Abstract
NMDA receptors comprised of different NR2 subunits exhibit strikingly unique biophysical and pharmacological properties. Here, we report that the extracellular amino-terminal domain (ATD) of the NR2 subunit controls pharmacological and kinetic properties of recombinant NMDA receptors, such as agonist potency, deactivation time course, open probability (POPEN), and mean open/shut duration. Using ATD deletion mutants of NR2A, NR2B, NR2C, NR2D, and chimeras of NR2A and NR2D with interchanged ATD [NR2A-(2D-ATD) and NR2D-(2A-ATD)], we show that the ATD contributes to the low glutamate potency of NR2A-containing NMDA receptors and the high glutamate potency of NR2D-containing receptors. The ATD influences the deactivation time courses of NMDA receptors, as removal of the ATD from NR2A slows the deactivation rate, while removal of the ATD from NR2B, NR2C and NR2D accelerates the deactivation rate. Open probability also is influenced by the ATD. Removal of the ATD from NR2A or replacement of the NR2A-ATD with that of NR2D decreases POPEN in single-channel recordings from outside-out patches of HEK 293 cells. In contrast, deletion of the ATD from NR2D or replacement of the NR2D ATD with that of NR2A increases POPEN and mean open duration. These data demonstrate the modular nature of NMDA receptors, and show that the ATD of the different NR2 subunits plays an important role in fine-tuning the functional properties of the individual NMDA receptor subtypes.
Introduction
NMDA receptors, one functional class of the ionotropic glutamate receptors, have unique features including the requirement of simultaneous binding of glutamate and glycine for activation, high Ca2+ permeability, and voltage-dependent Mg2+ block. NMDA receptors play key roles in physiological processes, such as neuronal development, synaptic plasticity, and learning (Dingledine et al., 1999; Petrie et al., 2000; Riedel et al., 2003), as well as in a number of pathophysiological conditions including epilepsy, ischemic damage, neurodegenerative diseases, and psychiatric disorders (Cull-Candy et al., 2001; Tsai and Coyle, 2002; Hallett and Standaert, 2004; Hynd et al., 2004; Cowan and Raymond, 2006; Mehta et al., 2007).
Functional NMDA receptors are composed of two glycine-binding NR1 subunits and two glutamate-binding NR2 subunits. There are four types of NR2 subunits (NR2A-D), which show developmental and regional differences in expression levels (Watanabe et al., 1992; Ishii et al., 1993; Monyer et al., 1994; Laurie et al., 1997; Sun et al., 2000). In addition to differences in their distribution in the brain, the four NR2 subunits endow NMDA receptors with different agonist potencies, deactivation time courses, open probabilities, single-channel conductances, as well as sensitivities to Mg2+ and extracellular modulators (Erreger et al., 2004). For example, the EC50 for glutamate activating heterodimeric NR1/NR2D receptors (0.5 μm) is lower (i.e., more potent) than that for NR1/NR2A receptors (3–5 μm), with NR1/NR2B and NR1/NR2C subunits showing intermediate glutamate potencies (Erreger et al., 2007). The NMDA receptors that contain different NR2 subunits show >50-fold variation in the deactivation time course of current responses, which govern the duration of a slow component of the EPSC (Lester et al., 1990). The time constants of deactivation of current responses to glutamate are ∼50 ms for NR1/NR2A, ∼400 ms for NR1/NR2B, ∼300 ms for NR1/NR2C, and >1 s for NR1/NR2D (Monyer et al., 1994; Vicini et al., 1998; Wyllie et al., 1998; Dravid et al., 2008). NMDA receptors subtypes also have widely varying (50-fold) open probabilities that depend on the NR2 subunit, which in outside-out patches is ∼0.5 for recombinant NR1/NR2A, ∼0.1 for NR1/NR2B (Erreger et al., 2005a), 0.01 for NR1/NR2C (Dravid et al., 2008), and 0.04 for NR1/NR2D (Wyllie et al., 1998). In addition, NR1/NR2A and NR1/NR2B have higher channel conductances than NR1/NR2C and NR1/NR2D receptors (Stern et al., 1992; Farrant et al., 1994; Béhé et al., 1999; Dravid et al., 2008).
