SUMMARY
An encounter between a DNA translocating enzyme and a DNA-bound protein must occur frequently in the cell but little is known about its outcome. Here, we developed a multi-color single molecule fluorescence approach to simultaneously monitor single stranded (ss) DNA translocation by a helicase and the fate of another protein bound to the same DNA. Distance-dependent fluorescence quenching by the iron-sulfur cluster of the archaeal XPD (Rad3) helicase was used as a calibrated proximity signal. Despite the similar equilibrium DNA binding properties, the two cognate ssDNA binding proteins, RPA1 and RPA2, differentially affected XPD translocation. RPA1 competed with XPD for ssDNA access. In contrast, RPA2 did not interfere with XPD-ssDNA binding but markedly slowed down XPD translocation. Mechanistic models of bypassing DNA-bound proteins by the Rad3 family helicases and their biological implications are discussed.
INTRODUCTION
Central to many DNA-repair machineries are DNA helicases, ubiquitous molecular motors that use the energy of nucleoside triphosphate (NTP) hydrolysis to move directionally along nucleic acids (von Hippel and Delagoutte, 2003; Wu and Hickson, 2006). Among these enzymes, superfamily II (SF2) helicases comprise the largest group of structurally related molecular motors and conformational switches involved in virtually every aspect of cellular nucleic acid metabolism (Gorbalenya and Koonin, 1993; Jankowsky and Fairman, 2007; Lohman et al., 2008; Pyle, 2008; Singleton et al., 2007). Consequently, defects in SF2 helicases are associated with a broad spectrum of disorders including premature aging, susceptibility to cancers, immunodeficiency and mental retardation (van Brabant et al., 2000). This is also true for the five human Rad3 helicases that comprise a subset of SF2 enzymes. Two of them, XPD and FancJ (Bach1), are involved in multiple DNA repair pathways. Mutations in XPD (ERCC2) helicase cause defects in nucleotide excision repair (NER) and result in hyperphotosensitivity and increased risk of skin cancer (van Brabant et al., 2000), while mutations in FancJ interfere with double stranded breaks repair causing predisposition to breast and ovarian cancers or lead to Fanconi Anemia due to an inability to repair interstrand crosslinks (Cantor et al., 2001; Gupta et al., 2006; Levitus et al., 2005).
All characterized members of the Rad3 family are bona fide DNA helicases that move along ssDNA with 5’→3’ polarity (Cantor et al., 2004; Farina et al., 2008; Hirota and Lahti, 2000; Pugh et al., 2008a; Rudolf et al., 2006; Sung et al., 1993; Voloshin et al., 2003). Similar to other helicases and helicase-like motors, translocation by Rad3 enzymes is the central ATP-powered activity that may drive dsDNA unwinding and remodeling of protein-nucleic acid complexes. Enzymes in this family differ from the other 24 identified families of SF2 helicases (Flaus et al., 2006) by the two unique insertions in helicase domain 1: one insertion is stabilized by an iron-sulfur (FeS) cluster (Pugh et al., 2008a; Rudolf et al., 2006), while the second forms an Arch domain characterized by a unique structural fold (Fan et al., 2008; Liu et al., 2008; Wolski et al., 2008). The FeS cluster containing insertion forms an auxiliary domain important for coupling of ATP hydrolysis to ssDNA translocation. Additionally, the FeS domain forms a secondary ssDNA binding site, which positions the helicase in an orientation consistent with duplex unwinding (Pugh et al., 2008a). Furthermore, these enzymes readily bind to and translocate along ssDNA which may allow them to carry out other physiologically important functions in addition to duplex unwinding such as resolution of G-quadruplex structures by the FancJ helicase (Wu et al., 2008). Duplex unwinding by FancJ (Gupta et al., 2007) and XPD (Pugh et al., 2008b), as well as remodeling of the G-quadruplexes by FancJ helicase (Wu et al., 2008) is greatly facilitated by ssDNA-binding proteins (RPAs) suggesting an important functional interaction between the RPA-coated lattice and a Rad3 family helicase.
Here we describe a single molecule approach which allowed us to follow translocation by the archaeal Rad3 family helicase, Ferroplasma acidarmanus XPD, in isolation and in the presence of two cognate ssDNA-binding proteins, RPA1 and RPA2. We observed that translocation by XPD along ssDNA is differentially affected by the presence of either of its two cognate ssDNA-binding proteins, RPA1 and RPA2. XPD translocation along RPA2-ssDNA complex may represent a generalized mechanism employed by the Rad3 family helicases for targeting the cognate DNA processing intermediates.
RESULTS
The FeS of XPD helicase quenches Cy3 fluorescence in a distance-dependent manner
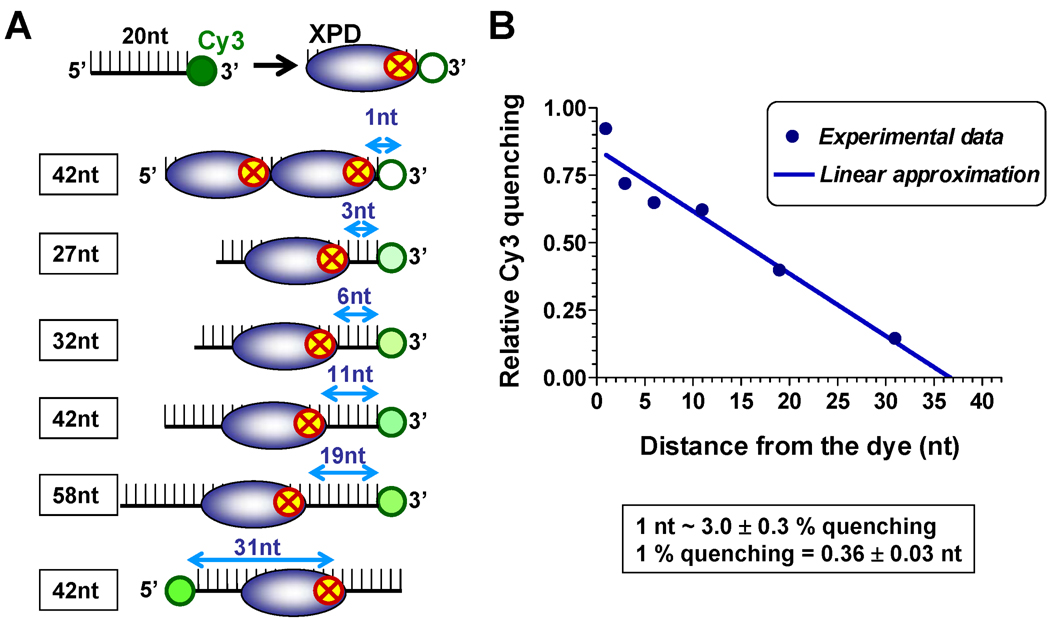
We recently demonstrated that binding of XPD to a DNA substrate proximal to the Cy5 dye results in significant fluorescence quenching mediated by the FeS cluster present in this helicase (Pugh et al., 2008a). In this study, we substituted the more photo-stable Cy3 fluorophore for Cy5 and examined the relationship between Cy3 quenching, and the distance between the FeS cluster and Cy3 (Figure 1). Upon binding to ssDNA, XPD occluded approximately 20 nucleotides of ssDNA with the FeS cluster positioned near the 3’-end of occluded region (Figure S1). Cy3 has proven useful in monitoring protein-nucleic acid interaction by tracking an increase in its fluorescence intensity, usually 10–50%, upon binding of a protein in close proximity (Fischer et al., 2004; Luo et al., 2007; Myong et al., 2009). This effect is markedly different from the almost 100% Cy3 quenching observed in our study (Figure 1B and Figure S1). Although the mechanism by which fluorescence is quenched by FeS cluster has not been reported, the iron atom in hemin was shown to quench GFP fluorescence by a mechanism involving intermolecular long-range dipole-dipole coupling (Takeda et al., 2003). An energy transfer similar to this may be responsible for the FeS cluster-dependent Cy3 quenching property of XPD. Regardless of the exact mechanism, the change in Cy3 fluorescence could be linearly approximated to the change in the position of the helicase (Figure 1B). A change in the helicase position by 1 nucleotide corresponded to a 3.0 ± 0.3% change in Cy3 fluorescence relative to the original signal. Therefore, Cy3 quenching can be used as a specific and sensitive probe for XPD-ssDNA interaction and for accurately tracking the position of XPD on the DNA lattice.
