Abstract
Foxl2 is a forkhead transcription factor required for ovary development and ovarian follicle maturation. In this report, we identified and characterized a functional relationship between Foxl2 expression and estrogen receptor (ER)-α signaling. We show that Foxl2 has no effect on classical ERα-mediated transcription, which occurs through canonical estrogen response elements. However, Foxl2 suppresses ERα signaling through nonclassical tethered transcriptional pathways. Specifically, the selective ER modulator tamoxifen stimulates activator protein-1 (AP1)-dependent transcription via the ERα, and this enhancement is blocked by Foxl2. Two lines of evidence suggest that Foxl2 suppression is mediated by physical interactions with ERα rather than direct action at AP1 binding sites. First, ERα is coimmunoprecipitated with Foxl2. Second, activation of a upstream activating sequence (UAS) reporter by Gal4-cJun in the presence of ERα and tamoxifen was blocked by Foxl2, demonstrating suppression in the absence of an AP1 site. Cyclooxygenase-2 (COX2), which is required for ovulation, was identified through expression profiling as a candidate physiological target for nonclassical ERα signaling and thus modulation by ERα/Foxl2 interactions. This possibility was confirmed by two sets of experiments. COX2 protein levels were induced by ERα in the presence of tamoxifen, and protein expression was suppressed by Foxl2. In addition, ERα stimulation of the COX2 promoter was repressed by Foxl2. We conclude that ERα and Foxl2 interact and that Foxl2 selectively suppresses ERα-mediated transcription of AP1-regulated genes. These data provide a potential point of convergence for ERα and Foxl2 to regulate ovarian development and function.
The forkhead transcription factor, Foxl2, which is required for ovary development, interacts with estrogen receptor α (ERα), and suppresses ERα signaling through tethered transcription pathways.
Estrogen plays an important role in the development and differentiation of the reproductive system and in the function of a number of adult tissues (1). Under most circumstances, the physiological effects of estrogen are mediated by the estrogen receptors (ERs), ERα and ERβ (2,3). ERα and ERβ have different biological functions, as indicated by their specific expression patterns, different target genes, and the distinct phenotypes observed in ERα and ERβ knockout mice. In the reproductive system, ERβ knockout females have a relatively mild phenotype characterized by smaller ovaries, some arrested follicular development, and a reduced number of corpora lutea (4). By comparison, ERα knockout females are acyclic and infertile and possess hyperemic ovaries devoid of corpora lutea (1).
In the classical model of ER action, ligand-activated receptors bind to estrogen response elements (EREs), where they recruit transcriptional cofactors to activate or suppress estrogen-responsive genes. In addition, there are nonclassical pathways, which do not involve EREs. These include a rapid membrane-associated ER pathway (5), interactions of the ER with signaling molecules downstream of transmembrane receptors (e.g. epithelial growth factor receptor) (5), and a tethered pathway in which the ER interacts with other transcription factors bound to their response elements. In the latter category, ER has been shown to interact with activator protein-1 (AP1), SP1, and nuclear factor-κB to transactivate or suppress various genes (6), and it is likely that ER interacts with other transcription factors as well. An ERα DNA binding domain mutant that eliminates binding to EREs but retains tethered transcriptional activity has revealed physiologic actions mediated by nonclassical signaling. These include estrogen-mediated feedback on the female reproductive axis (7) and estrogen actions on the testis-associated ducts in males (8). However, the specific genes and transcription factors that mediate the nonclassical physiologic actions remain unclear.
Foxl2 is a member of the forkhead family of transcription factors. Its features include a winged helix domain important for DNA binding and a conserved polyalanine tract at the C terminus that is involved in transcriptional repression (9). Expression of murine Foxl2 has been reported in the embryonic eyelids, pituitary, and follicle cells of the ovary (10,11). Consistent with this expression pattern, Foxl2 knockout mice exhibit eyelid/craniofacial malformation in both sexes (11) and impaired ovarian development and function (12). Specifically the granulosa cells of Foxl2 homozygous mutant mice do not complete the squamous to cuboidal transition, leading to an absence of secondary follicles, oocyte atresia, and premature ovarian failure. Sterility in Foxl2 null mice is restricted to females.
Inactivating mutations in human FOXL2 cause blepharophimosis-ptosis-epicanthus inversus syndrome and affected patients exhibit eyelid malformations that can be observed with or without premature ovarian failure (type I or type II, respectively) (13). A FOXL2 missense point mutation, [402C–>G, (C134W)] is also present in 97% of adult-type granulosa cell tumors of the ovary, suggesting that it is capable of driving the pathogenesis of these tumors (14). Despite the importance of Foxl2 in ovarian development, few direct gene targets of Foxl2 are known. Those that have been identified include the glycoprotein hormone α-subunit, steroidogenic acute regulatory protein (15), GnRH receptor (16), and aromatase (17). Notably, these genes are all important in reproduction.
Foxl2 is expressed in phylogenetically distant vertebrate groups, including species that have distinct mechanisms of sex determination (18). It is thus considered to be a highly conserved regulator of ovarian development. In rainbow trout, FoxL2b is expressed during oocyte entry into meiosis. Notably, meiotic entry is a hallmark event that commits embryonic germ cells to the female pathway in mammals. Of further interest, the female expression pattern of trout Foxl2a is recapitulated by exposure of the male gonad to 17α-ethynylestradiol (19). In turtles, in which sex determination is temperature dependent, ERα expression emerges in parallel with Foxl2 as part of a group of genes in a core ovary-determining pathway (20). Based on these data, we hypothesized that Foxl2 might interact with one or more of the ERα signaling pathways. In the present study, we explored this hypothesis in mammalian cells.