Each NR2 subunit consists of a large extracellular amino-terminal domain (ATD), a bilobed agonist-binding domain, a transmembrane domain, and an intracellular C-terminal domain (see Fig. 1A,B). The ATD is composed of approximately the first 350 aa of the protein and appears to form a clamshell-like structure, which interacts with various extracellular allosteric modulators such as zinc for NR2A (Paoletti et al., 1997; Choi and Lipton, 1999; Fayyazuddin et al., 2000; Low et al., 2000; Choi et al., 2001), and ifenprodil for NR2B (Perin-Dureau et al., 2002; Malherbe et al., 2003; Wong et al., 2005; Ng et al., 2008). Therefore, understanding how the ATD affects the function of NR2 subunits will provide valuable insight into the mechanisms of channel activation for NMDA receptors. In this study, we investigate the role of the NR2 ATD on the functional properties of NMDA receptors. We show that the identity and presence of the ATD influences agonist potency, deactivation time course, mean open duration, and open probability.
Figure 1.

Schematic representation of the NR2 ATD deletion constructs and NR2A-NR2D chimeras. A, Hypothetical arrangement of two of the four subunits that comprise NMDA receptors (left). The diagram on the right illustrates the key features of a single subunit. The ATD shown in green was generated from a homology model based on the mGluR1 crystal structure (Yuan et al., 2009), the agonist-binding domains shown in blue are the crystal structure of the NR1-NR2A heterodimer (PDB code 2A5T) (Furukawa et al., 2005), and the transmembrane domains shown in orange are the M1, M2, and M3 regions aligned with the structure of the P-loop region of KcsA (PDB code 1BL8). The C-terminal domain and the transmembrane helix M4 from NR1 and NR2A is omitted from the hypothetical arrangement (left). B, Linear schematic organization of NR2 subunits. Each subunit consists of the ATD, segments S1 and S2, three transmembrane helices (M1, M3, and M4), one cytoplasmic re-entrant pore loop (M2), and an intracellular C-terminal domain (CTD). Together, the S1 and S2 segments form the agonist-binding domain. C, The ATD deletion constructs of the NR2 subunits were developed from wild-type NR2A, NR2B, NR2C, NR2D, and NR2B-(ΔS28-M394). All ATD deletion constructs encode the first 28 residues of NR2B, including the signal peptide. The residue following Ser 28 of NR2B was His 405 of NR2A, His 405 of NR2B, His 415 of NR2C, and His 428 of NR2D. D, Constructs of NR2A-NR2D chimeras were developed by interchanging the ATD of NR2A and NR2D. The residue following Gln 427 of NR2D was His 405 of NR2A, and the residue following Asn 404 of NR2A was His 428 of NR2D. Amino acid numbering is done according to full-length protein, including the signal peptide.
Materials and Methods
Molecular biology.
Site-directed mutagenesis was performed using the QuikChange protocol (Stratagene) as described previously (Low et al., 2000). cDNAs for NR1-1a (GenBank accession numbers U11418 and U08261; hereafter NR1), NR2A (D13211), NR2B (U11419), NR2C (M91563), and NR2D (L31611) were provided by Drs. S. Heinemann (Salk Institute, San Diego, CA), S. Nakanishi (Kyoto University, Kyoto, Japan), and P. Seeburg (University of Heidelberg, Heidelberg, Germany). The NR2 ATD deletion constructs were developed by inserting a restriction enzyme site into wild-type NR2 subunits at the junction between the ATD and the S1 segment without changing the amino acid sequence. Subsequently, the downstream portion of these constructs was subcloned into the vector of the NR2B-(ΔS28-M394) construct (provided by Dr. Chian-Ming Low, National University of Singapore, Singapore). The junction between the ATD and the S1 segment was determined using the crystal structure of the NR2A ligand binding domain as a guide (Furukawa et al., 2005). NR2A-NR2D chimeras were developed by interchanging the ATD from NR2A at Asn 404 and NR2D at Gln 427. All insertions and deletions are identified in Figure 1. Amino acid numbering is done according to full-length protein, including the signal peptide. The vector pCI-neo was used for all constructs, except for NR2C-related constructs used for recordings from HEK 293 cells (pRK).
Two-electrode voltage-clamp recordings from Xenopus oocytes.
