations indicate that overexpression of OsGLK1 ectopically induces light-dependent development of chloroplasts.
Fig. 4.
Chloroplast and proplastid ultrastructures in control rice (A, C, E) and OsGLK1-FOX rice (B, D, F). Leaf sheath cells of 4-day-old T1 seedlings grown on hormone-free medium with 30 mg l−1 Hyg (A, B). T1 calli grown on N6D + Hyg medium for 14 d under light (C, D) and dark (E, F) conditions. Scale bars = 1 μm. CP, chloroplast; P, proplastid.
Fig. 5.
Expression levels of OsGLK1-upregulated genes in OsGLK1-FOX rice calli under light and dark conditions. Total RNA was isolated from calli of the OsGLK1-FOX (AI109) and control lines grown on N6D medium with 30 mg l−1 Hyg for 14 d after sowing under continuous light and dark conditions, and subjected to real-time PCR analysis using primers that specifically amplify OsGLK1, RbcS (accession No. AK068266) and plastid-type GAPDH (AK067755) cDNAs. RNA levels were quantified and normalized to the levels of corresponding Actin1 mRNA, which was assigned a value of 1. Error bars indicate SD for three biological replicates.
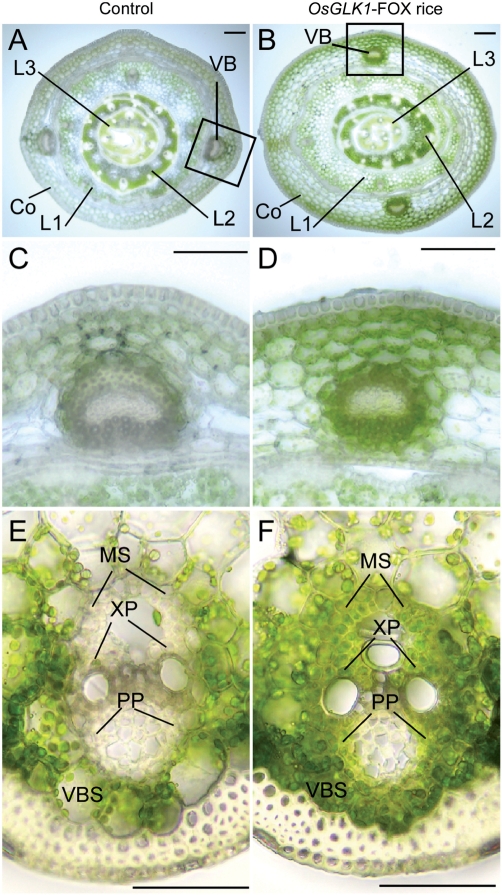
As described above, the green color in the germinated shoots from OsGLK1-FOX seeds was darker than that in control shoots during the early growth stage (Fig. 1D). We prepared cross-sections of leaves from AQ190 OsGLK1-FOX and control lines, and observed them using an optical microscope. The darker green color was observed in the coleoptiles and young leaves of 4-day-old OsGLK1-FOX rice seedlings (Fig. 6A, B). In addition, chloroplast development was prominent in the VBs of OsGLK1-FOX coleoptiles (Fig. 6D), while few chloroplasts were observed in the control coleoptiles (Fig. 6C). In 14-day-old OsGLK1-FOX rice plants, xylem and phloem parenchyma cells of VB, and VBS and mestome sheath cells in leaf sheaths exhibited a green color (Fig. 6F), while the corresponding cells in wild-type plants exhibited little green coloration (Fig. 6E). In addition, it is noteworthy that the OsGLK1-FOX rice accumulated significant levels of Chl in roots (Fig. 7). However, we did not detect obvious differences in the Chl content of the uppermost leaf blades between the OsGLK1-FOX rice and control rice at this age (data not shown). Furthermore, developed chloroplasts observed in the VB and VBS cells of 14-day-old OsGLK1-FOX rice plants could not be detected in those cells of 2-month-old plants (data not shown). These observations demonstrate that overexpression of the OsGLK1 gene induces the development of chloroplasts at least in coleoptiles and leaf sheaths of young plants both ectopically and orthotopically.
Fig. 6.
Cross-sections of leaves from control (A, C, E) and OsGLK1-FOX (B, D, F) rice. (A, B) Cross-sections of leaves of 4-day-old T1 seedlings grown on hormone-free medium with 30 mg l−1 Hyg. (C, D) Magnified views of VB in coleoptiles surrounded by rectangular boxes in (A) and (B), respectively. (E, F) VB and VBS cells of leaf sheaths of 14-day-old T1 shoots. Scale bars = 100 μm (A–D) and 50 μm (E, F). VB, vascular bundle; VBS, vascular bundle sheath; MS, mestome sheath; XP, xylem parenchyma cells; PP, phloem parenchyma cells; Co, coleoptile; L1, first leaf; L2, second leaf; L3, third leaf.