Figure 1. Calibration of Cy3 fluorescence quenching by the FeS cluster of XPD helicase.
(A) Experimental design: The quenching experiments were carried out under stoichiometric binding conditions. When XPD and the ssDNA substrate were present in equimolar concentrations, XPD binding positions were uniquely dispersed along the 42mer oligonucleotide labeled with Cy3 at one end. The ensemble-average position was expected to coincide with the center of the oligonucleotide. When XPD was present at two-fold excess concentration over the DNA substrate, we expected two helicase molecules to be bound to each oligonucleotide occluding its complete length. Thus, for each helicase: ssDNA ratio, we predicted the distance between the front edge of the helicase and Cy3. (B) Quantification of the quenching magnitudes (blue circles). The solid line represents linear approximation of Cy3 quenching as a function of the distance between the leading edge of XPD and the dye.
Visualization of XPD binding and translocation on individual ssDNA molecules
Using Cy3 quenching as a proximity indicator, we visualized XPD binding to and translocation along the Cy3-labeled ssDNA at the level of individual nucleo-protein complexes. Oligonucleotides biotinylated at one end and labeled with the Cy3 dye at the opposite end were immobilized on the surface of the total internal reflection microscopy (TIRM) flow chamber. This approach yielded single molecule fluorescence trajectories with distinct quenching patterns that were dependent on both the length of the oligonucleotide and its polarity (Figure 2A–C). When Cy3 was placed near the 5’-end of a 42mer ssDNA, binding of XPD was observed as an instantaneous quenching followed by gradual recovery of Cy3 fluorescence, which reflected XPD translocation away from the dye (Figure 2A). When Cy3 was incorporated at the 3’-end of a 42mer ssDNA, translocation by XPD towards the dye manifested in a gradual decrease in fluorescence, and its dissociation was observed as an instantaneous recovery of fluorescence (Figure 2B). For both types of ssDNA substrates labeled with Cy3 at the 5’ or 3’-end, the binding and translocation initiation sites did not have a preferred location along ssDNA (Figure S2). To avoid loss of the substrate associated with XPD translocation towards the surface and disrupting biotin-avidin anchoring, all subsequent experiments were carried out with the substrate immobilized through the 5’ end and labeled with Cy3 dye at the 3’ end.
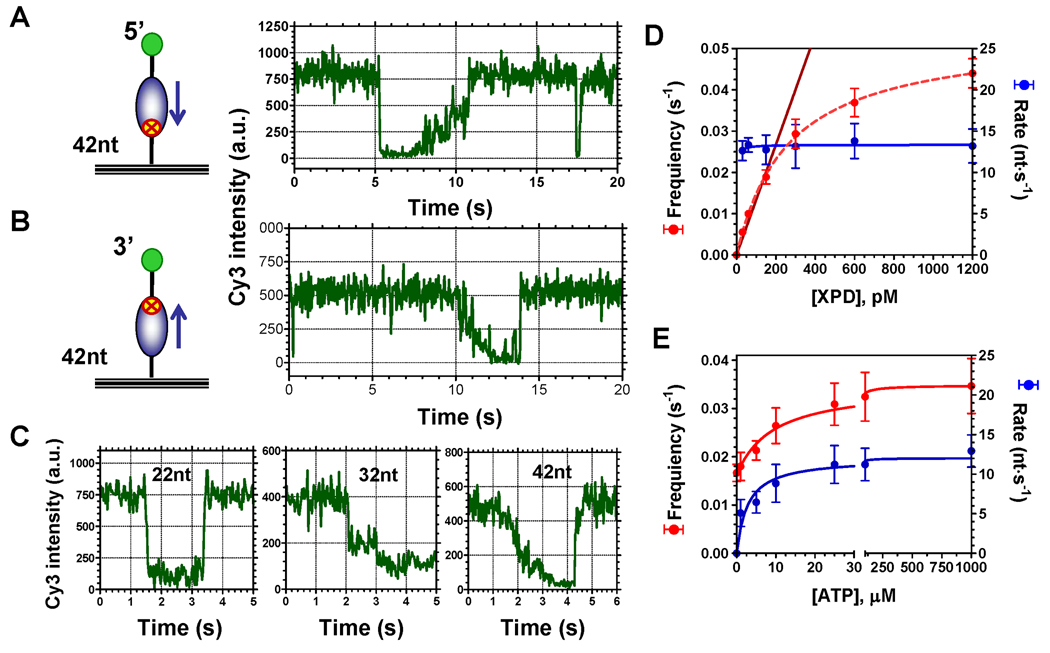
Figure 2. Cy3 quenching allows visualization and analysis of XPD binding to and translocation along ssDNA.
(A) Schematic depiction of the immobilized ssDNA (42mer) with Cy3 at the 5’-end and a representative single-molecule trajectory of XPD-dependent Cy3-quenching in the presence of ATP (right panel). Cy3 fluorescence intensity abruptly decreased as the result of XPD binding and then gradually recovered as the helicase translocated away from the dye in the 5’→3’ direction. (B) Surface-tethered ssDNA (42mer) labeled with Cy3 at the 3’-end. A representative fluorescence trajectory shows that XPD translocation along this substrate resulted in gradual quenching of Cy3 intensity followed by its full recovery when XPD dissociated from the substrate. (C) Representative fluorescence trajectories for the substrates of 22, 32 and 42 nucleotides long carrying the Cy3 dye at the 3’-end depict the length-dependence of the binding/translocation patterns. (D) Effect of XPD concentration on the frequency of the observed events (red) and on the rate of XPD translocation (blue). (E) Both frequency of observed translocation events (red) and translocation rate (blue) were functions of ATP concentration.
The gradual quenching signal attributed to translocation was detected only in the presence of ATP, whereas binding events were observed in the absence of ATP or in the presence of non-hydrolyzable ATP analogues (Figure S3). In agreement with our estimated binding site size of ~ 20 nt, only binding and no translocation was observed on almost all of the 22 mer ssDNA molecules (Figure 2C, left). When the length of the substrate was increased to 32 nucleotides we observed Cy3 quenching, which corresponded to XPD binding at a distance between 0 and 10 nucleotides from the dye, followed by translocation towards the Cy3 labeled 3’-end (Figure 2C, middle). Binding of XPD to and its translocation along the 42 nucleotide ssDNA yielded trajectories similar to that described for 22-mer and 32-mer ssDNA. In addition, a significant fraction of observed events displayed gradual fluorescence decrease without an initial quantal drop in fluorescence seen for shorter DNA molecules. We attributed trajectories of this type to events where initial binding of XPD positioned its FeS cluster beyond the limit of the fluorescence quenching detection (Figure 2C, right).