Materials and Methods
Plasmids
A mouse Foxl2-HA expression vector was created by PCR amplifying the Foxl2 coding sequence using a 3′ primer that incorporated the hemagglutinin (HA) coding sequence. Primers were as follows: sense, 5′-ATG ATG GCC AGC TAC CCC-3′ and antisense, 5′-GAT CGC GGC CGC TCA AGC GTA GTC TGG GAC GTC GTA TGG GTA GAG ATC CAG ACG CGA GTG-3′ (HA coding sequence is underlined). The PCR product was subcloned into pcDNA3.0 (Invitrogen, Carlsbad, CA) and sequence verified using GenBank NM_012020 as a reference. The Gal4-DBD expression vector, Gal4-DBD/c-Jun expression vector, ERE2-TK109-Luc reporter, AP1-7-Luc reporter, −73Col-Luc reporter, and UAS-E1b-TATA-Luc reporter have been described previously (21). The Cox2-Luc reporter (pGS459; containing −459/+9 from the human Cox2 promoter) was provided by Dr. Lee-Ho Wang (University of Texas, Houston, TX).
Cell culture
293FT cells (ERα negative human embryonic kidney) and MDA-MB-231 cells (ERα negative human breast cancer; subclone 10A) were cultured at 37 C in a humidified atmosphere of 5% CO2 in DMEM/F12 supplemented with 10% fetal bovine serum. Four days before transfection, cells were changed to estrogen-depleted media, prepared without phenol red, and supplemented with sera extracted three times with dextran-coated charcoal.
Transfections and luciferase assays
Cells were transferred to 24-well plates in estrogen-depleted medium 1 d before transfection. 293FT cells were transfected with calcium phosphate and MDA-MB-231 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The ERE, AP1, −73 collagenase, cyclooxygenase-2 (COX2), and UAS-E1b-TATA-Luc reporter plasmids were transfected at a concentration of 500 ng/well. The ERα, mFoxl2, ERαAA, and Gal4-c-Jun expression vectors or empty vector pcDNA3.0 was transfected at a concentration of 50 ng/well. 17β-Estradiol and tamoxifen (Sigma-Aldrich, St. Louis, MO) were reconstituted in EtOH and diluted into media at final concentrations of 1 nm 17β-estradiol and 100 nm tamoxifen. Equal volumes of ethanol alone were added to control wells. Luciferase activity was determined 48 h after transfection using a Biotek Clarity luminometer (Winooski, VT).
Western blots
MDA-MB-231and 293FT cells were transfected with the indicated expression vectors and cultured for 2 d in estrogen-depleted medium. Nuclear and cytoplasmic extracts were prepared as described previously (22) or using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. Extracts were run on 10% sodium dodecyl sulfate-polyacrylamide gels and transferred onto Hybond membranes (Amersham Biosciences, Piscataway, NJ). Immunodetection was performed using rabbit polyclonal human COX-2 antibody H-62, mouse monoclonal human ERα antibody D-12 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal Foxl2 antibody PA1–802 (Affinity Bioreagents, Rockford, IL), and antirabbit or antimouse horseradish peroxidase-conjugated IgG. Primary antibodies were diluted 1:1000 in 1% nonfat milk containing 1× Tris-buffered saline and 0.1% Tween 20 and incubated for overnight at 4 C. Proteins were visualized using an ECL Plus kit (Amersham Biosciences) according to the manufacturer’s instructions.
Immunofluorescence
293FT cells were grown on poly-d-lysine-coated coverslips in six-well plates and transfected with 500 ng of each expression construct using Lipofectamine 2000 (Invitrogen), following the manufacturer’s recommendations. After 48 h, cells were treated with tamoxifen or vehicle for 1 h, washed in PBS, fixed 10 min with 4% paraformaldehyde, permeabilized 5 min with 0.1% Triton X-100, and blocked in 1.5% BSA. Cells transfected with ERα were incubated with mouse anti-ERα primary at 1:50 in blocking buffer, followed by goat antimouse-AlexaFluor 568 (Invitrogen) secondary at 1:500, whereas cells transfected with Foxl2 were incubated with rabbit anti-Foxl2 serum at 1:2000 and goat antirabbit-AlexaFluor 488 (Invitrogen) secondary at 1:500. Antibody incubations were each performed for 1 h at room temperature. All cells were also stained with 4′,6′-diamidino-2-phenylindole (DAPI).
Immunoprecipitation
293FT cells were mock transfected or transfected with Foxl2-HA and ERα overnight and then treated for 48 h with ligands. Extracts were prepared by suspending cells in Nonidet P-40 lysis buffer [50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40] containing minicomplete protease inhibitor mixture (Roche Applied Science, Indianapolis, IN). Protein concentrations were measured by the Bradford assay (Bio-Rad, Hercules, CA). Reactions were started by adding 10 μl of mouse preimmune serum. After incubation at 4 C for 30 min, 10 μl of mouse anti-HA (Cell Signaling, Danvers, MA) were added to 400 μg of whole-cell extract resuspended in 1× immunoprecipitation buffer (Sigma) to a total volume of 600 μl. Reactions were incubated at 4 C overnight on a rocking table, 30 μl of protein A-agarose was added to each tube, and tubes were again incubated overnight at 4 C. After incubation, samples were centrifuged at 12,000 × g for 30 sec at 4 C, washed six times with 600 μl of 1× immunoprecipitation buffer, and resuspended in 60 μl of 1× Laemmli buffer. For detection of ERα, samples were subjected to electrophoresis on 12% sodium dodecyl sulfate-polyacrylamide gels and transferred onto Hybond membranes. Blots were probed with mouse monoclonal ERα antibody D-12 (Santa Cruz) and antimouse horseradish peroxidase-conjugated IgG. Proteins were visualized using an ECL Plus kit according to the manufacturer’s instructions.