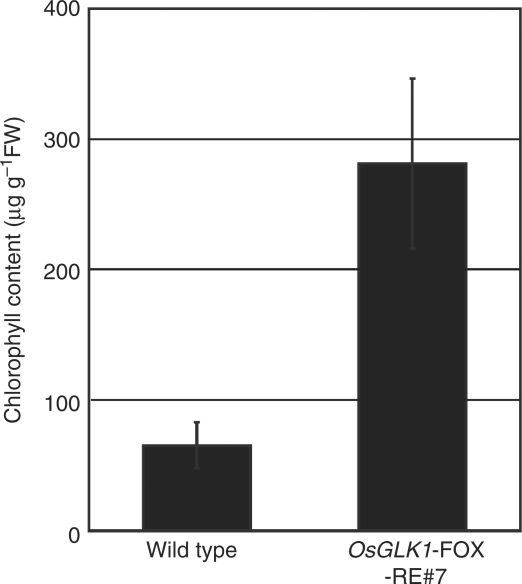
Fig. 7.
Chl content in roots of control and OsGLK1-FOX lines. Seedlings were grown on solid hormone-free medium for 15 d under continuous illumination, before roots were collected for determination of the Chl content. The data are mean ± standard error (n = 3).
Chl content and photosynthetic activity of OsGLK1-FOX calli
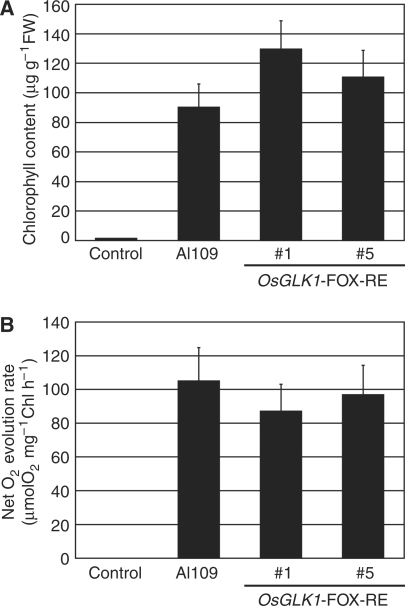
Substantial amounts of Chl were present in the green calli of OsGLK1-FOX lines, whereas the Chl content in the control calli was extremely low (Fig. 8A). Photosynthetic activity, measured as light-dependent O2 evolution, was detected in the OsGLK1-FOX calli, but not in the control calli (Fig. 8B).
Fig. 8.
Chl content and photosynthetic activity in calli of the OsGLK1-FOX and control lines. (A) Chl content of calli of wild-type, OsGLK1-FOX lines (AI109, RE #1 and #5) lines. The data are mean ± SD of at least three independent measurements. (B) The net O2 evolution rates under saturated light and saturated CO2 concentrations in calli of the control and OsGLK1-FOX lines. The data are mean ± SD of at least three independent experiments.
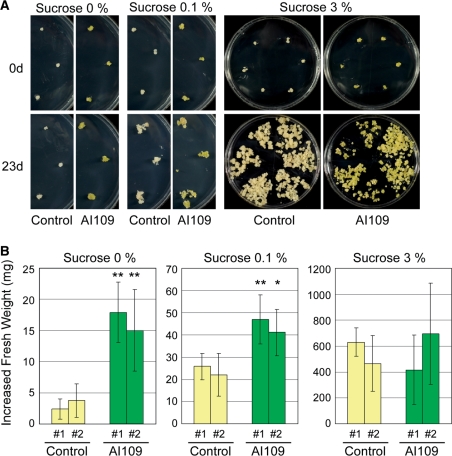
Although no obvious difference in the growth of cultured cells was observed between OsGLK1-FOX and control lines when cultivated on N6D agar medium containing a standard concentration of 3% sucrose, the OsGLK1-FOX calli grew better than the control calli on N6D medium containing only 0.1% sucrose (one-thirtieth of the standard). Moreover, the fresh weight of OsGLK1-FOX calli was significantly higher than that of the control calli, even on N6D medium without sucrose after 2 weeks of tissue culture (Fig. 9A, B). These results indicate that the OsGLK1-FOX cultured cells have the ability to proliferate photoautotrophically, and that their developed chloroplasts induced by the overexpression of OsGLK1 are both functional and active.
Fig. 9.
Autotrophic growth of OsGLK1-FOX calli. (A) Rice calli of the AI109 and control lines were grown on N6D medium with 30 mg l−1 Hyg for 2 weeks at 28°C and then transferred to fresh N6D medium with or without sucrose at concentrations indicated in the figure. Photographs are representative of the calli grown in fresh N6D medium for the indicated period. (B) Fresh weights of calli were measured at 0 and 23 d after transfer to fresh medium, and the difference between the two weights was calculated. The bars are the mean ± SD of six blocks of calli of indicated lines. Each asterisk indicates that the increase in fresh weight was significantly different from that of the Control #1 line (Student’s t-test; *P < 0.05, **P < 0.005).
Discussion
OsGLK1 positively regulates chloroplast development in rice
Calli of the OsGLK1-FOX line grown under white light on medium containing 2,4-D exhibited a pale green color and had developed chloroplasts, while the control calli did not (Figs. 1, 4). Similar results were obtained in VB and VBS cells in coleoptiles and leaf sheaths of the OsGLK1-FOX and control lines (Fig. 6). The OsGLK1-FOX calli had a high Chl content, high photosynthetic activity level (Fig. 8), and were able to grow on a sucrose-depleted medium (Fig. 9), suggesting that the developed chloroplasts were functional and active. Rossini et al. (2001) showed that OsGLK1 transcripts do not accumulate in roots, but accumulate in leaf sheaths and leaf blades, speculating that OsGLK1 is involved in photosynthetic development. Consistent with their speculation, we showed that the expression of OsGLK1 was synchronized with greening of calli (Fig. 2). Considering that chloroplast development was severely impaired in Arabidopsis double mutants of Atglk1 and Atglk2 and in the moss Physcomitrella patens double mutants of Ppglk1 and Ppglk2 (Fitter et al. 2002, Yasumura et al. 2005), we strongly suggest that GLKs are positive regulators of chloroplast development in the plant kingdom.