Preparation and injection of cRNA, as well as two-electrode voltage-clamp recordings from Xenopus laevis oocytes, were performed as previously described (Traynelis et al., 1998). Briefly, oocytes were injected with 5–10 ng of cRNAs synthesized in vitro from linearized template cDNA. Following injection, the oocytes were stored at 15°C in Barth's solution containing (in mm): 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, 5 Tris/HCl (pH 7.4 with NaOH). The ratio of injected NR1 to NR2 cRNA was 1:2. Two-electrode voltage-clamp recordings were performed 2–4 d postinjection at room temperature (23°C). The recording solution contained (in mm) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2, 0.01 EDTA (pH 7.4 with NaOH). Solution exchange was computer controlled through an 8-modular valve positioner (Digital MVP Valve). Voltage and current electrodes were filled with 0.3 and 3.0 m KCl, respectively, and current responses were recorded at a holding potential of –40 mV. Data acquisition and voltage control were accomplished with a two-electrode voltage-clamp amplifier (OC725, Warner Instrument). Glutamate (100 μm) and glycine (100 μm) were used in all oocyte experiments unless otherwise stated. 200 μm methanethiosulfonate ethylammonium (MTSEA) was prepared fresh and used within 30 min.
Transfection of HEK 293 cells.
HEK 293 cells (CRL 1573, ATCC) were plated on glass coverslips (Warner Instruments) coated in 100 μg/ml poly-d-lysine and were incubated at 37°C in humidified 5% CO2 in culture media (Dulbecco's Modified Eagle Medium with GlutaMax; Invitrogen) supplemented with 10% dialyzed fetal bovine serum, 10 units/ml penicillin, and 10 μg/ml streptomycin. Cells were transiently co-transfected using the Fugene 6 transfection reagent (Roche Diagnostics) with plasmid cDNA encoding GFP, NR1, and either wild-type NR2 subunits, NR2 ATD deletion mutants, or NR2A-NR2D chimeric receptors. After transfection, the cells were incubated overnight in culture media supplemented with 200 μm D, l-2-amino-5-phosphonovalerate and 200 μm 7-chlorokynurenic acid. Following transfection and overnight incubation, the cells were used for whole-cell voltage-clamp recordings and single-channel recordings in outside-out patches.
Whole-cell voltage-clamp recording.
Whole-cell voltage-clamp recordings were performed using transfected HEK 293 cells at −60 mV using an Axopatch 200B amplifier (Molecular Devices) at room temperature (23°C). Recording electrodes (3.5–7 ΜΩ) were made from thin wall glass micropipettes (TW150F-4, World Precision Instruments) pulled using a vertical puller (Narishige PP-830). The electrodes were filled with internal solution containing (in mm) 110 d-gluconate, 110 CsOH, 30 CsCl, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, and 0.3 NaGTP (pH 7.35). The external solution was composed of (in mm) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, 0.01 EDTA at 23°C and pH 7.4. Rapid solution exchange was achieved with a two-barreled theta-glass pipette controlled by a piezoelectric translator (Burleigh Instruments) with <1 ms open tip exchange times for the solutions around the cell. Once a gigaseal and breakthrough were achieved, the whole cell was lifted and first exposed to 50 μm glycine, followed by a one-second jump into 50 μm glutamate in 50 μm glycine, pH 7.4. The deactivation time course was fitted by ChanneLab (Synaptosoft) to the equation: Response = Amplitude · exp (−time/τ), where τ is the time constant.
Single-channel recording.
Single-channel recordings with outside-out patches from transfected HEK 293 cells were recorded at –80 mV (digitized at 40 kHz, filtered at 8 kHz) using an Axopatch 200B amplifier (Molecular Devices) at room temperature (23°C). Recording electrodes were made from thick-wall glass pipettes (G150F-4, Warner Instruments) pulled using a vertical puller (Narishige PP-830), coated with Sylgard (Dow Corning), and then fire-polished to 8.5–11 ΜΩ. The internal solution was the same as used for whole-cell voltage-clamp recording. The channel was activated by 50 μm glycine and 1 mm glutamate in an external solution composed of (in mm) 150 NaCl, 10 HEPES, 30 d-mannitol, 3 KCl, 0.5 CaCl2, and 0.01 EDTA at pH 8.0. For analysis, the recordings were prefiltered at 8 kHz (−3 dB), digitized at 40 kHz, and analyzed using the time course fitting method (SCAN software provided by Dr. David Colquhoun, University College London, London, UK, http://www.ucl.ac.uk/Pharmacology/dcpr95.html). Open and closed duration histograms were constructed with an imposed resolution for open and shut durations of 53 and 31 μs, respectively (Colquhoun and Hawkes, 1990). The amplitude distributions were fitted to the sum of multiple Gaussian components and apparent open and closed duration distributions fitted to the sum of multiple exponential components using the maximum likelihood (EKDIST, http://www.ucl.ac.uk/Pharmacology/dcpr95.html; ChanneLab, Synaptosoft). The number of channels in the patch was determined statistically using methods described previously (Colquhoun and Hawkes, 1990; Dravid et al., 2008). Only recordings with a high probability (p < 0.001) of containing only one active channel were used for data analysis.