Translocation experiments were carried out at several XPD and ATP concentrations to analyze kinetic parameters of XPD translocation on ssDNA. Recorded fluorescence trajectories yielded the frequency of binding/translocation events. The translocation rates were calculated from analysis of the individual events (Figure S4) using the quenching vs. distance calibration shown in Figure 1. Distributions of the rates for different XPD and ATP concentrations are shown in the Figure S5. Nearly all helicase molecules reached the ssDNA end without detectable pauses or dissociation. The translocation rate was not affected by the enzyme concentration and was 13 ± 2 nt/s for the tested range of conditions (between 30 pM and 1.2 nM XPD) (Figure 2D). In contrast, the frequency of XPD binding/translocation events increased linearly with increasing XPD concentration in the range of 0–200 pM XPD. In this range, each observed event represented binding and translocation by an individual helicase molecule. Therefore, most of the subsequent single-molecule data were collected at 150 pM XPD, which is well within the linear range. However, to compensate for a low frequency of XPD translocation events at low ATP concentrations, ATP titration experiments (Figure 2E) were carried out at 600 pM XPD. Under these conditions some of the observed events may be caused by more than one enzyme. In most cases, such events are obvious (such as shown in Figure S4D) and were excluded from analysis. Both the frequency of observed events and the average translocation rates were functions of ATP concentration (Figure 2E). Dependence of the translocation rate on ATP concentration yielded kcat = 12 ± 1 nt/s and KM = 2.8 ± 1.0 µM. The frequency of observed events was fitted to the following equation: where f1 and (f1+f 2) are frequencies of XPD and XPD-ATP complex binding to ssDNA, respectively. This analysis resulted in the KATP = 9.4 ± 2.1 µM, f1 = 0.016 ± 0.001 s−1 and f2 = 0.019 ± 0.001 s−1, indicating that the ATP bound form of XPD displays two-fold increase in binding rate to the DNA relative to ATP-free form. All individual binding events resulted in a single round of translocation on the same ssDNA in contrast to the superfamily I helicase, E. coli Rep, which translocates on the same segment of ssDNA multiple times without dissociating (Myong et al., 2005).
Two cognate ssDNA-binding proteins differentially affect XPD-ssDNA interactions
ssDNA-binding proteins are ubiquitous and indispensable for numerous aspects of cellular DNA metabolism and are known to affect activities of DNA helicases (Cadman et al., 2005; Cadman and McGlynn, 2004; Cui et al., 2004; Doherty et al., 2005; Gupta et al., 2007; Shen et al., 2003; Shereda et al., 2007). F. acidarmanus has two distinct ssDNA-binding proteins, RPA1 and RPA2 (Robbins et al., 2005). Each subunit of the homodimeric RPA1 contains 2 oligonucleotide/oligosaccharide binding (OB) folds and a Zn-finger with an overall binding site size of 20 nucleotides per dimer. Monomeric RPA2 contains a single OB fold and occludes approximately 5 nucleotides (Pugh et al., 2008b). The two proteins display different ssDNA binding modes: RPA1 stretches ssDNA, while RPA2 binding results in DNA bending (Robbins et al., 2005). Under our experimental conditions, both RPA1 and RPA2 bind ssDNA with affinity in the low nM range (Figure S6A). Therefore, 100 nM RPA used in our single-molecule assays should fully saturate ssDNA. Under the steady-state conditions, neither of the two proteins completely prohibited XPD translocation on ssDNA as evident from its ability to facilitate streptavidin dissociation from biotin conjugated to the 3’ end of ssDNA (Figure S6B).
We first probed whether either of the two proteins affected XPD binding to and translocation along ssDNA. FeS cluster dependent fluorescence quenching propitiously allowed us to distinguish XPD binding from RPAs binding, and therefore to analyze XPD translocation on ssDNA coated with RPA1 or RPA2. Although RPA2 binding in the vicinity results in a small increase in Cy3 fluorescence, calibration of the Cy3 quenching by XPD in the presence of RPA2 showed that RPA2 has little or no effect on the quenching magnitude at several tested distances (Figure S6C). In addition, RPA2 did not impair XPD binding to ssDNA (Figure S6D). In contrast, XPD affinity for ssDNA was reduced in the presence of RPA1 complicating calibration of the quenching signal. However, since the quenching magnitudes at saturating amounts of XPD were the same with and without RPA1, we used the same conversion value (1nt ~ 3% quenching) for protein-free and for RPA-coated DNA.
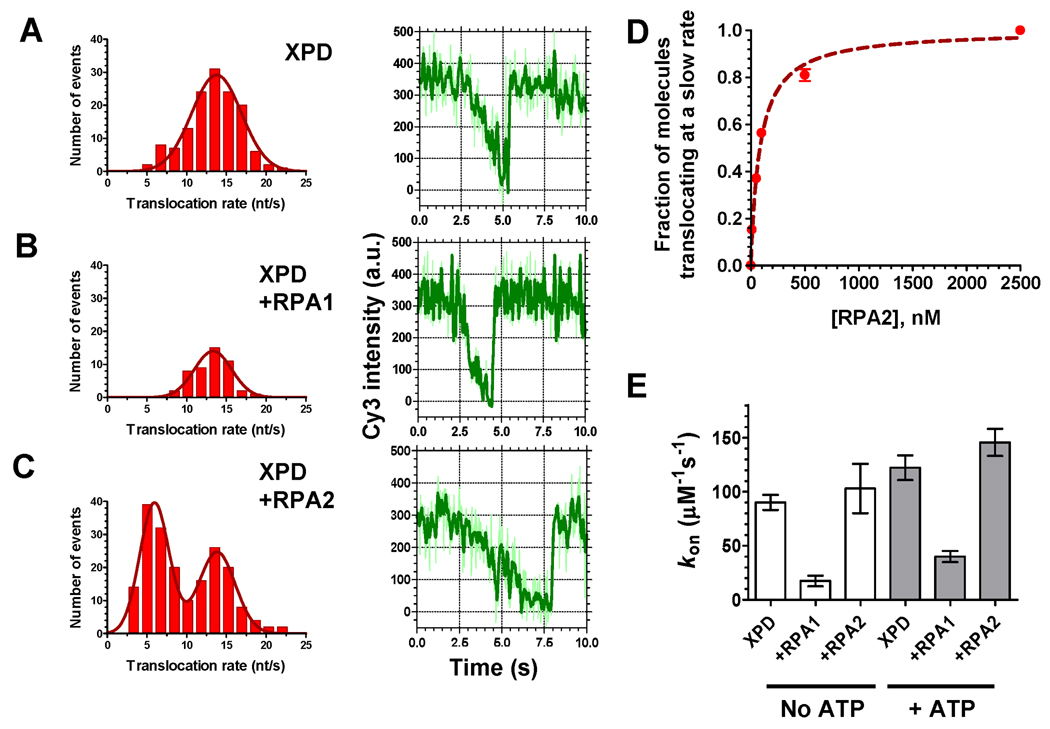
The translocation rate of XPD alone on free ssDNA was compared with that on RPA1 or RPA2 coated-ssDNA. Figure 3A–C shows distributions of the translocation rates obtained by analysis of 600 fluorescence trajectories recorded at each condition (left) and representative time trajectories of Cy3 fluorescence intensity (right). Analysis of the translocation events observed on protein-free ssDNA yielded a Gaussian distribution with the mean translocation rate of 13.8 ± 3.1 nt/s (Figure 3A). The mean translocation rate (13.3 ± 2.4 nt/s) in the presence of RPA1 (Figure 3B) was similar to that on the protein-free ssDNA. Given the competition between RPA1 and XPD for access to ssDNA and their comparable binding site sizes, the observed XPD translocation is likely to occur along naked ssDNA as a result of spontaneous or XPD-facilitated dissociation of RPA1 within the detection range of the iron-dependent Cy3 quenching. Interestingly, the distribution of XPD translocation rates in the presence of RPA2 displayed two maxima, one identical to that observed in the absence of RPA (13.8 ± 2.4 nt/s) and the second at a >2-fold reduced rate, 5.9 ± 1.9 nt/s (Figure 3C).
Figure 3. The two F. acidarmanus RPAs differentially affect XPD binding to and translocation along ssDNA.