OsGLK1 upregulates plastid-encoded genes possibly through enhanced expression of nucleus-encoded plastid genes
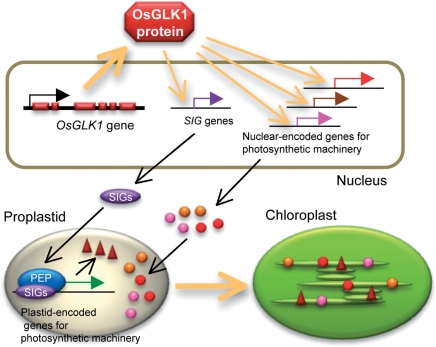
Microarray analysis showed that transcript levels of a large number of genes related to chloroplast functions were elevated in calli from the OsGLK1-FOX line compared with those from the control (Tables 1, 2 and Supplementary Table S1). It is notable that the expression levels of genes encoding sigma factors, which regulate the expression of plastid-encoded genes, were higher in the microarray data (Table 1). In addition, Northern blot analysis revealed that the transcript levels of three plastid-encoded genes, psaA, psbA and rbcL, were also higher in OsGLK1-FOX calli than in control calli (Fig. 3). These data indicate that OsGLK1 regulates chloroplast development by elevating transcript levels of nuclear-encoded genes for chloroplast functions, and also plastid-encoded genes for photosynthetic machinery by activating expression of nuclear-encoded sigma factor genes (summarized in Fig. 10).
Fig. 10.
A schematic model illustrating the role of OsGLK1 in expression of nuclear- and plastid-encoded genes for photosynthetic machinery and chloroplast development. OsGLK1 upregulates expression of sigma factor (SIG) genes and other nuclear-encoded genes for photosynthetic machinery. Then SIGs, together with PEP core subunits, induce expression of plastid-encoded genes for photosynthetic machinery. Consequently, proplastids develop into chloroplasts and acquire photosynthetic function.
In particular, overexpression of OsGLK1 caused a significant increase in the expression level of OsSIG6 among sigma factor genes (Table 1). In Arabidopsis, SIG6 plays a key role in chloroplast development in young seedlings, and transcript levels of PEP-dependent genes, including rbcL, psbA and psaA, are significantly decreased in 4-day-old seedlings of Arabidopsis sig6 mutant (Ishizaki et al. 2005, Loschelder et al. 2006). We observed a dark-green color in young shoots of OsGLK1-FOX rice (Fig. 1D). Moreover, PEP-dependent genes (rbcL, psbA and psaA) were upregulated in green calli from OsGLK1-FOX rice (Fig. 3). These observations may be mediated by the upregulation of OsSIG6 expression, though it is not known whether OsSIG6 functions in young seedlings similarly to Arabidopsis SIG6.
Contrary to the enhanced expression of sigma factor genes, expression levels of genes for PEP core subunits were not increased in the OsGLK1-FOX calli (Supplementary Fig. S1), indicating that transcription of the PEP core subunit genes is independent of OsGLK1. The result further suggested that the amounts of PEP core subunits in rice calli are sufficient to generate photosynthetically active plastids (Fig. 8B) in combination with OsGLK1 overexpression, and that transcript levels of the PEP-dependent genes are largely affected by the activity of sigma factors. However, comparing the grana stacks in plastids between leaf-sheath cells (Fig. 4A, C) and light-grown OsGLK1-FOX cultured cells (Fig. 4D), chloroplasts in the OsGLK1-FOX green calli were not fully developed. Accordingly, it is likely that the amounts of PEP core subunits in the plastids are still insufficient to induce fully developed chloroplasts. Expression of rpoB, encoding a PEP core subunit, is shown to be under the control of a nuclear-encoded plastid RNA polymerase (NEP) (Silhavy and Maliga 1998, Liere and Maliga, 1999, Shiina et al. 2005). In our microarray analysis, the expression level of OsRpoTp gene (accession No. AK069977) encoding an NEP enzyme (Kusumi et al. 2004) was not enhanced by the overexpression of OsGLK1 (data not shown). This inability of OsGLK1 to enhance the NEP transcription system could be an explanation of why the chloroplasts were not fully developed in the OsGLK1-FOX calli. Our findings also implied a role of transcription factor(s) other than OsGLK1 in the accumulation of OsRpoTp transcripts.
