Abstract
HBV cccDNA, the template for transcription of all viral mRNAs, accumulates in the nucleus of infected cells as a stable episome organized into minichromosomes by histones and non-histone viral and cellular proteins. Using a cccDNA-specific chromatin immunoprecipitation (ChIP)-based quantitative assay, we have previously shown that transcription of the HBV minichromosome is regulated by epigenetic changes of cccDNA-bound histones and that modulation of the acetylation status of cccDNA-bound H3/H4 histones impacts on HBV replication. We now show that the cellular histone acetyltransferases CBP, p300, and PCAF/GCN5, and the histone deacetylases HDAC1 and hSirt1 are all recruited in vivo onto the cccDNA. We also found that the HBx regulatory protein produced in HBV replicating cells is recruited onto the cccDNA minichromosome, and the kinetics of HBx recruitment on the cccDNA parallels the HBV replication. As expected, an HBV mutant that does not express HBx is impaired in its replication, and exogenously expressed HBx transcomplements the replication defects. p300 recruitment is severely impaired, and cccDNA-bound histones are rapidly hypoacetylated in cells replicating the HBx mutant, whereas the recruitment of the histone deacetylases hSirt1 and HDAC1 is increased and occurs at earlier times. Finally, HBx mutant cccDNA transcribes significantly less pgRNA. Altogether our results further support the existence of a complex network of epigenetic events that influence cccDNA function and HBV replication and identify an epigenetic mechanism (i.e., to prevent cccDNA deacetylation) by which HBx controls HBV replication.
Keywords: histone acetylation, HATs, HDACs
Hepatitis B virus (HBV) infection is a major health problem, with ≈400 million people chronically infected worldwide who are at high risk of developing liver cirrhosis and hepatocellular carcinoma (HCC) (1). The epidemiological evidence linking HBV infection to HCC is very strong, and despite the mechanisms underlying HBV-associated carcinogenesis remain to be fully defined, a growing number of studies support a direct role of HBV in the process (2–5). The HBV-encoded regulatory protein hepatitis B virus X protein (HBx) is thought to contribute to HBV oncogenicity (5, 6). HBx transforms SV40-immortalized murine hepatocytes, induces cell cycle progression within the regenerating liver, causes liver cancer in some transgenic mice, and acts as a cofactor to accelerate cancer development in other mouse models (6–11). HBx is a 154-amino acid protein with an N-terminal negative regulatory domain and C-terminal transactivation or coactivation domain that has been detected both in the cytoplasm and in the nuclei of infected hepatocytes (6, 12, 13). Studies in transfected cells have shown that HBx expression affects several cellular functions such as cytoplasmic calcium regulation, cell signaling, transcription, cell proliferation, DNA repair, and apoptosis (11, 13–16). To perform its multiple functions, HBx interacts with many cellular partners including the tumor suppressor p53, the UV-stimulated E3-ubiquitin ligase DDB1, the nuclear export protein CRM1, a number of nuclear proteins involved in the regulation of transcription such as the RPB5 subunit of RNA polymerase II, TFIIB, TFIIH, the TATA binding protein (TBP), the basic domain-leucine zipper (bZIP) family transcription factors ATF2, CHOP, and cAMP-response element (CRE)-binding protein (CREB) (6, 14). Recently, it has been shown that HBx interacts and cooperate with the CREB-binding protein (CBP)/p300 to modify chromatin dynamics of target genes and to synergistically enhance CREB activity (17).
The lack of homology of the X ORF to host protein and its high conservation to other mammalian hepadnaviruses genomes strongly suggest that HBx play a role in viral life cycle (6). Although initial studies suggested that HBx was not required for virus replication in cell culture (18), experiments with the highly related woodchuck hepatitis virus (WHV) system indicate that the WHV X protein (WHx) is required for virus replication in vivo (19–22). Studies performed using a plasmid-based replication assays that use greater-than-unit-length HBV genomes transfected in HCC cell lines or injected via the mouse tail vein under hydrodynamic conditions have repeatedly confirmed that HBx potentiate HBV replication (23–27).