Statistical analysis
Individual transfection experiments were carried out in quadruplicate, and experiments were repeated two to four times. Data for each experiment were scaled to the mean of all values in that experiment before comparison because luminometer units are relative and vary between experiments. Differences between treatments were determined using ANOVA followed by Neumann-Keuls post hoc testing, with a threshold for statistical significance of P < 0.05.
Results
Foxl2 inhibits AP1-dependent, but not ERE-dependent, ERα activity
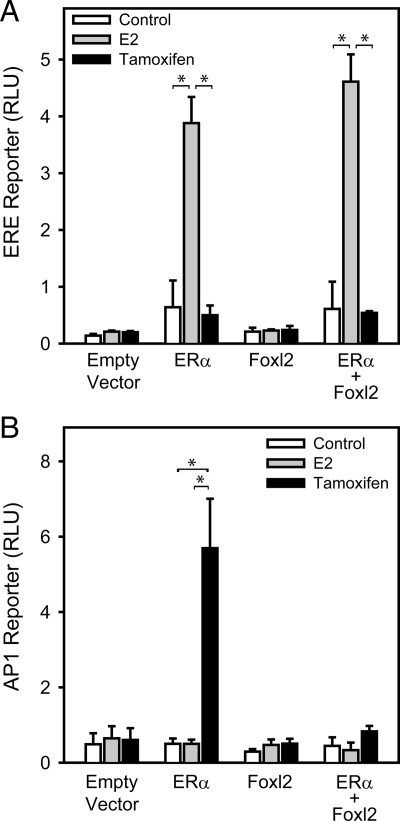
Reporter constructs were used to determine whether Foxl2 has activity on, and specificity for, different ERα signaling pathways. On an ERE-containing reporter (classical pathway), 17β-estradiol (E2; 1 nm) stimulated activity 6-fold relative to control in the presence of ERα [3.88 vs. 0.64 relative light units (RLU)], whereas tamoxifen (100 nm) was without effect (Fig. 1A). Foxl2 alone conveyed no response to either ligand, nor did it alter the ERα-mediated response to E2.
Figure 1.
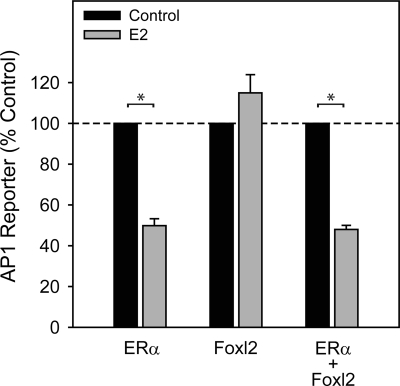
Effect of Foxl2 expression on ERE- and AP1-mediated transactivation. 293FT cells were transfected with human ERα and/or mouse Foxl2 expression vectors and treated for 24 h with vehicle control (ethanol), E2, or tamoxifen. A, Response of a reporter construct containing two copies of the vitellogenin ERE upstream of a 109-bp fragment of the thymidine kinase promoter. B, Response of a reporter construct containing multiple AP1 sites upstream of a basal promoter. Bars, mean ± sem of two to three experiments. *, P < 0.05.
On an AP1 reporter (nonclassical pathway), E2 was without effect in the presence of ERα (Fig. 1B), but tamoxifen stimulated reporter activity 11.3-fold. These data are consistent with the previous observation that tamoxifen acts as an ERα agonist at AP1 sites (21). Foxl2 again conveyed no response to either ligand. Unexpectedly, however, tamoxifen stimulation of the AP1 reporter through ERα was completely abolished by coexpression of Foxl2. Thus, Foxl2 selectively represses ERα stimulation of a nonclassical estrogen signaling pathway.
This effect is not a consequence of ERα degradation, which can occur in response to certain stimuli. Overexpression of Foxl2 did not alter levels of exogenously expressed ERα protein in 293FT cells or endogenous ERα protein in MCF-7 breast epithelial cells (not shown).
Foxl2 blocks AP1-mediated activity on the native collagenase promoter
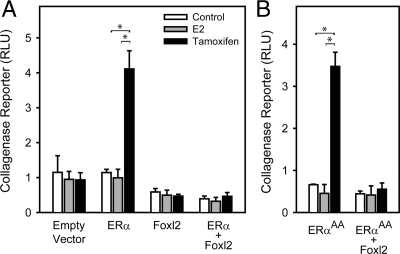
The proximal collagenase promoter (−73 to +63), which contains a single AP1 site, was used to evaluate whether Foxl2 can act via ERα and AP1 in a native promoter context. In the presence of ERα, tamoxifen induced collagenase promoter activity 3.6-fold (Fig. 2A). Expression of Foxl2 blocked tamoxifen induction, similar to the results on the synthetic AP1 reporter. E2 had no effect under these conditions.
Figure 2.
Effect of Foxl2 expression on non-ERE-dependent transactivation of the collagenase promoter. A, 293FT cells were transfected with human ERα and/or mouse Foxl2 expression vectors and treated for 24 h with vehicle control (ethanol), E2, or tamoxifen. Response of the −73Col-luciferase reporter is shown. B, Same as panel A, except the mERαAA contains two point mutations in the DNA-binding domain (E207A/G208A) that preclude binding to DNA. Bars, mean ± sem of two experiments. *, P < 0.05.