Upregulation by OsGLK1 of genes encoding the late steps of Chl biosynthesis
We found that calli from OsGLK1-FOX lines synthesized substantial levels of Chl (Fig. 8A). In plants and algae, it is widely accepted that the rate-limiting step in the Chl biosynthesis pathway is the initial step, i.e. the synthesis of 5-aminolevulinic acid (ALA) (Papenbrock and Grimm 2001, Tanaka and Tanaka 2007). ALA is synthesized from glutamate in three steps of catalysis, by glutamyl-tRNA synthetase (GltX), glutamyl-tRNA reductase (HemA) and glutamate-1-semialdehyde aminotransferase (HemL). It has been reported that transcription of HEMA1 is upregulated by light during de-etiolation in Arabidopsis (McCormac et al. 2001). However, the expression levels of rice GLTX (accession No. AK099931) and HEMA (AK099393) in greening OsGLK1-FOX calli were almost the same as those in wild-type calli (mean difference 1.13:1 and 1.07:1, respectively), while the expression level of HEML (AK064826) was only slightly higher (mean difference 1.60:1) in greening OsGLK1-FOX calli (Table 2). In addition, the expression levels of genes related to the late steps of Chl biosynthesis, such as the reduction of protochlorophyllide to chlorophyllide, were significantly elevated in OsGLK1-FOX rice (Table 2). These results suggest that expression of genes involved in the early steps of Chl biosynthesis are regulated by factor(s) other than OsGLK1, whereas the genes involved in the late steps of Chl biosynthesis are under the control of OsGLK1.
Possible involvement of GLK1 in the defense response
Savitch et al. (2007) showed a subset of Arabidopsis genes that were highly upregulated by overexpression of AtGLK1, and designated them the ‘GLK1 regulon’. However, almost all known chloroplast-related genes were not included in the GLK1 regulon. The AtGLK1 gene was expressed at a low level in leaves of wild-type Arabidopsis (Savitch et al. 2007). We assume that the level of endogenous AtGLK1 expression was sufficient for the induction of the expression of chloroplast-related genes, and further expression of AtGLK1 did not affect their expression.
Savitch et al. (2007) also reported that overexpression of AtGLK1 upregulated the expression of a subset of genes encoding disease defense-related genes and conferred resistance against Fusarium graminearum, a broad host pathogen. The expression of only one rice homolog among them, which encodes isochorismate synthase (ICS), a key enzyme in salicylic acid biosynthesis and required for systemic acquired resistance (SAR) (Wildermuth et al. 2001), was markedly upregulated by the overexpression of OsGLK1 in our microarray analysis (mean fold change = 24.7, Supplementary Table S1), while others were not (data not shown). Meanwhile, despite the upregulation of the ICS-homolog gene, no transcript accumulations of PR-1a or PR-1b, both of which are molecular markers of SAR (Agrawal et al. 2000a, Agrawal et al. 2000b), were observed. Regardless, we cannot exclude the possibility that OsGLK1 regulates gene expression of disease defense-related genes in leaves or other rice organs because we only used calli during our microarray analysis. It would be interesting to investigate whether OsGLK1-FOX rice is resistant to pathogen invasion, as observed in AtGLK-overexpressing Arabidopsis plants.
Overlapped functions of OsGLK1 and OsGLK2 in plastid differentiation
Although we have shown that ectopic overexpression of OsGLK1 can induce chloroplast development in calli as well as in VB and VBS cells in young shoots, it remains unclear where and when the native OsGLK1 acts in the rice plant. Our current suggestion is that OsGLK1 facilitates chloroplast development during the early stages of shoot growth in a light-dependent manner because shoots and embryos were dark green in seedlings of the OsGLK1-FOX lines and reverted to a normal color as they grew. This suggestion is supported by the observation that the transcript level of OsGLK1 is higher in leaves of young seedlings than in mature leaves (Rossini et al. 2001).
As mentioned above, Fitter et al. (2002) showed that Atglk1 Atglk2 double mutant plants were pale green throughout development and had reduced granal thylakoids in chloroplasts, whereas no phenotype specific to Atglk1 single mutant plants was observed, suggesting that AtGLK1 and AtGLK2 are functionally redundant. Furthermore, overexpression of either AtGLK1 or AtGLK2 suppressed various phenotypes of Atglk1 Atglk2 double mutants, such as pale green leaves and siliques, low levels of LHCB6 transcript and early flowering under long-day conditions, confirming that AtGLK1 and AtGLK2 are functionally equivalent (Waters et al. 2008). Rice also has two GLK genes, OsGLK1 and OsGLK2, and these OsGLKs show overlapping expression patterns, suggesting that the roles of OsGLK1 and OsGLK2 may be similar (Rossini et al. 2001). Consistent with these results, we observed that calli of OsGLK2-FOX lines show a green color on N6D medium, and OsGLK2 exhibits similar expression patterns during regeneration of the wild-type callus (data not shown). To assess the roles of OsGLKs further, we are producing single and double knockout/knockdown lines of OsGLK genes.
Post-transcriptional regulation of OsGLK1 function under different light conditions
The transcript levels of ZmG2, ZmGLK1, OsGLK1, OsGLK2, AtGLK1 and AtGLK2 are regulated by light (Rossini et al. 2001, Fitter et al. 2002), while the transcript levels of Brassica GLK1 and GLK2 are induced by cold stress (Savitch et al. 2005). In rice, development of membrane structures in chloroplasts of OsGLK1-FOX calli was not observed in the dark (Fig. 4), even though OsGLK1 was highly expressed. In addition, expression of a subset of chloroplast-related genes was not markedly affected by overexpression of OsGLK1 in darkness (Fig. 5). These results strongly suggest that the function of OsGLK1 might be post-transcriptionally regulated by light.