The HBV replicative intermediate cccDNA (covalently closed circular HBV DNA), which serves as a template for the transcription of all viral transcripts including the pregenomic RNA (pgRNA), is organized into a minichromosome in the nuclei of infected hepatocytes by histone e non-histone proteins (28). The HBV core protein is a structural component of the HBV minichromosome, and its binding results in a reduction of the nucleosomal spacing of the HBV nucleoprotein complexes (29). Recently, we have developed a ChIP-based quantitative technique to study the recruitment in vivo of cellular and viral proteins onto the HBV minichromosome (30). Chromatin immunoprecipitation (ChIP) techniques allow the identification in vivo of the DNA binding sites of virtually any chromosome component (31). The principle relies upon the fixation of protein-DNA and protein-protein interactions in vivo by the cross-linking agent formaldehyde, and the immunoprecipitation of the cross-linked protein of interest with specific antibody directed against immunoprecipitated DNA is used as a template in real-time PCR. The HBV cccDNA ChIP assay combines a cccDNA ChIP step with a sensitive and specific real-time PCR protocol for cccDNA quantification (30). Using an anti-acetylated-H3 or -H4 cccDNA ChIP assay, we found that HBV replication is regulated, both in HBV replicating Huh7 cells and in the liver of HBV chronically infected patients by the acetylation status of H3/H4 histones bound to the viral cccDNA (30). Here we investigated the recruitment of HBx onto the HBV minichromosome and its ability to regulate the epigenetic control of cccDNA function in HBV replicating cells.
Results
HBx Potentiates HBV Replication.
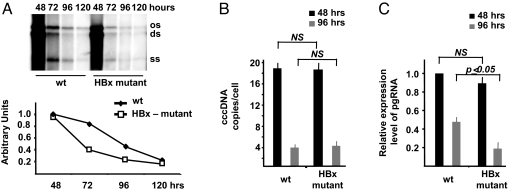
The HBV regulatory protein HBx has been shown to potentiate viral replication in plasmid-based replication assays that use greater-than-unit-length HBV genomes transfected in HCC cell lines or injected via the mouse tail vein under hydrodynamic conditions (23–27). We have recently shown that transfection of plasmid free linear HBV DNA into Huh7 and HepG2 HCC cell lines (32) allows to fully recapitulate the HBV replication cycle, including the nuclear generation of viable cccDNA (30) (Fig. S1). To examine the contribution of HBx to viral replication in our cccDNA-driven replication system, equivalent numbers of HepG2 cells were transfected with either linear wild-type (WT) HBV genomes or with a mutant HBV DNA carrying a stop codon for amino acid 8 of the HBx ORF. Cells were harvested at the indicated times posttransfection, cytoplasmic viral core particles were isolated, and HBV DNA was extracted, analyzed by hybridization with a 32P-labeled full-length HBV DNA probe, and quantitated by densitometric analysis. As shown in Fig. 1A, the level of capsid associated HBV DNA in cells transfected with the HBx mutant DNA was reduced as compared to the levels measured in cells transfected with WT HBV DNA. Whereas the peak replication at 48 h is only marginally affected by the lack of HBx, the HBx mutant virus displays a faster decrease in replication at 72 and 96 h posttransfection as compared to WT HBV. These results confirm that HBx potentiates HBV replication and suggest that HBx activity is required to sustain replication in our assay.
Fig. 1.
HBx is involved in viral replication, and it is required for HBV pregenomic RNA transcription. (A) HepG2 cells were transfected with monomeric linear full-length WT or HBx mutant HBV adw genomes, and cytoplasmatic HBV core particles were isolated at the indicated time points after transfection. (Upper) Southern blot analysis of HBV DNA replicative intermediates. (Lower) Densitometric quantification of HBV replicative intermediates. Signal intensity of the single-strand (SS) band underneath the linear double-stranded (DS) HBV DNA band was quantified with Quantity One 1-D Analysis Software (BioRad Laboratories). The band corresponding to the DS HBV DNA was not included in the quantitative analysis, because this DNA may be partially derived from transfected input DNA. The optical density value of the SS band at 48 h in WT replicating cells is set at 1.0. Data are expressed as relative arbitrary units (mean + SD) from three independent experiments. OC, open circular duplex HBV DNA; DS, double-strand; and SS, single-strand HBV DNA replicative intermediates. (B) cccDNA accumulation in HepG2 cells transfected with WT or HBx mutant HBV genomes. Real-time quantitative PCR analysis was performed using selective cccDNA primers to amplify cccDNA and beta-globin primers to normalize the DNA samples. Results are expressed as number of cccDNA copies per cell (mean + SD) from four independent experiments. (C) HepG2 were transfected with WT or with HBx mutant HBV genomes and mRNAs were extracted 48 and 96 h posttransfection as described in the Materials and Methods section. Specific primers were used to quantitate the HBV pregenomic RNA, and GAPDH amplification was used to normalize for equal loading of each RNA sample. Results are expressed as number of cccDNA copies per cell (mean + SD) from three independent experiments. pgRNA, HBV pregenomic RNA.