(A) In the case of XPD translocation on protein-free ssDNA, 138 individual translocation events were observed in 600 analyzed trajectories. Individual rates were binned in 5% fluorescence change per second intervals, plotted and analyzed by fitting the resulting histogram to normal distribution. The average and standard deviation were used to calculate the average translocation rate. (B) Only 48 events were observed and analyzed per 600 trajectories recorded in the presence of 100 nM RPA1. (C) In the presence of 100 nM RPA2, the frequency of observed events slightly increased (193 events were observed and analyzed per 600 traces). The histogram displayed two peaks and was fitted with double normal distribution resulting in the two characteristic rates. Representative fluorescence trajectories for each condition are shown on the right of respective histograms. (D) Fractions of XPD molecules translocating at slow rates were calculated as the ratio of area under the left hump of double Gaussian distribution to the total area under the distribution curve. (E) Kinetic association constant, kon was calculated from the frequency of the quenching events recorded at 150 pM XPD. Error bars represent standard deviation for three sets of 100 randomly selected trajectories.
It is possible that the slower translocation rates were observed when XPD had to bypass RPA2 by sliding it along the ssDNA, sharing the lattice with RPA, or displacing it. The faster rate may represent XPD translocation events where RPA2 has dissociated from or was distributed less densely along ssDNA. Indeed, fraction of XPD molecules translocating with slow rates increased in an RPA2 concentration-dependent manner, while the fraction of molecules translocating at fast rates decreased (Figures 3D). The RPA2 dependent transition of fast to slower translocation rate was bimodal (Figure S7A). Additionally, the lifetime of XPD on ssDNA in the presence of RPA2 increased due to slower translocation rate (Figure S7B). In addition to trajectories displaying fast or slow translocation rates, some of the trajectories recorded in the presence of RPA2 exhibited a biphasic behavior where the translocation of the helicase suddenly accelerated or slowed down (Figure S8).
Next, we compared the association constants (kon) for the XPD-ssDNA interactions with or without 100 nM RPA1 and RPA2 at 150 pM XPD. As expected from the equilibrium binding assays (Figure S6D), coating ssDNA with RPA1 interfered with XPD binding to the immobilized ssDNA substrates (Figure 3E) with ~5-fold decrease in the apparent kon. In contrast, binding of XPD to ssDNA was unchanged or slightly increased in the presence of the smaller, monomeric RPA2. Combined, our observations demonstrated that XPD binds efficiently to and translocates along RPA2-coated ssDNA while RPA1 competes XPD for ssDNA binding.
Two distinct mechanisms of XPD translocation along RPA coated ssDNA
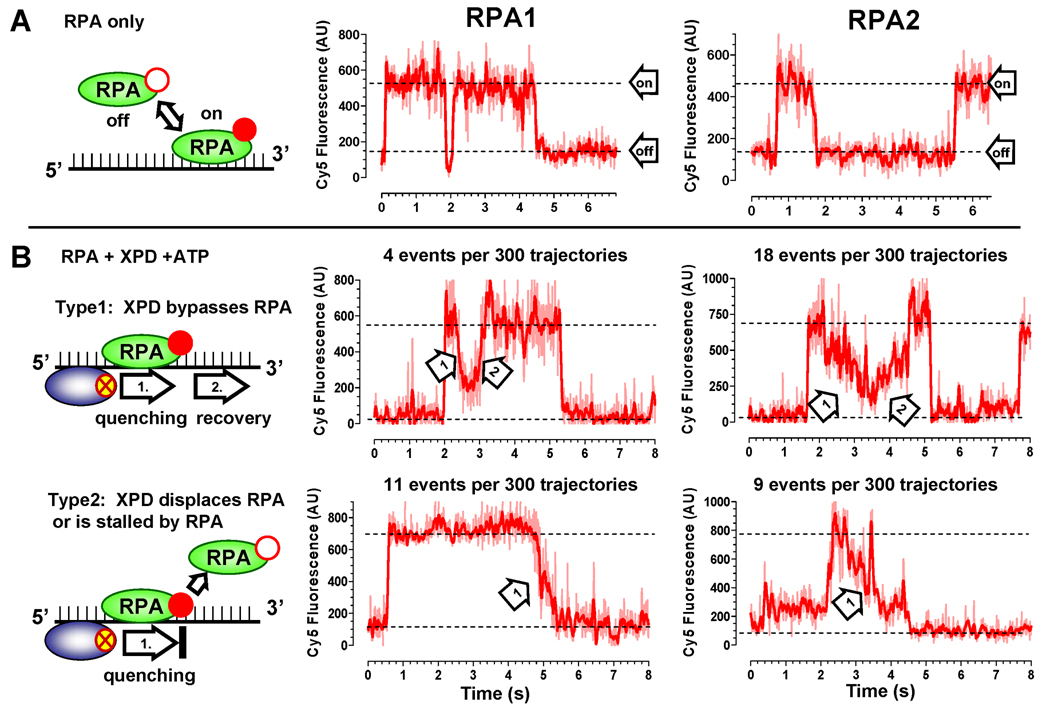
Elucidating the relationship between XPD translocation and the presence of RPA on ssDNA required simultaneous observation of RPA and XPD. To simultaneously detect RPA and XPD, we combined the FeS cluster-dependent quenching with single-molecule fluorescence resonance energy transfer (smFRET). Both RPAs were site-specifically labeled with Cy5 at the N-terminus. Incorporation of approximately 1 Cy5 dye per monomer of RPA2 did not impair binding to ssDNA while RPA1 binding was only slightly affected (Figure S9). Binding of RPA to and its presence on ssDNA was monitored by measuring FRET between Cy3 (donor) incorporated into 3’-end of ssDNA and Cy5 (acceptor) incorporated into RPA (Figure 4A). When saturating amounts (100 nM) of Cy5 labeled RPA1 or RPA2 were added to the flow chamber containing immobilized Cy3-ssDNA, all recorded trajectories displayed a combination of high and low FRET states that reflected RPA binding to and dissociation from ssDNA (the “on” and “off” states are indicated for the representative trajectories shown in Figure 4A). While several RPA molecules may be bound to each ssDNA substrate, our experimental scheme follows the protein most proximal to Cy3.
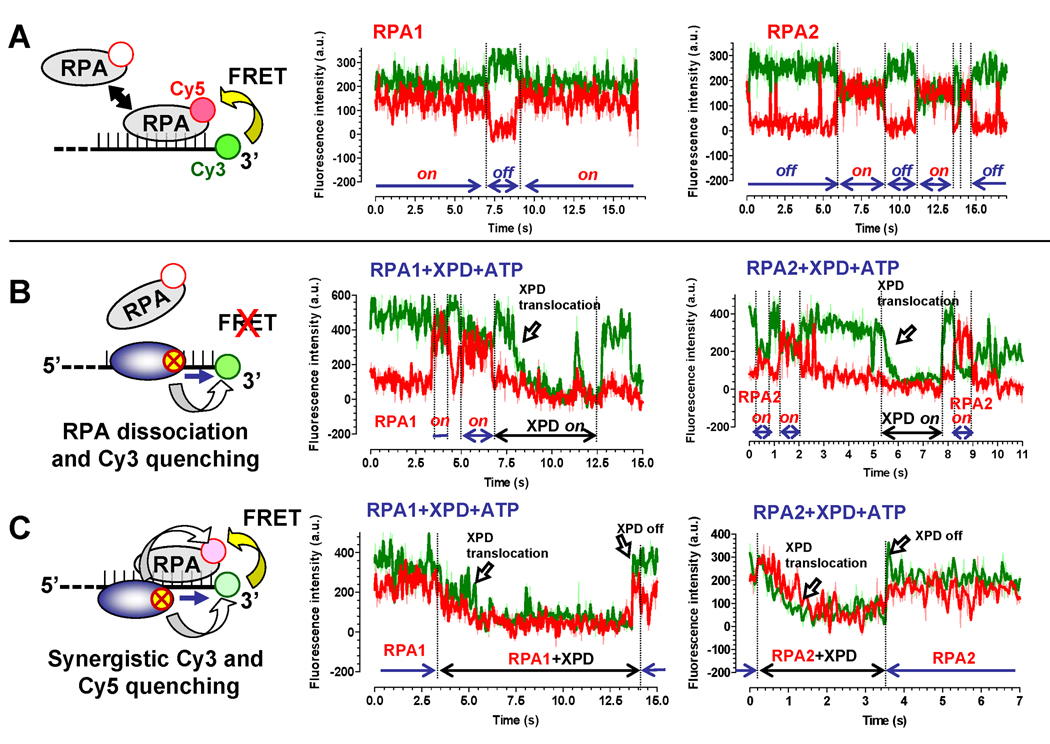
Figure 4. Two observed modes of XPD translocation along RPA coated ssDNA.