The mechanism by which light enables OsGLK1 to function is unclear; however, we propose the following hypotheses: (i) signaling from light activates OsGLK1 by modifications such as phosphorylation; (ii) OsGLK1 requires a light-regulated accompanying factor(s); (iii) in darkness, the function of OsGLK1 is downregulated by signals, e.g. COP1 signals like HY5, LAF1 and HFR in Arabidopsis (Osterlund et al. 2000, Seo et al. 2003, Jang et al. 2005); (iv) the function of GLK1 is suppressed by a factor(s) that is/are downregulated by light (e.g. phytochrome interacting factors; Castillon et al. 2007). To investigate these hypotheses, we are producing an anti-OsGLK1 antibody and a yeast two-hybrid system to analyze factors that accompany OsGLK1.
Niwa et al. (2006) produced Arabidopsis green callus by overexpressing CES101, which encodes a receptor-like kinase. Furthermore, AtGLK1 may possibly regulate or be regulated by CES101 in Arabidopsis. Although we did not find rice cDNA significantly similar to CES101 in our BLAST search, it would be interesting to investigate whether OsGLK1 and OsCES101 facilitate the same signaling pathway in the induction of chloroplast development.
OsGLK1 upregulates a set of genes for chloroplast function similarly to AtGLK1
During preparation of this paper, Waters et al. (2009) reported that GLKs coregulate and synchronize expression of a suite of nuclear photosynthetic genes in Arabidopsis. AtGLK1- and AtGLK2-regulated genes in their microarray data are similar to the OsGLK1-regulated genes in our microarray data, suggesting the existence of common roles of GLKs in chloroplast development between these two distantly related species. Furthermore, they showed that GLKs directly bind to promoters of several photosynthesis genes, implying that GLKs directly upregulate expression of a set of photosynthesis-related genes. It is likely that OsGLKs also elevate transcript levels of chloroplast-related genes by binding directly to their promoters. However, there are differences between the results in these two studies. In Arabidopsis, the transcript levels of RbcS1 (a nucleus-encoded small subunit of RuBisCO) were not affected by GLK overexpression or glk1 glk2 mutants, whereas expression of nucleus-encoded RbcS genes were highly elevated by OsGLK1 overexpression (Table 1). GLK-upregulation profiles of the Chl biosynthesis pathway were similar, with the exception that GLK1 overexpression markedly promoted the transcript level of CAO, encoding the chlorophyllide a oxygenase, with a 13.6-fold increase in Arabidopsis (Waters et al. 2009). OsGLK1 overexpression, however, promoted expression of CAO1, transcription of which is positively regulated by light (Lee et al. 2005), only by 1.92-fold. Interestingly, expression of CAO2, the second CAO gene in rice whose expression is repressed under light and higher in darkness (Lee et al. 2005), was downregulated by OsGLK1 overexpression (Table 2).
Among the progenies of our OsGLK1-FOX lines, we are currently selecting lines with a single copy of the transgene insertion per haploid genome and homozygous for the insertion. We also have been producing transgenic rice plants bearing the antisense OsGLK1 construct and those with the chimeric OsGLK1 repressor (Hiratsu et al. 2003) to further characterize the unique roles of rice GLK transcription factors in chloroplast differentiation. We expect that these lines may contribute not only to the basic aspects (e.g. uncovering mechanisms of chloroplast development), but also to applied aspects (e.g. genetic engineering for crop improvement), since rice is one of the most important crops and is recognized as the best model plant among the monocot crops (Khush 2005).
During the preparation of this paper, Kakizaki et al. (2009) reported that GENOMES UNCOUPLED 1 (GUN1), a chloroplastic pentatricopeptide repeat protein, downregulates expression of photosynthesis-related nuclear genes through suppression of AtGLK1 expression. Then, the authors proposed that AtGLK1 acts as a positive regulator not only for the expression of photosynthesis-related nuclear genes, but in a plastid-to-nucleus signaling pathway that coordinates plastid protein import and nuclear photosynthetic gene expression. Since a putative GUN1 ortholog has been found in rice (Koussevitzky et al. 2007), it may be interesting to know whether the provisional GUN1 could play a similar role in plastid-to-nucleus signaling by regulating OsGLK1 gene expression in rice.
Materials and Methods
Plant materials, growth conditions and rice transformation
Cultured cells, regenerated plants and seedlings of rice (Oryza sativa L., cultivar ‘Nipponbare’) were grown aseptically in growth chambers under constant white fluorescent light (70 μmol photons m−2 s−1) at 30°C. When transgenic rice plants were further grown in soil, the plants were transferred to growth chambers with irradiation of white light (∼300 μmol m−2 s−1) at a cycle of 14 h light (28°C)/10 h dark (25°C). Rice transformation was performed and growth media were prepared as previously described (Toki et al. 2006).
Plasmid construction
To prepare pRiceFOX-OsGLK1, a fragment of full-length OsGLK1 cDNA was excised with SfiI from pFLC1B-OsGLK1, the OsGLK1 FL-cDNA (AK098909) clone in λ-FLC-1-B vector (Rice Full-Length cDNA Consortium 2003), and then inserted into compatible SfiI sites of pRiceFOX (Nakamura et al. 2007) in the forward orientation.