HBx Is Required for HBV Pregenomic RNA Transcription.
To investigate the mechanism involved in HBx-induced potentiation of HBV DNA replication, we measured the size of the cccDNA pool and the levels of pgRNA in HepG2 cells replicating either the WT or the HBx mutant HBV virus. The median number of cccDNA copies per hepatocyte (33) was similar in cells replicating WT or HBx mutant viruses both at 48 h (replication peak) and at 96 h posttransfection, when both cccDNA levels and HBV replication are sharply reduced (30) (Fig. 1B). On the opposite, the levels of pgRNA were slightly lower at 48 h and reduced by 50% to 70% at 96 h in HepG2 cells replicating the HBx mutant virus (P ≤ 0.05) (Fig. 1C). Altogether, these results indicate that HBx is not required for cccDNA maturation but modulates pgRNA transcription from cccDNA.
The HBx Viral Protein Is Recruited onto the cccDNA In vivo.
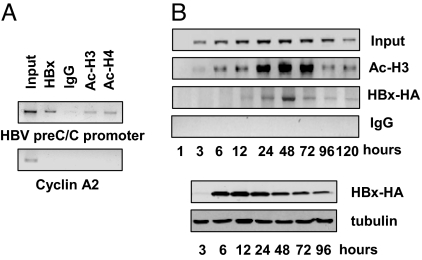
Nuclear cccDNA molecules are organized into a chromatine like structure by histone and non-histone proteins. Using the cccDNA ChIP assay, we have been able to confirm that the HBV core protein is recruited onto the viral chromatin (29, 30). HBx does not bind specific DNA sequences, but it is recruited onto the cellular chromatin through its ability to interact with cellular transcription factors and cofactors (17). To assess whether HBx interacts with the cccDNA to modulate its transcription, we have performed an anti-HBx cccDNA ChIP assay in HBV replicating HepG2 cells. We found that endogenous HBx binds to the cccDNA (Fig. 2A) at 48 h posttransfection. The kinetics of HBx recruitment onto the cccDNA was assessed in HepG2 cells cotransfected with WT linear HBV DNA and an HA-tagged HBx expression vector. Using an anti-HA antibody to perform the cccDNA ChIP assay, we found that the recruitment of exogenously expressed HA-tagged HBx onto the cccDNA closely parallels the kinetics of cccDNA bound H3 acetylation (Fig. 2B). Altogether, these results suggest that HBx might interact with the cccDNA to modulate its transcription.
Fig. 2.
The viral transactivator HBx is recruited onto the cccDNA in HBV replicating cells. (A, Upper) Cross-linked chromatin from HepG2 cells transfected with monomeric linear full-length HBV DNA was immunoprecipitated with the relevant control IgG or specific anti-HBx, anti-AcH3, and anti-AcH4 antibodies and analyzed by PCR with HBV cccDNA selective primers. (Lower) ChIPed samples were analyzed by PCR using primers specific for the cyclin A2 coding region as a negative control. (B) Kinetics of HBx recruitment onto the HBV cccDNA. (Upper) ChIP analysis was performed on chromatin from HepG2 cells cotransfected with monomeric linear full-length WT HBV DNA and an HA-tagged HBx expression vector using the relevant control IgG or specific anti-HA tag and anti-AcH3 specific antibodies. (Lower) Exogenously expressed HBx is detected by anti-HA immunoblotting. Tubulin levels detected by immunoblotting were used to normalize equal loadings from lysate samples.