(A) Binding of the Cy5-labeled RPAs to the Cy3-labeled ssDNA (schematically depicted on the left) can be detected by following FRET between the two dyes. Fluorescence of the Cy3 and Cy5 dyes is shown in green and red, respectively. (B) If RPA (100 nM) is displaced by the translocating XPD helicase (150 pM) or dissociates from ssDNA, we expected to observe uncoupled fluorescence trajectories. Examples of the fluorescence trajectories displaying such a behavior are shown for ssDNA coated with both RPA1 (middle) and RPA2 (right). (C) If XPD (150 pM) translocates over the bound RPA (100 nM) without displacing it from ssDNA, we expected to observe a synergistic quenching followed by the recovery of Cy3 and Cy5 fluorescence.
Next, we added 150 pM of XPD and 1 mM ATP to the reaction and expected to observe one of the two possible types of fluorescent trajectories. If XPD translocation promotes RPA displacement, we expected gradual Cy3 quenching associated with XPD translocation toward Cy3-labeled ssDNA end (arrow labeled “XPD translocation” in Figure 4B) to coincide with or to be immediately preceded by a sudden disappearance of the Cy5 signal indicative of dissociation of Cy5-labeled RPA (Figure 4B). In this scenario, eventual dissociation of XPD would result in recovery of Cy3 fluorescence but not that of Cy5. If the helicase bypasses the bound RPA molecule without displacing it, Cy5 emission, which is due to FRET from Cy3, should decrease concomitantly with that of Cy3 (arrow labeled “XPD translocation” in Figure 4C). The two effects that may be responsible for this fluorescence change are: (i) reduced excitation of the Cy5 dye due to the quenched intensity of the Cy3 dye, and (ii) direct quenching of the Cy5 emission by the translocating XPD. When XPD dissociates, both Cy3 and Cy5 should recover their fluorescence simultaneously (arrow labeled “XPD off” in Figure 4C). Among 50 analyzed translocation events in the presence of RPA1 and XPD, 78% events displayed patterns characteristic of XPD translocation with RPA displacement from ssDNA (Figure 4B), and only 22% translocation patterns displayed synergistic Cy3 and Cy5 quenching/recovery patterns (Figure 4C). Among 71 trajectories recorded in the presence of RPA2, 59% of events displayed patterns characteristic of RPA2 dissociation and 41% displayed synergistic quenching.
Notably, the average XPD translocation rate associated with events we attributed to RPA dissociation (14.3 ± 2.6 and 13.3 ± 2.4 nt/s in the presence of Cy5-labeled RPA1 and RPA2, respectively) was similar to that observed on the protein-free ssDNA (13.8 ± 2.4 nt/s), in the presence of unlabeled RPA1 (13.3 ± 2.4 nt/s) and to the average “fast” translocation rate in the presence of RPA2 (13.8 ± 2.4 nt/s). Although we could not use our calibration parameter to unambiguously calculate the XPD translocation rate in the synergistic quenching/recovery patterns (such as shown in Figure 4C) due to a possibly complex interplay between FeS cluster-mediated quenching and FRET between Cy3 and Cy5 dyes, it took noticeably longer for XPD to achieve complete quenching of both dyes from the initial FRET state than expected for the protein-free ssDNA (if take into consideration only Cy3 signal, the rates of XPD translocation deduced from synergistic quenching trajectories were 8.9 ± 4.3 and 6.4 ± 2.9 nt/s for Cy5-RPA1 and Cy5 RPA2 respectively, which correlated well 5.9 ± 1.9 nt/s “slow” average rate for XPD translocation on the RPA2-coated ssDNA). This suggests that the slower translocation rate observed in the presence of unlabeled RPA2 (Figure 3C) might reflect the scenario depicted in Figure 4C.
Evidence that XPD can bypass bound RPA
When the Cy3 and Cy5 dyes displayed a synergistic behavior, we attributed this behavior to translocation of XPD over the bound RPA. However, since the dye is located at the very terminus of the ssDNA molecule, we could not completely rule out the possibility that if the helicase is to stop before the last bound RPA2 (i.e. 4 – 5 nucleotides from the 3’-end), we may still observe simultaneous quenching and recovery of the Cy3 and Cy5 dyes. To test if XPD indeed bypasses bound RPAs, we monitored Cy5 fluorescence associated with ssDNA bound RPAs by directly exciting Cy5 (Figure 5). In this experiment, RPA concentration was lowered to 5 nM. In this range of RPA concentration, appearance of the Cy5 signal represented an individual RPA molecule binding to an immobilized ssDNA. In the absence of XPD helicase, we observed two-state fluctuations in the Cy5 intensity (Figure 5A), which were attributed to binding and dissociation of RPA. When XPD and ATP were present in the reaction mixture, we observed two distinct types of fluorescence trajectories (Figure 5B). Type 1 trajectories displayed gradual Cy5 quenching events immediately followed by gradual fluorescence recovery. We attributed these quenching/recovery (indicated as arrow 1 and arrow 2) patterns to the FeS cluster-dependent quenching of the Cy5 fluorescence. In this scenario, the gradual fluorescence quenching by an approaching XPD helicase would reach its maximum quenching when the distance between the RPA-linked Cy5 dye and the FeS cluster was the shortest, and the subsequent translocation of XPD away from the dye would result in recovery of the fluorescent signal, indicating that RPA remained bound to ssDNA through the reaction. In the type 2 trajectories, XPD translocation only caused gradual quenching of Cy5 and no recovery. These types of trajectories may reflect events where XPD displaced RPA, pushed RPA off the edge of the molecule, or was stopped by encountering a bound RPA (indicated as arrow 1). Among 300 randomly selected traces collected in the presence of RPA1 and XPD, only 4 events displayed Type 1 patterns characteristic of XPD bypassing bound RPA without displacement and 11 events displayed Type 2 trajectories characteristic of XPD translocation with RPA displacement. The small number of events observed in these experiments was limited by the low concentration of RPAs. Cy5-RPAs were the only species that produced fluorescence signal upon red laser excitation. We could only observe XPD translocation when XPD and RPA binding to ssDNA coincided both spatially and temporarily. Under selected experimental conditions both RPA1 and RPA2 spent only a small fraction of time on DNA, yielding only a few events when both ssDNA binding protein and the helicase were present on DNA spatially proximal and simultaneously. Among 300 traces collected in the presence of RPA2, 18 events displayed Type 1 patterns and 9 events displayed Type 2. The higher frequency of events where XPD bypassed RPA2 compared to RPA2 dissociation in the middle of the translocation event and a higher frequency of RPA1 dissociations were consistent with results shown in Figure 4.
Figure 5. Translocation of XPD observed through quenching of directly excited Cy5- RPA1 and 2.