The pRiceFOX and the pRiceFOX-OsGLK1 plasmids were transferred into Agrobacterium tumefaciens strain EHA105 (Hood et al. 1993) by electroporation.
RNA preparation and RT–PCR
An RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) was used according to the manufacturer’s instructions to extract total RNA from the leaf blades of each transgenic plant grown for 2 weeks after their transfer to soil. First-strand cDNAs were synthesized from each RNA preparation (1 μg/reaction) with an oligo(dT) primer using an ExScript RT reagent kit (Takara Bio Inc., Otsu, Japan) in a total volume of 20 μl, in accordance with the manufacturer’s instructions.
The specific sequences of the primers for amplifying cDNA for OsGLK1 (5′-AGCTGCGAGATTTCCTGCTC-3′ and 5′-ATAGCTGCGTCGATGCTCTC-3′), for RbcS (5′-CCC GGATACTATGACGGTAGG-3′ and 5′-AACGAAGGCATCA GGGTATG-3′), and for GAPDH (5′-GTGGCCAACATTAT CAGCAA-3′ and 5′-GGTCATGGTTCCCTTTACGA-3′) were used in semi-quantitative and quantitative RT–PCR. The primer pairs for amplifying Actin1 cDNA (AK100267; 5′-CTTCATAGGAATGGAAGCTGCGGGTA-3′ and 5′-TTCCT GTGCACAATGGATGG-3′) or UBQ5 cDNA (AK061988; 5′-ACCACTTCGACCGCCACTACT-3′ and 5′-ACGCCTAAGCC TGCTGGTT-3′) were used as loading controls for semi-quantitative RT–PCR. The Actin1 primer pair was also used as an internal control for normalization of quantitative RT–PCR (Jain et al. 2006).
Semi-quantitative RT–PCR was performed with 1 μl of cDNA template per 50 μl reaction with TaKaRa Ex Taq for 30 s at 94°C, followed by 28 cycles of 10 s at 98°C, 30 s at 60°C and 30 s at 72°C. Quantitative RT–PCR was performed with a Thermal Cycler Dice Real Time system (Takara Bio) using SYBR Premix Ex Taq (Takara Bio) and 1 μl of cDNA template per 25 μl reaction, in accordance with the manufacturer’s instructions. The threshold cycle (Ct) was auto-calculated by the analysis software that accompanied the system. The expression level, normalized to that of the endogenous control gene (Actin1), was calculated by relating the measured Ct to a standard curve obtained by diluting PCR-amplified DNA for which the exact DNA concentration was known.
Microarray analysis
T1 seeds of the AI109 and vector control lines were sown and grown for 13 d on callus-induction medium containing 2,4-D at 2 mg l−1. Total RNA was isolated from each callus using an RNeasy plant mini kit (Qiagen). The RNAs (400 ng aliquots) were labeled with a Low RNA Input Linear Amplification/Labeling Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. Aliquots of Cy5-labeled cRNA (1 μg each) of the AI109 sample and Cy3-labeled cRNA (1 μg each) of the vector control samples were used for hybridization in a Rice Gene Expression Microarray (4 × 44k; Agilent Technologies). Four biological replicate sample sets were analyzed. After hybridization, microarray slides were scanned (scanner model G2505B with G2565BA software; Agilent Technologies) and the data were analyzed using Feature Extraction software (version 9.1; Agilent Technologies) at the default settings. All microarray procedures and data analyses were performed according to the manufacturer’s manual.
Northern blot analysis
Northern blot hybridization was performed according to standard protocols (Sambrook et al. 1989). Total RNA was isolated from 7-day-old calli using a Total RNA Isolation Kit (Cartagen Molecular Systems Inc., San Carlos, CA, USA) according to the manufacturer’s instructions. Total RNA (2–8 μg) was fractionated by electrophoresis on a 1.2% (w/v) agarose gel with 0.6 M formaldehyde and blotted onto a nylon membrane (Hybond-N+; GE Healthcare UK Ltd., Little Chalfont, Buckinghamshire, UK). Equal loading of total RNA in all lanes was checked by methylene blue staining. Hybridization and detection were performed according to the standard protocol for digoxigenin (DIG) hybridization (Roche Diagnostics, Rotkreuz, Switzerland).
To generate gene-specific probes, the following primers were used to obtain PCR fragments of each gene from the genomic DNA of rice: psaA forward (5′-ATGATGATTCG TTCGCCGGA-3′) and reverse (5′-ACATGGATTTGGTGCC CCGC-3′), psbA forward (5′-ATGATCCCTACCTTATTGAC-3′) and reverse (5′-TTACCAAGGAACCATGCATA-3′), and rbcL forward (5′-AAGCTGGTGTTAAGGATTAT-3′) and reverse (5′-TTACCAAGGAACCATGCATA-3′). To use the PCR products as direct templates for in vitro transcription, the T7 polymerase recognition site (5′-TAATACGACTCACTA TAGGGCGA-3′) was added to the reverse primers at their 5′-termini. The conditions for PCR using ‘PrimeSTAR HS DNA Polymerase with GC Buffer’ (Takara Bio) were 35 cycles of 10 s at 98°C for denaturation, 15 s for annealing and 5 s at 72°C for elongation. Annealing temperatures for amplification of psaA, psbA and rbcL were 56°C, 52°C and 50°C, respectively. In vitro transcription reactions were carried out with a DIG RNA labeling kit (Roche) according to the manufacturer’s instructions.