To confirm that Foxl2 acts on ERα that is not bound directly to DNA, we used a modified ERα (ERαAA) that contains two point mutations in the DNA binding domain (E207A/G208A). The mutations eliminate ERα binding to DNA but preserve ERα interactions with other proteins, including cJun (21). Tamoxifen stimulated collagenase reporter activity 5.3-fold via ERαAA (Fig. 2B), similar to the response with WT ERα. This activity was suppressed by Foxl2, confirming that Foxl2 acts on ERα that is signaling via the tethered nonclassical mechanism.
Foxl2 binds to ERα
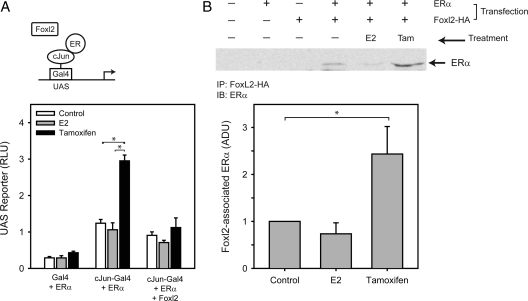
Transcriptional control by Foxl2 and ERα was examined further using a one-hybrid reporter system. In this assay, a cJun-Gal4 hybrid binds to the upstream activating sequence (UAS) recognition sequence to stimulate transcription, allowing cJun-mediated transactivation to be studied in the complete absence of AP1 or other transcription factor recognition sequences (Fig. 3A, schematic). The Gal4 DNA-binding protein alone was unresponsive to ligands despite the presence of ERα, as expected (Fig. 3A). By comparison, the cJun-Gal4 hybrid conferred tamoxifen responsiveness when coexpressed with ERα, indicating an association of ERα with cJun. Importantly, tamoxifen activation was blocked by coexpression of Foxl2, confirming that Foxl2 interacts with other proteins rather than with DNA response elements to suppress ERα-mediated transcriptional activation.
Figure 3.
Physical interactions between Foxl2, ERα, and cJun. A, 293 FT cells were transfected with Gal4, cJun-Gal4, human ERα, and/or mouse Foxl2 expression vectors and treated for 24 h with vehicle control (ethanol), E2, or tamoxifen. Activation of an upstream activating sequence (UAS)-E1b-TATA-luciferase reporter is shown. Bars, mean ± sem of two experiments. B, 293FT cells were transfected with human ERα and/or HA-mouse Foxl2 expression vectors and treated for 24 h with vehicle control, E2, or tamoxifen (Tam). Cells lysates were immunoprecipitated (IP) with anti-HA antibody and immunoblotting (IB) blot was performed for human ERα. The background in the representative image was lightened for clarity of illustration; raw images were used for quantitation. Bars, mean ± sem of four experiments. ADU, Arbitrary densitometric units; RLU, relative luciferase units. *, P < 0.05 (A and B).
Immunoprecipitation assays were performed to assess whether Foxl2 and ERα interact directly. HA-tagged Foxl2 was immunoprecipitated using an HA antibody, followed by Western blot with an antibody for ERα. ERα was not detected after HA immunoprecipitation in the absence of HA-Foxl2 (Fig. 3B, top panel, lanes 1 and 2) or absence of transfected ERα (lanes 1 and 3). When both ERα and HA-Foxl2 were transfected, ERα was detected in HA-Foxl2 immunoprecipitates, demonstrating that the two proteins are associated (lane 4). Levels of detected ERα were 2.4-fold higher after tamoxifen treatment [Fig. 3B, lane 6 (top panel) and graph (bottom panel)], suggesting that tamoxifen binding may induce conformational changes in ERα that enhance the interaction with Foxl2. In similar experiments, Foxl2 did not bind to cJun directly (not shown). Thus, Foxl2 appears to bind ERα, which is in turn tethered to cJun.
Foxl2 does not alter ERα cellular localization
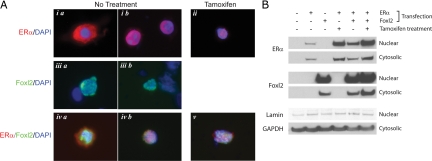
Other members of the Fox family are inactivated by phosphorylation, which causes them to be exported from the nucleus (23). The cellular localization of Foxl2 and ERα were examined to assess whether Foxl2 silencing of ERα reflects relocation of Foxl2 to the cytoplasm (Fig. 4). Immunohistochemical detection is presented in Fig. 4A. When ERα alone is transfected, the localization of the unliganded receptor is variable, being cytoplasmic (Fig. 4, i.a) in some cells and nuclear (Fig. 4, i.b) in other cells. However, after tamoxifen treatment ERα localization is exclusively nuclear (Fig. 4, ii), suggesting that ERα in the nonclassical conformation is transported to and/or retained in the nucleus. Foxl2, by contrast, was found predominantly in the nucleus (Fig. 4, iii.a and iii.b).
Figure 4.
Cellular localization of ERα and Foxl2. A, HEK293FT cells were transfected with human ERα and/or mouse Foxl2 expression vectors and treated for 1 h with vehicle control (ethanol) or tamoxifen. Immunodetection of ERα (red) and/or Foxl2 (green) is shown; all slides were also stained for DNA using 4′,6′-diamino-2-phenylindole (DAPI; blue). B, Cells were transfected as above and treated for 2 h before fractionation and Western blot using ERα and Foxl2 antibodies. Blots containing nuclear and cytoplasmic fractions were also stained for lamin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively, to confirm equal loading of the lanes.