(A) Visualization of ssDNA binding by 5 nM Cy5-RPA. The ssDNA binding/dissociation of either RPA1 (middle) or RPA2 (right) was monitored by following change in Cy5 fluorescence intensity (the “on” and “off” states are indicated by respective dotted lines). (B) The effect of XPD translocation on DNA bound RPAs. Two distinct types of Cy5 fluorescence trajectories were observed in the presence of 150 pM XPD and 1 mM ATP. In the type 1, the Cy5 fluorescence was quenched and recovered resulting in a symmetric trajectory (Type 1). Gradual decrease and increase of Cy5 intensity reflected XPD approaching (arrow 1) and moving away (arrow 2) from Cy5 labeled RPA. In the second type of trajectories we observed only gradual quenching of Cy5 and no recovery (Type 2).
XPD translocation facilitates RPA1 dissociation from ssDNA
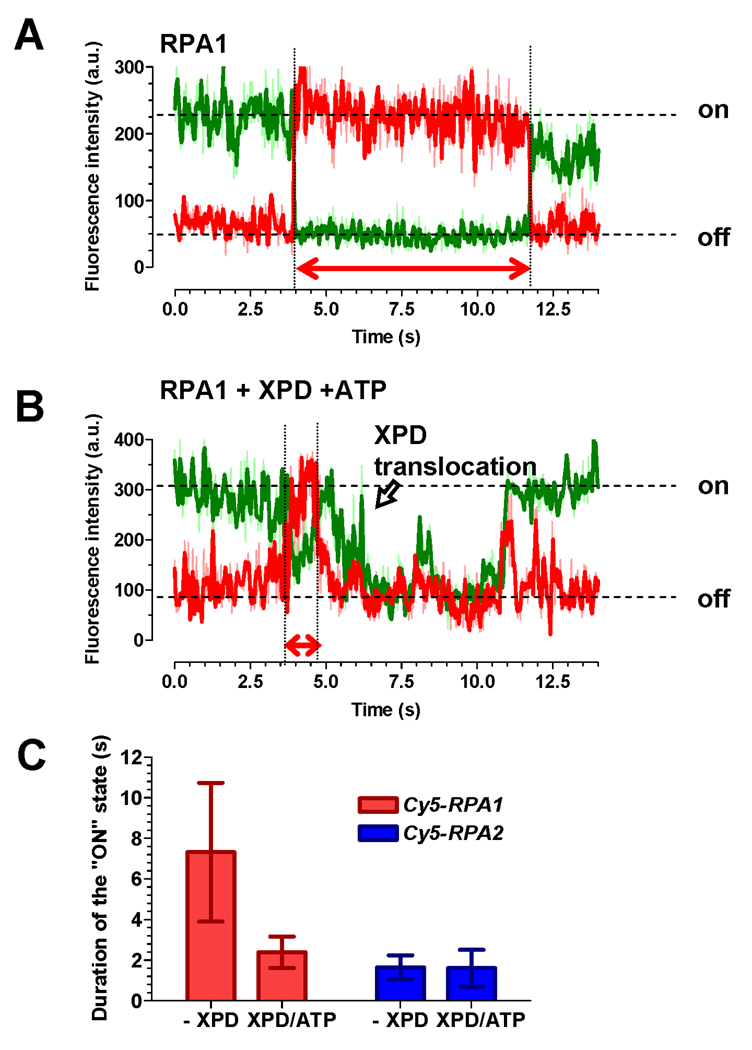
Although the data above indicate that XPD can bypass RPA without displacing it, the events in which RPA dissociation immediately preceded XPD translocation represented the majority of events for RPA1 and were observed for RPA2 as well. Dissociation of the RPAs could result from XPD-facilitated displacement or from spontaneous dissociation of RPA from the ssDNA lattice. To determine whether XPD translocation actively promotes RPA dissociation from ssDNA, we calculated the life time of both RPA1 and RPA2 on ssDNA immediately preceding XPD translocation and compared it to that calculated in the absences of XPD. In these experiments, we followed RPA1 or RPA2 binding by monitoring FRET between Cy3 dye on ssDNA and Cy5-labeled RPA. The Cy5-RPA concentration was decreased to 5 nM to allow observation of binding and dissociation by individual RPA molecules (Figure 6A). Thus, we were able to evaluate the effect of XPD translocation on the lifetime of RPAs on ssDNA. For this we selected and measured the dwell times of RPA “on” states directly preceding XPD translocation (Figure 6B). The average lifetime of RPA1 on ssDNA just before XPD translocation (1.65 ± 0.77 s) was ~5 times lower than in the absence of XPD and ATP (7.32 ± 3.42 s). In contrast, the lifetime of RPA2 on ssDNA was not affected by XPD translocation (Figure 6C).
Figure 6. XPD facilitates displacement of RPA1 but not RPA2.
(A) Representative FRET trajectory of Cy5-RPA1 binding and dissociating from Cy3-labeled ssDNA. The “on” and “off” states for the Cy5 signal (red) are indicated on the right. The duration of the ssDNA bound state of Cy5-RPA1 is indicated by the arrow. (B) Representative FRET trajectory of Cy5-RPA1 recorded in the presence of 150 pM XPD and 1 mM ATP. The duration of the ssDNA-bound state of Cy5-RPA1 just before XPD translocation was measured as indicated by the arrow. (C) Average duration of the XPD “on” state. The error bars represent standard deviation for 25 individual events.
DISCUSSION
DNA helicases frequently encounter ssDNA binding proteins during various DNA processing events. Using several complementary single-molecule fluorescence approaches we have analyzed binding and translocation of the archaeal SF2 helicase, XPD, along ssDNA decorated with ssDNA-binding proteins. Our approaches allowed for real-time observation of the interplay between the two proteins on individual DNA molecules. Our models interpreting the observed fluorescence trajectories and the differential effects of the two F. acidarmanus RPAs (RPA1 and RPA2) on XPD-ssDNA interaction are summarized in Figure 7. The illustration depicts distinct modes with which RPA1 extends ssDNA upon binding, while RPA2 bends the DNA strand pre-configuring it for XPD binding. RPA1 strongly competes with XPD for binding to ssDNA, whereas, RPA2 does not impair, but rather slightly facilitates ssDNA binding by XPD. In the presence of RPA2, translocation by XPD along ssDNA was slower than that observed for protein-free ssDNA. Our results further indicate that an encounter by XPD of a bound RPA may result in one of the two possible outcomes: it may bypass RPA without displacing it from the DNA or may facilitate RPA dissociation. In a majority of the observed events, XPD translocation was coupled to RPA1 dissociation (Figure 4 and Figure 5). Analysis of the dwell-times for RPA1 bound to ssDNA directly preceding XPD translocation suggested that its dissociation was actively facilitated by XPD (Figure 6). This facilitated displacement may allow XPD to overcome obstacles encountered during translocation. In contrast, RPA2 is not displaced from ssDNA during translocation by XPD helicase in a majority of events. Predominantly, we observed coexistence of RPA2 and XPD helicase. In a smaller fraction of events where XPD translocation coincided with RPA2 dissociation, its dissociation was spontaneous and not facilitated by the helicase.