Anatomical and ultrastructural studies
Microscopic observation was performed as described (Ueno 2004). Calli grown for 14 d on N6D medium in the dark, or under white light, were fixed in 3% (v/v) glutaraldehyde in 50 mM sodium phosphate buffer (pH 6.8) at room temperature for 3 h. Subsequently, they were washed with phosphate buffer and post-fixed in 2% (w/v) OsO4 in phosphate buffer. They were then dehydrated through an acetone series and embedded in Spurr’s resin (Spurr 1969). Ultrathin sections were taken from the prepared calli, stained with uranyl acetate and lead citrate, and observed under an electron microscope (Model HU7000; Hitachi, Tokyo, Japan).
Determination of the photosynthetic rate in calli
Photosynthetic O2 evolution of 1-week-old calli was measured with a Clark-type O2 electrode (Rank Brothers Ltd., Bottisham, Cambridge, UK) supported by 2 mM NaHCO3 under saturated light from a tungsten projector lamp. The net photosynthetic O2 evolution under illumination was defined as the difference between respiration rate in the dark and gross photosynthetic rate under saturated light.
Determination of Chl content in roots and calli
Roots and calli were homogenized in 80% acetone. The suspensions were held in the dark at 4°C for 1 week, followed by centrifugation at 3000 × g for 3 min. Chl concentrations in the supernatant were determined spectrophotometrically (Beckman DU 640 spectrophotometer; Beckman Coulter, Fullerton, CA, USA) using the equation of Arnon (1949).
Funding
The Ministry of Agriculture, Forestry and Fisheries of Japan (Green Technology Project EF-1004, Genomics for Agricultural Innovation GPN-0004).
Acknowledgments
We thank Shigeko Ando, Hiroko Abe and Mariko Kajikawa for their technical assistance. We also thank Drs Mitsue Miyao and Noritoshi Inagaki for their technical advice on the measurement of photosynthetic activity, the Rice Genome Resource Center of the National Institute of Agrobiological Sciences for providing the rice AK098909 FL-cDNA plasmid and allowing us to use the rice microarray analysis facility. In addition, we thank Ritsuko Motoyama for her technical support during the microarray experiment, and Dr Tetsuo Meshi for his useful comments on the function of GARP transcription factors.
Supplementary Material
Glossary
Abbreviations
- ALA
5-aminolevulinic acid
- CAO
chlorophyllide a oxygenase
- Chl
chlorophyll
- DIG
digoxigenin
- FL-cDNA
full-length cDNA
- FOX
full-length cDNA overexpresser
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GltX
glutamyl-tRNA synthetase
- HemA
glutamyl tRNA reductase
- HemL
glutamate-1-semialdehyde aminotransferase
- Hyg
hygromycin B
- ICS
isochorismate synthase
- NEP
nuclear-encoded plastid RNA polymerase
- PEP
plastid-encoded plastid RNA polymerase
- PSI
photosystem I
- PSII
photosystem II
- RT–PCR
reverse transcription–PCR
- SAM
shoot apical meristem
- SAR
systemic acquired resistance
- VB
vascular bundle
- VBS
vascular bundle sheath.
Footnotes
A complete set of microarray data was deposited to the Gene Expression Omnibus (GEO) repository under accession number GSE11451.
References
- Agrawal GK, Jwa NS, Rakwal R. A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphate inhibitors. Biochem. Biophys. Res. Commun. 2000a;274:157–165. doi: 10.1006/bbrc.2000.3114. [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Rakwal R, Jwa NS. Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signal. Biochem. Biophys. Res. Commun. 2000b;278:290–298. doi: 10.1006/bbrc.2000.3781. [DOI] [PubMed] [Google Scholar]
- Allison LA. The role of sigma factors in plastid transcription. Biochemie. 2000;82:537–548. doi: 10.1016/s0300-9084(00)00611-8. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt U, Grimm B, Hörtensteiner S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 2004;56:1–14. doi: 10.1007/s11103-004-2331-3. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31:713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Mizukado S, Fujita Y, Ichikawa T, Nakazawa M, Seki M, et al. Identification of stress-tolerance-related transcription-factor genes via mini-scale full-length cDNA over-expressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 2007;364:250–257. doi: 10.1016/j.bbrc.2007.09.124. [DOI] [PubMed] [Google Scholar]
- Hall LN, Rossini L, Cribb L, Langdale JA. GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell. 1998;10:925–936. doi: 10.1105/tpc.10.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Holm M, Ma L, Qu L, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, et al. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. HFR is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Fang-Sik C, Terauchi R, Inaba T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009 doi: 10.1104/pp.109.145987. in press [epub ahead of print] (DOI:10.1104/pp.109.145987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005;59:1–6. doi: 10.1007/s11103-005-2159-5. [DOI] [PubMed] [Google Scholar]
- Kobayashi H. Differentiation of amyloplasts and chromoplasts. In: Bogorad L, Vasil IK, editors. The Photosynthetic Apparatus: Molecular Biology and Operation, Volume 7B, Cell Culture and Somatic Cell Genetics of Plants. San Diego: Academic Press; 1991. pp. 395–415. [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009;57:883–894. doi: 10.1111/j.1365-313X.2008.03733.x. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Kubota Y, Miyao A, Hirochika H, Tozawa Y, Yasuda H, Tsunoyama Y, et al. Two novel nuclear genes, OsSIG5 and OsSIG6, encoding potential plastid sigma factors of RNA polymerase in rice: tissue-specific and light-responsive gene expression. Plant Cell Physiol. 2007;48:186–192. doi: 10.1093/pcp/pcl050. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Yara A, Mitsui N, Tozawa Y, Iba K. Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol. 2004;45:1194–1201. doi: 10.1093/pcp/pch133. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J-H, Yoo ES, Lee C-H, Hirochika H, An G. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005;57:805–818. doi: 10.1007/s11103-005-2066-9. [DOI] [PubMed] [Google Scholar]