When ERα and Foxl2 were coexpressed, unliganded ERα could be detected in the perinuclear region of some cells (Fig. 4, iv.a), but this expression was much less than the cytoplasmic staining observed in the absence of Foxl2 (Fig. 4, i.a). In the presence of tamoxifen, both ERα and Foxl2 were observed only in the nucleus (Fig. 4, v). These data argue against the possibility that Foxl2 suppresses ERα signaling by transporting the receptor from the nucleus.
Western blot analysis is presented in Fig. 4B and supports this conclusion. Cells were fractionated into the nuclear and cytoplasmic fractions and analyzed for Foxl2 and ERα proteins. Consistent with the immunohistochemistry, Foxl2 was located predominantly in the nucleus. Levels were unchanged by cotransfection with ERα alone but were slightly elevated in both the nucleus and cytoplasm in the presence of ERα and tamoxifen. Levels of ERα were higher in the nucleus than in the cytoplasm and were elevated in both fractions after tamoxifen treatment, in the absence or presence of Foxl2 expression. The ratio of ERα in the nucleus vs. cytoplasm was relatively stable across treatments, again suggesting that Foxl2 suppression does not involve modulation of ER nuclear export.
On AP1 sites, Foxl2 is specific for transcriptional activation
We and others have shown that in addition to tamoxifen stimulation, E2 modestly suppresses ERα-mediated transcription at AP1 sites (21). Because this suppression can be difficult to detect relative to inherently low basal luciferase activity, repeated independent experiments were performed to assess whether Foxl2 altered this suppression. Using the AP1 reporter, luciferase activity was reduced by 50% in the presence of E2 and ERα (Fig. 5). Foxl2 alone had no effect on the AP1 reporter. Moreover, Foxl2 had no impact on E2 suppression. This finding suggests that Foxl2 action is selective for the complexes associated with ERα stimulation of AP1 reporters.
Figure 5.
E2 suppression in the presence of Foxl2. 293FT cells were transfected with human ERα and/or mouse Foxl2 expression vectors and treated for 24 h with vehicle control (ethanol) or E2. Response of the AP1-luciferase reporter is shown. Bars, mean ± sem of five experiments. *, P < 0.01.
COX2 is a physiological target of Foxl2/ERα interactions
Microarray experiments using the ERαAA mutant, which selectively acts through tethered transcriptional pathways, identified the gene for prostaglandin-endoperoxide synthase 2 (COX2) as a target of nonclassical ERα signaling (24). COX2 plays an important role in follicle maturation and ovulation (25), and the proximal COX2 promoter has at least one functional AP-1 site (26), suggesting that COX2 could be a physiological target of Foxl2/ERα interactions.
Using the COX2 promoter, transfection with ERα conferred a 6.3-fold response to tamoxifen (Fig. 6A), similar to that seen with the AP1 reporter genes. Foxl2 alone conferred no response to either ligand, but expression of Foxl2 with ERα strongly suppressed tamoxifen stimulation, although a small amount of residual induction remained.
Figure 6.
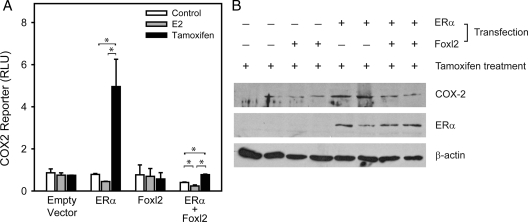
Foxl2 regulation of the COX2 promoter and COX2 protein expression. A, 293FT cells were transfected with human ERα and/or mouse Foxl2 expression vectors and treated for 24 h with vehicle control (ethanol), E2, or tamoxifen. Response of the −459COX2-luciferase reporter is shown. Bars, Mean ± sem of two experiments. *, P < 0.05. B, MDA-MB-231 human breast cancer cells were transfected with human ERα and/or mouse Foxl2 expression vectors as indicated and treated for 24 h with tamoxifen. COX2, ERα, and β-actin were detected by Western blot. RLU, Relative luciferase units.
To confirm that the native COX2 gene is regulated by Foxl2 and ERα, we measured protein levels in cells that are ERα-negative but that express COX2 (Fig. 6B). Compared with controls (Fig. 6B, lanes 1 and 2), expression of ERα conferred tamoxifen responsiveness as reflected by increased levels of endogenous COX2 protein (Fig. 6B, lanes 5 and 6), whereas expression of Foxl2 in the absence of ERα was without effect (Fig. 6B, lanes 3 and 4). When Foxl2 was coexpressed with ERα, COX2 protein returned to control levels despite the continued presence of tamoxifen-activated ERα (Fig. 6B, lanes 7 and 8). These data demonstrate that a native target gene of ERα nonclassical signaling is sensitive to the expression of Foxl2.