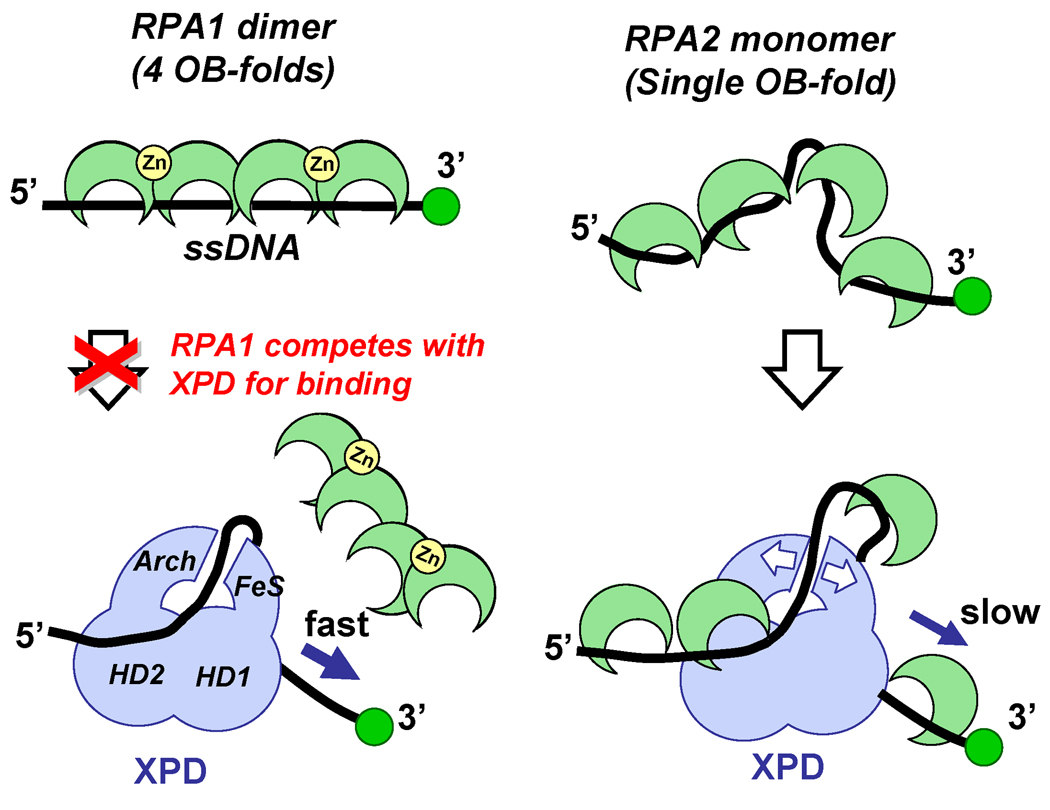
Figure 7. Model for XPD targeting to RPA2 coated ssDNA and its translocation.
XPD is schematically represented by the light blue figure. OB-folds (RPA1 and RPA2) and Zn-finger (RPA1) domains are depicted as green crescents and yellow circles, respectively. Stretching of the ssDNA molecule by RPA1 may inhibit XPD–ssDNA interaction, while ssDNA that is pre-bent due to the interaction with RPA2 represents a substrate that can be accommodated between the two ssDNA binding sites on XPD. Subsequent progression of XPD along a ssDNA lattice has to occur either by displacing bound RPA or translocation over it. The small binding site size of RPA2 and its dynamic association with ssDNA may result in ssDNA continuously occupied by ssDNA-binding proteins during XPD translocation.
The most attractive model explaining how XPD bypasses RPA2 is that XPD coexists on the same ssDNA lattice with and translocates over the RPA proteins. Although unprecedented, this proposition is not entirely counterintuitive because SF2 helicases are known to translocate along the phosphodiester backbones of nucleic acids (Singleton et al., 2007), whereas ssDNA binding by eukaryotic and archaeal OB folds involves primarily nucleic acid bases (Bochkarev et al., 1999; Kerr et al., 2003; Lei et al., 2004). While the structure of XPD bound to DNA is not available, the proposed DNA binding groove may be sufficiently large to accommodate RPA coated ssDNA (Fan et al., 2008; Liu et al., 2008; Wolski et al., 2008). However, the donut-like shape formed by helicase domain 2 and the domains unique to Rad3 family of helicases, the Arch and the FeS domains (Fan et al., 2008; Wolski et al., 2008) provides only a 15–20 Å wide hole for ssDNA passage. This is too narrow to accommodate a 30 Å wide ssDNA-RPA complex (Bochkarev et al., 1999). Since DNA helicases are known to undergo large conformational changes and domain swiveling upon binding to DNA (Korolev et al., 1997; Lee and Yang, 2006; Soultanas et al., 1999), we envision that interaction with DNA may result in a larger opening. Moreover, if ssDNA is indeed threaded through the hole between the Arch and FeS domains, the two domains have to separate to allow XPD to bind to its natural substrate. This is because, as an integral part of the TFIIH complex involved in nucleotide excision repair and transcription initiation (Boulikas, 1996), the physiological substrate for XPD binding is expected to be a bubble structure where the two complementary strands are transiently separated providing no free ssDNA end for initial entry. It is also possible that the contacts between the Arch and FeS cluster domains are dynamic promoting transient separation of the two domains, which can be further facilitated by XPD binding to the RPA-ssDNA complex. Such conformational rearrangements may represent the rate limiting step resulting in the slower translocation rate observed when XPD bypasses bound RPA2.
RPA2 has been found to aid XPD-mediated DNA unwinding to a greater extent than RPA1 (Pugh et al., 2008b). The difference between the two RPAs may represent a mechanism for targeting the archaeal NER helicase to its cognate DNA processing intermediates. XPD has at least two DNA binding sites (Pugh et al., 2008a) which are likely located on the opposite faces of the helicase (Wolski et al., 2008) (schematically depicted in Figure 7). The small binding site size (5 nt) of RPA2 and its ssDNA bending property (Robbins et al., 2005) may promote XPD binding by providing a preferred ssDNA configuration. In contrast, inhibition of XPD–ssDNA interaction by RPA1 may stem from its binding site size (20nt per homodimer) and binding mode that results in an extended and likely inflexible RPA1-ssDNAcomplex (Robbins et al., 2005). It is possible that each of the Rad3 family helicases has its own cognate ssDNA binding protein that provides an ideal substrate for targeting and translocation by the respective helicase, similarly to F. acidarmanus RPA2 and XPD.
An ssDNA binding protein consisting of a single OB fold is widely conserved among metazoans, including hSSB1 in humans which reportedly plays a role in genomic maintenance (Richard et al., 2008). RPA2 from F. acidarmanus also has a single OB fold and this RPA2-specific functional interaction with XPD may be used by Rad3 family helicases from the eukaryotic organisms that possess several nuclear ssDNA binding proteins for targeting of the respective helicase to the desired DNA processing intermediates. We propose that translocation on a protein-coated DNA lattice may represent a generalized feature used by Rad3 family helicases, whether it is achieved by translocating over the bound ssDNA-binding protein or by facilitating its dissociation ahead of the translocating enzyme. Both scenarios would provide a mechanism for these helicases to bypass ssDNA-binding proteins in the course of respective genome maintenance events.
EXPERIMENTAL PROCEDURES
Proteins and DNA
F. acidarmanus XPD, RPA1 and RPA2 were purified as previously described (Pugh et al., 2008a; Pugh et al., 2008b). Fluorescent labeling of RPA1 and RPA2 were performed by coupling Cy5 monoreactive NHS esters (GE Healthcare) to the N-terminal amine group at pH 7.0 (Galletto et al., 2006). Briefly, RPA1 and RPA2 were mixed with a 10-fold molar excess of the Cy5 monoreactive NHS esters (GE Healthcare) in buffer L (50 mM K2HPO4/KH2PO4, pH 7.0, 100 mM NaCl and 1 mM DTT) for 30 min at room temperature. The reaction mixtures were then incubated for 12 h at 4 °C and terminated by addition of 50 mM Tris-HCl, pH 7.5. Cy5-labeled RPA1 and RPA2 were separated from the free dye using a Desalting column (GE healthcare). The ratio of dye incorporated per protein molecule was 1.17 mol Cy5 per mol RPA1 and 1.09 mol Cy5 per mol RPA2, which was consistent with a single label per protein molecule, utilizing ε650(Cy5) = 250,000 M−1cm−1, ε280(Cy5) = 12,5 00 M−1cm−1, ε280(RPA1) = 34,000 M−1cm−1, and ε280(RPA2) = 21,890 M−1cm−1. To confirm that the proximity of the protein surface did not affect Cy5 dye, we compared the absorbance spectra of the native and denatured proteins. Oligonucleotide substrates were purchased from IDT. The sequences and modifications for all substrates are shown in Table S1.