- Leister D, Schneider A. From genes to photosynthesis in Arabidopsis thaliana. Int. Rev. Cytol. 2003;228:31–83. doi: 10.1016/s0074-7696(03)28002-5. [DOI] [PubMed] [Google Scholar]
- Liere K, Maliga P. In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J. 1999;18:249–257. doi: 10.1093/emboj/18.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G. Photoregulated development of chloroplasts. In: Bogorad L, Vasil IK, editors. The Photosynthetic Apparatus: Molecular Biology and Operation, Volume 7B, Cell Culture and Somatic Cell Genetics of Plants. San Diego: Academic Press; 1991. pp. 365–394. [Google Scholar]
- López-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. Int. J. Dev. Biol. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol. 2006;142:642–650. doi: 10.1104/pp.106.085878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko EA. Plant sigma factors and their role in plastid transcription. Plant Cell Rep. 2007;26:845–859. doi: 10.1007/s00299-007-0318-7. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Fischer A, Kumar AM, Söll D, Terry MJ. Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 2001;25:549–561. doi: 10.1046/j.1365-313x.2001.00986.x. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ. Light-signaling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 2002;32:549–559. doi: 10.1046/j.1365-313x.2002.01443.x. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, et al. A genome-wide gain-of-function analysis of rice genes using the FOX-hunting system. Plant Mol. Biol. 2007;65:357–371. doi: 10.1007/s11103-007-9243-y. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Goto S, Nakano T, Sakaiya M, Hirano T, Tsukaya H, et al. Arabidopsis mutants by activation tagging in which photosynthesis genes are expressed in dedifferentiated calli. Plant Cell Physiol. 2006;47:319–331. doi: 10.1093/pcp/pci242. [DOI] [PubMed] [Google Scholar]
- Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, et al. The rice annotation project database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–467. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock J, Grimm B. Regulatory network of tetrapyrrole biosynthesis – studies of intracellular signaling involved in metabolic and development control of plastids. Planta. 2001;213:667–681. doi: 10.1007/s004250100593. [DOI] [PubMed] [Google Scholar]
- Possingham JV. Plastid replication and development in the life cycle of higher plants. Annu. Rev. Plant Physiol. 1980;31:113–129. [Google Scholar]
- Rice Annotation Project. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rossini L, Cribb L, Martin DJ, Langdale JA. The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell. 2001;13:1231–1244. doi: 10.1105/tpc.13.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. 2nd edn. [Google Scholar]
- Savitch LV, Allard G, Seki M, Robert LS, Tinker NA, Huner NPA, et al. The effect of overexpression of two Brassica CBF/ DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant Cell Physiol. 2005;46:1525–1539. doi: 10.1093/pcp/pci165. [DOI] [PubMed] [Google Scholar]
- Savitch LV, Subramaniam R, Allard GC, Singh J. The GLK1 ‘regulon’ encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochem. Biophys. Res. Commun. 2007;359:234–238. doi: 10.1016/j.bbrc.2007.05.084. [DOI] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;26:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Maliga P. Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet. 1998;33:340–344. doi: 10.1007/s002940050345. [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2074. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman JI, Mindrinos MN, Fankhauser C, Richards D, Lutes J, Chory J, et al. Cloning of the Arabidopsis RSF1 gene by using a mapping strategy based on high-density DNA arrays and denaturing high-performance liquid chromatography. Plant Cell. 2000;12:2485–2498. doi: 10.1105/tpc.12.12.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. A low viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Tamai H, Iwabuchi M, Meshi T. Arabidopsis GARP transcriptional activators interact with the Pro-rich activation domain shared by G-box-binding bZIP factors. Plant Cell Physiol. 2002;43:99–107. doi: 10.1093/pcp/pcf011. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Tanaka R. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Tozawa Y, Teraishi M, Sasaki T, Sonoike K, Nishiyama Y, Itaya M, et al. The plastid sigma factor SIG1 maintains photosystem I activity via regulated expression of the psaA operon in rice chloroplasts. Plant J. 2007;52:124–132. doi: 10.1111/j.1365-313X.2007.03216.x. [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba, G, et al. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl Acad. Sci. USA. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno O. Environmental regulation of photosynthetic metabolism in the amphibious sedge Eleocharis baldwinii and comparisons with related species. Plant Cell Environ. 2004;27:627–639. [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008;56:432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Börner T. Transcription and the architecture of promoters in chloroplasts. Trends Plant Sci. 1999;4:169–170. doi: 10.1016/s1360-1385(99)01407-7. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–571. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell. 2005;17:1894–1897. doi: 10.1105/tpc.105.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.