Epigenetics Changes in HepG2 Cells Replicating HBx Mutant HBV.
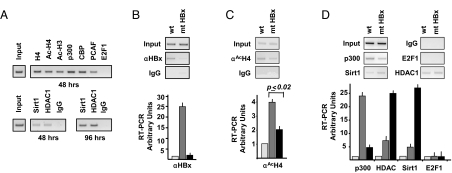
cccDNA transcriptional activity and HBV replication are modulated both in in vitro cellular systems and in vivo in the liver of chronic HBV carriers by epigenetic changes of cccDNA-bound H3 and H4 histones (30). Moreover, HBV replication in HuH7 cells can be modulated by substances that interfere with the enzymatic activity of well-known chromatin regulators, such as class I/II histone deacetylases (30). Finally, the presence of the class I deacetylase HDAC1 on the cccDNA correlates with the decline of HBV replication in our in vitro HuH7 cells based system and with a low replication state in vivo (30). First, to analyze the recruitment of chromatin-modifying enzymes on the cccDNA, HepG2 cells were transfected with full-length WT HBV and harvested 48 and 96 h posttransfection. Chromatin was immunoprecipitated using antibodies that specifically recognize acetylated histones H3 and H4, the acetyltransferases p300, CBP and PCAF, the histone deacetylases HDAC1 and hSirt1, and the transcription factor E2F1. As shown in Fig. 3A (upper panel), 48 h posttransfection, the histones H3 and H4 bound to the cccDNA are highly acetylated, and the coactivators p300, CBP, and PCAF bind the cccDNA. Interstingly, 48 h posttransfection, the recruitment hSirt1 and HDAC1 histone deacetylases onto the cccDNA is very low, whereas 96 h after transfection, when the HBV replication declines, their recruitment is higher (Fig. 3A, lower panel). The transcription factor E2F1 does not bind the cccDNA and serves as negative control. These results indicate that the differential recruitment of coactivators and corepressors onto the cccDNA correlates with HBV replication activity.
Fig. 3.
HBx modulates the epigenetic control of cccDNA function by affecting the recruitment of chromatin modifying enzymes. (A) HepG2 cells were transfected with monomeric linear full-length HBV DNA. (Upper) Chromatin prepared 48 h after transfection was immunoprecipitated with the relevant control IgG or specific anti-H4, anti-AcH3, anti-AcH4, anti-p300, anti-CBP, anti-PCAF, and anti-E2F1 antibodies and analyzed by PCR with HBV cccDNA selective primers. (Lower) Chromatin prepared 96 h posttransfection was immunoprecipitated with the relevant control IgG or specific antibodies to the class I HDAC1 and class III hSirt1 histone deacetylases. ChIPed DNA was analyzed by PCR with HBV cccDNA selective primers. (B–D) Chromatin was prepared from HepG2 cells transfected with WT or HBx mutant monomeric linear full-length genomes and immunoprecipitated with the relevant control IgG or a specific anti-HBx (B), anti-AcH4 (C), anti-p300, anti-HDAC1, anti-hSirt1, and anti-E2F1 (D) antibodies. Immunoprecipitated chromatin was analyzed by semiquantitative PCR (Upper) and real-time quantitative PCR (Lower) with HBV cccDNA selective primers. Light gray columns, mock; gray columns, WT HBV; black columns, mt HBx. Results are expressed as RT-PCR arbitrary units (mean + SD) from four independent experiments.
Next we investigated whether HBx contributes to the epigenetic regulation of HBV minichromosome functions. As expected, no HBx recruitment onto the cccDNA could be demonstrated in cells replicating the HBx mutant virus neither by semiquantitative PCR (Fig. 3B, left panel) nor by quantitative real-time PCR (Fig. 3B, right panel). Interestingly, in the absence of HBx, the acetylation of cccDNA-bound histone H4 is significantly reduced (P ≤ 0.02) (Fig. 3C). This result is in agreement with the observed reduction in both pgRNA transcription (Fig. 1C) and viral replication (Fig. 1A). To gain an insight into the mechanism by which HBx might influence cccDNA transcription, we assessed the impact of HBx on the recruitment of cofactors and corepressors onto the cccDNA in HepG2 cells replicating WT and HBx mutant HBV. As shown in Fig. 3D, the recruitment of the acetyltransferase p300 on the cccDNA is strongly reduced in the absence of HBx. Interestingly, the binding to the cccDNA of hSirt1 and HDAC1 deacetylases is strongly increased in cells replicating the HBx mutant virus (Fig. 3D) correlating with the acetylation of cccDNA-bound H4 histone (Fig. 3C), pgRNA transcription (Fig. 1C), and HBV replication (Fig. 1A).