Equilibrium Cy3-quenching assays
All bulk equilibrium binding assays were performed using a Cary Eclipse Fluorescence Spectrophotometer (Varian). To determine the distance dependence of XPD-mediated quenching, we monitored quenching of the Cy3 fluorescent dye incorporated at the 5’ or 3’-end of the DNA substrates schematically depicted in Figure 1A. The Cy3 dye was excited at 550 nm and its emission was detected at 568 nm. Binding reactions were carried out at 20 °C in reaction mixtures containing 50 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 1 mM DTT, 100 µg/mL BSA. The reaction mixtures were incubated at 20 °C for 2 min and the baseline fluorescence was recorded before addition of 10 nM Cy3-labeled DNA (5Cy3-42, 3Cy3-42, 3Cy3-32, 3Cy3-27 or 3Cy3-58). Increase in the fluorescence was recorded and attributed to the fluorescence of the free DNA substrate. Upon the addition of each aliquot of XPD, the fluorescence signal was allowed to equilibrate, recorded over 1 min and averaged. The change in the fluorescence was plotted and analyzed using GraphPad Prism software.
Reaction conditions for the single-molecule assay
Cy3-labeled oligonucleotides were immobilized on the surface through biotin-neutravidin linkage. The quarts slides (Finkenbeiner) were coated with polyethyleneglycol in order to eliminate nonspecific surface adsorption of proteins (Ha et al., 2002; Roy et al., 2008). The immobilization was mediated by biotin-neutravidin interaction between biotinylated DNA, neutravidin (Pierce), and biotinylated polymer (PEG-MW 5,000, Nectar Therapeutics). Unbound DNA was then removed by washing the microscope slide with the reaction buffer. Typical translocation buffer contained 50 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 1 mM DTT, 100 µg/mL BSA and the oxygen scavenging system consisting of 1 mg/ml glucose oxidase (Sigma), 0.4% (w/v) D-glucose (Sigma), 0.04 mg/ml catalase (Roche) and 1% v/v 2-mercaptoethanol (Acros). Immobilization of 100 pM of each oligonucleotide allowed for detection of 100–600 individual molecules per slide. DNA density was confirmed by counting the surface-tethered Cy3 dyes excited with the 532 nm laser. XPD was then added and incubated for 5 min at 25°C in the standard translocation buffer in the presence of the indicated concentrations of ATP, RPA1, RPA2, Cy5-labeled RPA1 and Cy5-labeled RPA2. Unless otherwise indicated, typical assays were carried out in the presence of 150 pM XPD.
Single-molecule data acquisition
Total internal reflection was used to excite fluorophores linked to the surface tethered molecules. Cy3 fluorescence was excited by a frequency-doubled Nd:YAG laser (532 nm, 75mW, Crysta-Laser), and HeNe laser (633 nm, 30mW) was used for direct Cy5 excitation. The fluorescence signals from the Cy3 and Cy5 dyes were collected by a water immersion objective 60x (Olympus), passed through a 550 nm long-pass filter to block out laser scattering, separated by a 630 nm dichroic mirror and detected by EMCCD camera (Andor) with a time resolution of 30 ms. Fluorescence signals of both dyes were amplified before camera readout; therefore, the recorded fluorescence intensity is reported in arbitrary units (a.u.). The signal was recorded using in house software written in Visual C++. Single-molecule traces were extracted from the recorded video file by IDL software.
Single-molecule data analysis
Fluorescence trajectories extracted for the individual molecules were sorted based on visual examination of the quenching patterns. The translocation rates were then determined by fitting the experimental data to the models shown in Figure S4 using GraphPad Prism software.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the University of Illinois start-up funds to M.S. and by the NIH grant GM 065367 to T.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. Embo J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. Xeroderma pigmentosum and molecular cloning of DNA repair genes. Anticancer Res. 1996;16:693–708. [PubMed] [Google Scholar]
- Cadman CJ, Lopper M, Moon PB, Keck JL, McGlynn P. PriB stimulates PriA helicase via an interaction with single-stranded DNA. J Biol Chem. 2005;280:39693–39700. doi: 10.1074/jbc.M508521200. [DOI] [PubMed] [Google Scholar]
- Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci U S A. 2004;101:2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J Biol Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD Helicase Structures and Activities: Insights into the Cancer and Aging Phenotypes from XPD Mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina A, Shin J-H, Kim D-H, Bermudez VP, Kelman Z, Seo Y-S, Hurwitz J. Studies with the Human Cohesin Establishment Factor, ChlR1: ASSOCIATION OF ChlR1 WITH Ctf18-RFC AND Fen1. J. Biol. Chem. 2008;283:20925–20936. doi: 10.1074/jbc.M802696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent Translocation of E. coli UvrD Monomers Along Single-stranded DNA. Journal of Molecular Biology. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases - Amino-Acid-Sequence Comparisons and Structure-Function-Relationships. Curr Opin Struc Biol. 1993;3:419–429. [Google Scholar]
- Gupta R, Sharma S, Doherty KM, Sommers JA, Cantor SB, Brosh RM., Jr Inhibition of BACH1 (FANCJ) helicase by backbone discontinuity is overcome by increased motor ATPase or length of loading strand. Nucleic Acids Res. 2006;34:6673–6683. doi: 10.1093/nar/gkl964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–2398. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Lahti JM. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28:917–924. doi: 10.1093/nar/28.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Fairman ME. RNA helicases -- one fold for many functions. Curr Opin Struc Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Kerr ID, Wadsworth RI, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of Sulfolobus SSB protein. Embo J. 2003;22:2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA Repair Helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc Natl Acad Sci U S A. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IK, Spies M. The Iron-containing Domain Is Essential in Rad3 Helicases for Coupling of ATP Hydrolysis to DNA Translocation and for Targeting the Helicase to the Single-stranded DNA-Double-stranded DNA Junction. J Biol Chem. 2008a;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- Pugh RA, Lin Y, Eller C, Leesley H, Cann IK, Spies M. Ferroplasma acidarmanus RPA2 facilitates efficient unwinding of forked DNA substrates by monomers of FacXPD helicase. J Mol Biol. 2008b;383:982–998. doi: 10.1016/j.jmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Richard DJ, Bolderson E, Cubeddu L, Wadsworth RIM, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- Robbins JB, McKinney MC, Guzman CE, Sriratana B, Fitz-Gibbon S, Ha T, Cann IK. The euryarchaeota, nature's medium for engineering of single-stranded DNA-binding proteins. J Biol Chem. 2005;280:15325–15339. doi: 10.1074/jbc.M412870200. [DOI] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Shen JC, Lao Y, Kamath-Loeb A, Wold MS, Loeb LA. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech Ageing Dev. 2003;124:921–930. doi: 10.1016/s0047-6374(03)00164-7. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282:19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Soultanas P, Dillingham MS, Velankar SS, Wigley DB. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J Mol Biol. 1999;290:137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature. 1993;365:852–855. doi: 10.1038/365852a0. [DOI] [PubMed] [Google Scholar]
- Takeda S, Kamiya N, Nagamune T. A novel protein-based heme sensor consisting of green fluorescent protein and apocytochrome b562. Analytical Biochemistry. 2003;317:116–119. doi: 10.1016/s0003-2697(03)00096-4. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- Voloshin ON, Vanevski F, Khil PP, Camerini-Otero RD. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J Biol Chem. 2003;278:28284–28293. doi: 10.1074/jbc.M301188200. [DOI] [PubMed] [Google Scholar]
- von Hippel PH, Delagoutte E. Macromolecular complexes that unwind nucleic acids. Bioessays. 2003;25:1168–1177. doi: 10.1002/bies.10369. [DOI] [PubMed] [Google Scholar]
- Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.