Foxl2 binding and transcriptional coactivator binding colocalize on ERα
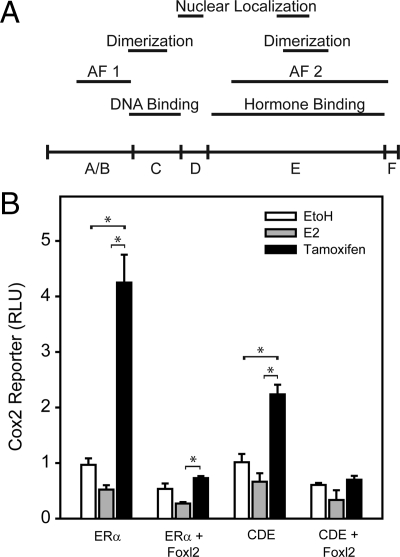
ERα mutants were also tested on the COX2 promoter to identify the ERα domains required for Foxl2 suppression. The schematic in Fig. 7A illustrates the human ERα domains. The graph in Fig. 7B compares the wild-type ERα to a truncation mutant, termed CDE, in which the A/B and F domains have been removed. This mutant eliminates the activator function (AF) 1 protein interaction domain but retains DNA binding, ligand binding, and the AF 2 domain. On the COX2 promoter, tamoxifen induction and its suppression by Foxl2 are both retained in the CDE mutant, demonstrating that AF 1 is dispensable for these actions. These data also demonstrate that coactivator binding and Foxl2 binding colocalize to the CDE domains of the ERα.
Figure 7.
ERα domains required for Foxl2 action. A, Schematic representation of the domains of the human ERα. B, 293FT cells were transfected with wild-type human ERα or a CDE truncation mutant and/or mouse Foxl2 expression vectors and treated for 24 h with vehicle control (EtOH, ethanol), E2, or tamoxifen. Response of the −459Cox2-luciferase reporter is shown. Bars, mean ± sem of two experiments. *, P < 0.05.
We attempted to narrow the active region further using the ERα point mutants I362R and K366D (21). These mutations are located in ERα helix 3, which together with helices 4, 5, and 12 form a hydrophobic cavity that is involved in cofactor interactions. Using mammalian two-hybrid assays, these mutant ERs lost most (I362R) or all (K366D) of their interaction with steroid receptor coactivator-1 (SRC-1) and glucocorticoid-interacting protein-1 (GRIP-1; data not shown), two ERα transcriptional cofactors. However, both mutations also abolished tamoxifen induction, precluding further analyses of the role of Foxl2 suppression of tamoxifen-induced transcription (data not shown).
Discussion
The complexity of estrogen signaling has become apparent as studies have revealed additional receptors, multiple ER-dependent signaling pathways, different ER binding sites on DNA, and interactions of the ER pathways with those of other transcription factors (27). These observations have stimulated interest in nonclassical ER pathways that do not involve binding of the ER to a classical ERE. Included in the nonclassical group are membrane-initiated ER signaling and tethering of ER to other transcription factors (28), including AP1 proteins, which are bound to their own DNA response elements.
Foxl2, a forkhead protein, and ERα are both critical to female reproductive development and function (1,12), raising the possibility that their actions converge in the ovary. We initially considered whether Foxl2 might regulate ERα expression, or vice versa. However, ERα did not alter Foxl2 promoter activity in transfection experiments (our unpublished data) and expression profiling of human granulosa cells overexpressing Foxl2 did not identify estrogen receptor as a target gene (29). Although these findings do not preclude interdependence of Foxl2 and ERα expression, we also considered the possibility that the ERα and Foxl2 pathways might intersect at the level of protein-protein interactions. In pull-down assays, we found that HA-tagged Foxl2 immunoprecipitates ERα. These interactions might be direct between ERα and Foxl2, or they might be indirect through additional proteins. In either case, this mechanism appears to be highly specific for ERα because the actions of other nuclear receptors (progesterone receptor, thyroid hormone receptor, peroxisome proliferator activated receptor) were unaffected by coexpression of Foxl2 (not shown).
Genome-wide screens have identified AP1 as a binding site for the ERα (30), and additional studies have shown that ERα can regulate minimal AP1 promoters (31). Of note, ERα ligands have different effects on AP1 sites when compared with EREs. E2 stimulates transcription at the ERE but represses transcription at AP1 sites. Tamoxifen represses transcription at the ERE but stimulates transcription at AP1 sites. These divergent actions are incompletely understood but have been attributed to the induction of distinct ERα conformations by E2 and tamoxifen (32), and the subsequent recruitment of alternate groups of coregulatory factors. Physiologically, selective estrogen receptor modulators, such as tamoxifen, mediate a spectrum of estrogen actions, ranging from agonist to antagonist actions (33). For example, tamoxifen acts as an antagonist in the breast but as an agonist in the uterus (34).
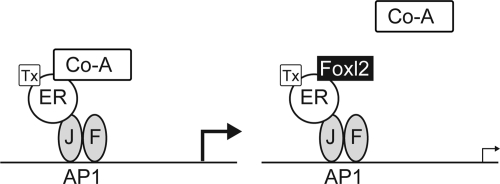
In this study, we found that Foxl2 repressed tamoxifen-mediated stimulation of AP1 regulated promoters. Notably, Foxl2 had no effect on E2-stimulated classical EREs and did not alter E2-mediated repression of AP1 promoters. Thus, the effect of Foxl2 appears to be restricted to the tamoxifen-induced ERα conformation. It is interesting to consider the potential mechanisms of Foxl2 action. One possibility would be for Foxl2 to occupy the ERα binding motif on the AP1 proteins. However, this model predicts that Foxl2 would also alter E2-mediated suppression of AP1, which we did not observe. A second possibility is that Foxl2 binds to a novel site on the ERα and prevents its interaction with the AP1 proteins. However, based on our data, the most likely possibility is that Foxl2 impedes coactivator binding to ERα (Fig. 8). In the presence of tamoxifen, helix 12 of the ER obscures a domain that otherwise interacts with coactivator proteins. Thus, for tethered AP1 signaling to occur, coactivators must interact with a domain of the ERα that is specifically available in the tamoxifen-induced conformation. Because the Foxl2 effects are also selective for the tamoxifen-induced conformation, it is plausible that the two ERα binding partners compete for the same site. This hypothesis is supported by the colocalization of Foxl2 suppression and coactivator binding to the CDE core of the ERα.