Discussion
The HBx protein behaves as a promiscuous transactivator of viral and cellular promoters (13, 14). Although the subcellular localization of HBx seems to be mainly cytoplasmic, a small variable fraction of the protein can be found in the nucleus, and the ability of HBx to activate transcription of host genes is thought to occur indirectly by interaction with nuclear transcription factors or by activation of different signal transduction pathways in the cytoplasm (12, 13). HBx has also been shown to modulate chromatin dynamics in HCC cells and tissues. HBx binds several nuclear proteins involved in the regulation of transcription, including components of the basal transcriptional machinery (RPB5, TFIIB, TBP, TFIIH), coactivators (CBP, p300, and PCAF), and transcription factors (ATF/CREB, ATF3, c/EBP, NF-IL-6, Ets, Egr, SMAD4, Oct1, RXR receptor, p53) (14, 34). HBx is bound in vivo to the promoters of a number of CREB-regulated genes and favors transcription by increasing the amount of CBP and p300 recruited on the same promoters (17). In addition to stimulate transcription, HBx can also repress gene expression by upregulating DNMT1, DNMT3A1, and DNMT3A2 levels, increasing total DNA methyltransferase (DNMT) activity, and selectively facilitating the regional hypermethylation of the promoters of certain tumor suppressor genes (35, 36). More recently, it has been shown, by ChIP and sequential-ChIP experiments that HBx recruits DNMT3A on promoters of the repressed genes MT1F and IL4R leading to promoter hypermethylation and suppression of gene expression (37). In the presence of HBx, DNMT3A dissociates from the promoter region of the upregulated genes IGFBP3 and CDH6, thus resulting in hypomethylation and transcriptional activation (37).
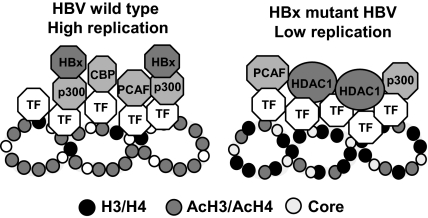
Here, coupling cccDNA quantification (38) and the cccDNA ChIP assay (30), we show that the HBx produced in HBV replicating cells is recruited onto the cccDNA minichromosome. HBx recruitment on the cccDNA parallels HBV replication and the recruitment of the PCAF/GCN5, p300, and CBP acetyltransferases. We also found that cccDNA-bound histones are more rapidly hypoacetylated in cells replicating the HBx mutant, and the recruitment of the p300 acetyltransferase is severely impaired, whereas the recruitment of the histone deacetylases hSirtl and HDAC1 is increased and occurs earlier (Fig. 4). Accordingly, we show that in cells replicating the HBx mutant, the pool of cccDNA in the nuclei is not reduced, but the HBx mutant cccDNA transcribes significantly less pgRNA. Although our observations imply that HBx exerts its role in potentiating HBV replication by acting directly on the cccDNA, we cannot exclude that Ca++ mobilization and src activation (13, 15) might also play a role. Importantly, it has been recently reported that HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically injected mice (39), thus supporting the importance of HBx interaction with the cccDNA in the nucleus. Altogether our results indicate that HBx influences the epigenetic control of nuclear HBV cccDNA function by modulating the recruitment of chromatin-modifying enzymes onto the viral minichromosome.
Fig. 4.