Figure 8.
Model for Foxl2 Action. AP1 reporters are shown binding Jun (J) and Fos (F). Tamoxifen (Tx) bound ERα is suggested to interact with the Jun/Fos complex and to recruit coactivator (CoA) proteins and stimulate transcription. In the presence of Foxl2, the ERα interaction with coactivator proteins is disrupted, thereby preventing transcriptional stimulation.
There is mounting evidence that nonclassical signaling is a key component of ERα actions in vivo, particularly in the reproductive system (35). In the ovary, although the theca cell is the predominant site of ERα expression, recent data demonstrate that ERα is expressed in mouse granulosa cells as well (36). Foxl2 is expressed in the granulosa cells of ovarian follicles at multiple stages of development (15). Thus, ERα and Foxl2 have the potential to interact functionally in differentiated somatic cells. The physiological implications of Foxl2 actions through nonclassical ERα signaling are unclear. However, a number of ovarian candidate genes, including many with AP1 sites, might be targets of concerted regulation by Foxl2 and ERα. For example, the steroidogenic acute regulatory protein gene, which is required for steroidogenesis in ovarian theca cells, is regulated by both AP1 proteins (37) and Foxl2 (15).
In an earlier study (24), we identified 268 targets of the nonclassical pathway in breast cancer cells. A review of these genes for potential ovarian targets revealed COX2, which is expressed in granulosa cells (38) and is required at multiple steps in female reproduction, including ovulation, fertilization, implantation, and decidualization (39). COX2 is also regulated through an AP1 site in gastric cells (26). In the current report, COX2 exhibited all of the responses collectively observed with the model AP1 reporter genes. The COX2 promoter and endogenous COX2 protein were induced by tamoxifen in the presence of ERα, and this induction was suppressed by Foxl2. Thus, COX2 may be representative of ovarian genes that are regulated by ERα and Foxl2.
In summary, we have demonstrated that Foxl2 is an ERα interacting protein that can disrupt nonclassical ERα signaling through AP1. We further identified COX2 as a target of Foxl2 activity, providing a direct link of this mechanism to ovarian function. Based on the ubiquity of AP1 action and ERα interaction, we predict that other targets of the Foxl2-ERα pathway remain to be discovered and will provide insight into estrogen action in the ovary.
Footnotes
This work was supported by National Institutes of Health Grant P01 HD21921.
Disclosure Summary: The authors have nothing to disclose.
First Published Online September 24, 2009
Abbreviations: AF, Activator function; AP1, activator protein-1; COX2, cyclooxygenase-2; E2, 17β-estradiol; ER, estrogen receptor; ERE, estrogen response element; HA, hemagglutinin; RLU, relative light unit.
References
- Couse JF, Korach KS 1999 Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev 20:358–417 [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Greene G, Krust A, Goffin C, Jensen E, Scrace G, Waterfield M, Chambon P 1986 Cloning of the human oestrogen receptor cDNA. J Steroid Biochem 24:77–83 [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA 1996 Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA 93:5925–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA, Smithies O 1998 Generation and reproductive phenotypes of mice lacking estrogen receptor β. Proc Natl Acad Sci USA 95:15677–15682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes SR, Levin ER 2007 Extranuclear steroid receptors: nature and actions. Endocr Rev 28:726–741 [DOI] [PubMed] [Google Scholar]
- Björnström L, Sjöberg M 2005 Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol 19:833–842 [DOI] [PubMed] [Google Scholar]
- Glidewell-Kenney C, Hurley LA, Pfaff L, Weiss J, Levine JE, Jameson JL 2007 Nonclassical estrogen receptor α signaling mediates negative feedback in the female mouse reproductive axis. Proc Natl Acad Sci USA 104:8173–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Bernhardt ML, Laronda MM, Hurley LA, Glidewell-Kenney C, Pillai S, Tong M, Korach KS, Jameson JL 2008 Estrogen actions in the male reproductive system involve estrogen response element-independent pathways. Endocrinology 149:6198–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baere E, Beysen D, Oley C, Lorenz B, Cocquet J, De Sutter P, Devriendt K, Dixon M, Fellous M, Fryns JP, Garza A, Jonsrud C, Koivisto PA, Krause A, Leroy BP, Meire F, Plomp A, Van Maldergem L, De Paepe A, Veitia R, Messiaen L 2003 FOXL2 and BPES: mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am J Hum Genet 72:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth BS, Egashira N, Haller JL, Butts DL, Cocquet J, Clay CM, Osamura RY, Camper SA 2006 FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol 20:2796–2805 [DOI] [PubMed] [Google Scholar]
- Uda M, Ottolenghi C, Crisponi L, Garcia JE, Deiana M, Kimber W, Forabosco A, Cao A, Schlessinger D, Pilia G 2004 Foxl2 disruption causes mouse ovarian failure by pervasive blockage of follicle development. Hum Mol Genet 13:1171–1181 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, Treier AC, Treier M 2004 The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 131:933–942 [DOI] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G 2001 The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 27:159–166 [DOI] [PubMed] [Google Scholar]
- Shah SP, Köbel M, Senz J, Morin RD, Clarke BA, Wiegand KC, Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E, Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y, Zeng T, Glover JN, Vanderhyden B, Zhao C, Parkinson CA, Jimenez-Linan M, Bowtell DD, Mes-Masson AM, Brenton JD, Aparicio SA, Boyd N, Hirst M, Gilks CB, Marra M, Huntsman DG 2009 Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med 360:2719–2729 [DOI] [PubMed] [Google Scholar]
- Pisarska MD, Bae J, Klein C, Hsueh AJ 2004 Forkhead l2 is expressed in the ovary and represses the promoter activity of the steroidogenic acute regulatory gene. Endocrinology 145:3424–3433 [DOI] [PubMed] [Google Scholar]
- Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, Clay CM 2003 The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol Cell Endocrinol 206:93–111 [DOI] [PubMed] [Google Scholar]
- Pannetier M, Fabre S, Batista F, Kocer A, Renault L, Jolivet G, Mandon-Pépin B, Cotinot C, Veitia R, Pailhoux E 2006 FOXL2 activates P450 aromatase gene transcription: toward a better characterization of the early steps of mammalian ovarian development. J Mol Endocrinol 36:399–413 [DOI] [PubMed] [Google Scholar]
- Loffler KA, Zarkower D, Koopman P 2003 Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology 144:3237–3243 [DOI] [PubMed] [Google Scholar]
- Baron D, Cocquet J, Xia X, Fellous M, Guiguen Y, Veitia RA 2004 An evolutionary and functional analysis of FoxL2 in rainbow trout gonad differentiation. J Mol Endocrinol 33:705–715 [DOI] [PubMed] [Google Scholar]
- Rhen T, Metzger K, Schroeder A, Woodward R 2007 Expression of putative sex-determining genes during the thermosensitive period of gonad development in the snapping turtle, Chelydra serpentina. Sex Dev 1:255–270 [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL 2001 Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem 276:13615–13621 [DOI] [PubMed] [Google Scholar]
- Shapiro DJ, Sharp PA, Wahli WW, Keller MJ 1988 A high-efficiency HeLa cell nuclear transcription extract. DNA 7:47–55 [DOI] [PubMed] [Google Scholar]
- Jackson JG, Kreisberg JI, Koterba AP, Yee D, Brattain MG 2000 Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene 19:4574–4581 [DOI] [PubMed] [Google Scholar]
- Glidewell-Kenney C, Weiss J, Lee EJ, Pillai S, Ishikawa T, Ariazi EA, Jameson JL 2005 ERE-independent ERα target genes differentially expressed in human breast tumors. Mol Cell Endocrinol 245:53–59 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Morrow JD, Wang H, Dey SK 2006 Cyclooxygenase-2-derived prostaglandin E(2) directs oocyte maturation by differentially influencing multiple signaling pathways. J Biol Chem 281:37117–37129 [DOI] [PubMed] [Google Scholar]
- Chang YJ, Wu MS, Lin JT, Chen CC 2005 Helicobacter pylori-Induced invasion and angiogenesis of gastric cells is mediated by cyclooxygenase-2 induction through TLR2/TLR9 and promoter regulation. J Immunol 175:8242–8252 [DOI] [PubMed] [Google Scholar]
- Hall JM, Couse JF, Korach KS 2001 The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem 276:36869–36872 [DOI] [PubMed] [Google Scholar]
- Pietras RJ, Márquez-Garbán DC 2007 Membrane-associated estrogen receptor signaling pathways in human cancers. Clin Cancer Res 13:4672–4676 [DOI] [PubMed] [Google Scholar]
- Batista F, Vaiman D, Dausset J, Fellous M, Veitia RA 2007 Potential targets of FOXL2, a transcription factor involved in craniofacial and follicular development, identified by transcriptomics. Proc Natl Acad Sci USA 104:3330–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M 2006 Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS 1997 Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science 277:1508–1510 [DOI] [PubMed] [Google Scholar]
- Egner U, Heinrich N, Ruff M, Gangloff M, Mueller-Fahrnow A, Wurtz JM 2001 Different ligands-different receptor conformations: modeling of the hERα LBD in complex with agonists and antagonists. Med Res Rev 21:523–539 [DOI] [PubMed] [Google Scholar]
- Sato M, Rippy MK, Bryant HU 1996 Raloxifene, tamoxifen, nafoxidine, or estrogen effects on reproductive and nonreproductive tissues in ovariectomized rats. FASEB J 10:905–912 [DOI] [PubMed] [Google Scholar]
- Jordan VC, Morrow M 1999 Tamoxifen, raloxifene, and the prevention of breast cancer. Endocr Rev 20:253–278 [DOI] [PubMed] [Google Scholar]
- McDevitt MA, Glidewell-Kenney C, Jimenez MA, Ahearn PC, Weiss J, Jameson JL, Levine JE 2008 New insights into the classical and non-classical actions of estrogen: evidence from estrogen receptor knock-out and knock-in mice. Mol Cell Endocrinol 290:24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipp JL, Kilen SM, Woodruff TK, Mayo KE 2007 Activin regulates estrogen receptor gene expression in the mouse ovary. J Biol Chem 282:36755–36765 [DOI] [PubMed] [Google Scholar]
- Manna PR, Stocco DM 2007 Crosstalk of CREB and Fos/Jun on a single cis-element: transcriptional repression of the steroidogenic acute regulatory protein gene. J Mol Endocrinol 39:261–277 [DOI] [PubMed] [Google Scholar]
- Joyce IM, Pendola FL, O'Brien M, Eppig JJ 2001 Regulation of prostaglandin-endoperoxide synthase 2 messenger ribonucleic acid expression in mouse granulosa cells during ovulation. Endocrinology 142:3187–3197 [DOI] [PubMed] [Google Scholar]
- Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, Dey SK 1997 Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 91:197–208 [DOI] [PubMed] [Google Scholar]