Schematic representation of cccDNA-bound histones acetylation status and the recruitment of chromatin modifying enzymes onto the viral minichromosome in relation to viral replication and HBx status. In cells replicating an HBx mutant HBV, cccDNA-bound histones are hypoacetylated, the recruitment of the p300 acetyltransferase is severely impaired, whereas the recruitment of the histone deacetylases hSirtl and HDAC1 is increased. This behavior is similar to what is observed in the later phase (96–144 h) of the in vitro viral replication cycle in our model system and mimicks the situation found in the liver of anti-HBe patients with low viremia (30).
Materials and Methods
Transient Transfection of Full-Length HBV DNA Genomes.
Full-length WT and HBx mutant HBV genomes of genotype A (adw) were released by EcoRI (New England Biolabs) digestion from replication-competent plasmids HTD and HBX-21 HTD (18) (a gift from Husseyn Sirma and Hans Will, Hamburg, Germany), containing complete WT and HBx mutant HBV DNA dimers, respectively. In particular, the pHBX-21 HTD carries a single nucleotide substitution that results in a termination site at the codon 8 of the X ORF. After digestion, the 3.2-kb fragments were recovered by gel purification using the QIAquick gel extraction kit (Qiagen) and inserted in the EcoRI site of pCRII-TOPO vector (Invitrogen) to generate the pCR.HBV.A.EcoRI and the pCR.mtHBx.A.EcoRI plasmids. Monomeric linear full-length WT and HBx HBV genomes (32) were released from the pCR.HBV.A.EcoRI and the pCR.mtHBx.A.EcoRI plasmids using EcoRI-PvuI (New England Biolabs) and transfected into HepG2 human hepatoma cells using the IT-LT1 reagent (Mirrus Biotrans). Briefly, HepG2 cells were seeded at a density of 2–3 millions of cells in 100-mm diameter Petri dishes and transfected 24 h later with 1–2 μg digested HBV DNA. Unless otherwise specified, culture medium was changed 1 day after transfection, and cells were harvested at the indicated times posttransfection. All transfection included 0.5 μg green fluorescence protein expression vector (GFP) to assess transfection efficiency (HepG2 cells range 28–32%).
Purification and Analysis of HBV DNA from Core Particles.
HBV DNA was purified from intracellular core particles and examined by Southern blot analysis as described (33) (SI Text).
HBV cccDNA and pgRNA Quantification.
Nuclear HBV cccDNA was isolated from HBV-transfected HepG2 cells and quantified by real-time PCR as described (33 and SI Text). Total RNA was extracted from HBV-transfected HepG2 cells using the TRIzol reagent (Invitrogen). RNA samples were treated with RQ1 RNase-Free DNase (Promega) for 30 min at 37 °C and stored until used. RNA quality and quantity were monitored by ethidium-bromide staining and by UV adsorbance. For pgRNA analysis, 2 μg DNase-treated RNA were reverse-transcribed and amplified by the ThermoScript RT-PCR system (Invitrogen). Two microliters of each cDNA were quantitated by real-time PCR analysis (Light Cycler; Roche Diagnostics) using the following pgRNA specific primers and probes: Forward primer 5′-GCCTTAGAGTCTCCTGAGCA-3′, reverse primer 5′-GAGGGAGTTCT TCTTCTAGG-3′, FRET hybridization probes 5′-AGTGTGGATTCGCACTCCTCCAGC FL-3′, Red640–5′ATAGACCACCAAATGCCCCTATCTTATCAAC-3′. The h-G6PDH housekeeping gene Light Cycler set (Roche Diagnostics) was used to normalize the RNA samples.
cccDNA Chromatin Immunoprecipitation Assays.
The cccDNA ChIP assays were performed as described, with minor modifications (30 and SI Text). Cross-linked chromatin was subjected to immunoprecipitation for 14–16 h at 4 °C using antibodies specific to H4, AcH4, AcH3, p300, PCAF, E2F1, hSirt1, HDAC1, HBV-HBx (listed in SI Text). Immunoprecipitations with relevant nonspecific immunoglobulins (Santa Cruz Biotechnology) were included in each experiment as a negative control. Immunoprecipitated chromatins were processed and analyzed by PCR and real-time PCR using cccDNA selective primers and probes that discriminate between cccDNA and the relaxed circular DNA (OC species) present in cytoplasmic HBV particles (30, 38).
Immunoblotting.
Cells were lysed in RIPA buffer (50 mM Tris, pH 7.6, 1% Nonidet P-40, 140 mM NaCl, 0.1% SDS), and the insoluble pellet was discarded after centrifugation. Protein concentration was determined by the BCA protein assay reagent (Bio-Rad Laboratories). Protein lysates were transferred to nitrocellulose membrane and incubated with HA antibodies from Santa Cruz Biotechnology (catalog no. sc-805). Autoradiography images were analyzed with a GS-800 Calibrated Densitometer (Bio-Rad Laboratories), and signal intensity was quantitated using the Quantity One 1-D Analysis Software (Bio-Rad Laboratories).
Statistical Analysis.
Continuous variables were expressed as median. Two group comparisons of continuous variables were performed using the nonparametric Wilcoxon rank sum test. P values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments.
M.L. and G.R. are supported by grants from Associazione Italiana per la Ricerca sul Cancro and Progetti di Ricerca di Interesse Nazionale del Ministero dell'Istruzione, dell'Universita‘ e della Ricerca. M.L. was also supported by the Vigilance Network for the Management of Antiviral Drug Resistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908365106/DCSupplemental.
References
- 1.Shepard CW, Simard EP, Finelli L, Fiore AE, Bell BP. Hepatitis B virus infection: Epidemiology and vaccination. Epidemiol Rev. 2006;28:112–125. doi: 10.1093/epirev/mxj009. [DOI] [PubMed] [Google Scholar]
- 2.Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77–100. doi: 10.1111/j.1365-2613.2001.iep0082-0077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 4.Buendia MA. Genetics of hepatocellular carcinoma. Semin Cancer Biol. 2000;10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 5.Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: Paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823–3833. doi: 10.1038/sj.onc.1209559. [DOI] [PubMed] [Google Scholar]
- 6.Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgson AJ, Keasler VV, Slagle BL. Premature cell cycle entry induced by hepatitis B virus regulatory HBx protein during compensatory liver regeneration. Cancer Res. 2008;68:10341–10348. doi: 10.1158/0008-5472.CAN-08-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 9.Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol. 2001;75:3851–3858. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slagle BL, Lee TH, Medina D, Finegold MJ, Butel JS. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Terradillos O, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 12.Doria M, Klein N, Lucito R, Schneider RJ. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger C, Mason WS, Zoulim F. Hepadnaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2977–3029. [Google Scholar]
- 15.Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 16.Chirillo P, et al. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cougot D, et al. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2007;282:4277–4287. doi: 10.1074/jbc.M606774200. [DOI] [PubMed] [Google Scholar]
- 18.Blum HE, et al. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HS, et al. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melegari M, Wolf SK, Schneider RJ. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J Virol. 2005;79:9810–9820. doi: 10.1128/JVI.79.15.9810-9820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sitterlin D, Bergametti F, Tiollais P, Tennant BC, Transy C. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene. 2000;19:4427–4431. doi: 10.1038/sj.onc.1203770. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Torii N, Hu Z, Jacob J, Liang TJ. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J Clin Invest. 2001;108:1523–1531. doi: 10.1172/JCI13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein NP, Bouchard MJ, Wang LH, Kobarg C, Schneider RJ. Src kinases involved in hepatitis B virus replication. EMBO J. 1999;18:5019–5027. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H, et al. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J Virol. 2005;79:5548–5556. doi: 10.1128/JVI.79.9.5548-5556.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J Virol. 2007;81:2656–2662. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newbold JE, et al. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bock CT, et al. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 30.Pollicino T, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- 32.Gunther S, et al. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollicino T, et al. Hepatitis B virus maintains its pro-oncogenic properties in the case of occult HBV infection. Gastroenterology. 2004;126:102–110. doi: 10.1053/j.gastro.2003.10.048. [DOI] [PubMed] [Google Scholar]
- 34.Levrero M, et al. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Park IY, et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 36.Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771–5778. doi: 10.1158/0008-5472.CAN-07-0529. [DOI] [PubMed] [Google Scholar]
- 37.Zheng DL, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol. 2009;50:377–387. doi: 10.1016/j.jhep.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 38.Werle-Lapostolle B, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology. 2009;390:122–129. doi: 10.1016/j.virol.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.