All reagents were purchased from Sigma, except MTSEA (Toronto Research Chemicals). Data are expressed as mean ± SEM and analyzed statistically using the unpaired t test and one-way ANOVA with Bonferroni's post hoc test. Significance for all tests was set at p < 0.05. Error bars in all figures are SEM.
Results
To investigate how the ATD of the NR2 subunit influences the functional properties of NMDA receptors, two sets of NR2 constructs were designed for comparison to wild-type receptors. First, ATD deletion constructs of all four NR2 subunits (NR2A-ΔATD, NR2B-ΔATD, NR2C-ΔATD, NR2D-ΔATD) (Fig. 1C) were generated. Second, two NR2A-NR2D chimeras, NR2A-(2D-ATD) and NR2D-(2A-ATD) (Fig. 1D) were developed. Coexpression of wild-type NR1 with these mutated NR2 subunits generated functional NMDA receptors. The agonist EC50, deactivation time course, degree of MTSEA potentiation, and single-channel properties were evaluated for these mutant and wild-type receptors.
The NR2 ATD controls EC50 values for glutamate and glycine
NMDA receptors that contain different NR2 subunits show a range of agonist potencies. Glutamate potency was quantified as the concentration of agonist that can produce a half-maximally effective response (EC50). We evaluated the EC50 values of recombinant NMDA receptors comprised of wild-type or mutant NR2 expressed in oocytes in the presence of a maximally effective concentration of glycine (100 μm). Deletion of the ATD in all four NR2 subunits did not change glutamate potency compared with that for the corresponding wild-type subunit (Fig. 2A). The EC50 of chimeric NR2A- (2D-ATD) (0.33 μm) was lower than that of NR2A, and was similar to that of NR2D (0.35 μm). In contrast, the EC50 of chimeric NR2D-(2A-ATD) (2.4 μm) was higher than that of NR2D and similar to that for NR2A (3.3 μm) (Fig. 2A; Table 1). Whereas data obtained from ATD deletion constructs suggest that the ATD does not markedly influence glutamate potency, data using ATD chimeras suggest that the ATD contributes to the high EC50 value for glutamate in NR2A-containing NMDA receptors and the low EC50 value in NR2D-containing NMDA receptors. We interpret these apparent conflicting results to suggest that in full-length receptors the ATD influences glutamate potency through complex actions on gating and possibly the stability of the glutamate-bound agonist-binding domain.
Figure 2.
The NR2 ATD controls the EC50 values for glutamate and glycine on recombinant NMDA receptors expressed in oocytes. A, ATD deletion in the NR2A, NR2B, NR2C, and NR2D subunits causes minimal change in glutamate EC50 values (in the presence of 100 μm glycine) compared with the corresponding wild type. However, the glutamate EC50 of NR2A-(2D-ATD) is similar to that of wild-type NR2D, and the EC50 of NR2D-(2A-ATD) is similar to that of wild-type NR2A. B, Glycine EC50 (in the presence of 100 μm glutamate) was increased following ATD deletion in NR2B, NR2C, and NR2D subunits.
Table 1.
Summary of concentration-response data
| Constructs | Amplitude (pA) | Glutamate |
Glycine |
||||
|---|---|---|---|---|---|---|---|
| EC50 (μm) | Hill slope | n | EC50 (μm) | Hill slope | n | ||
| NR2A | 150–2700 | 3.3 | 1.4 | 5 | 1.1 | 1.0 | 5 |
| NR2A-ΔATD | 50–2600 | 3.4 | 1.2 | 5 | 0.82 | 1.7 | 5 |
| NR2A-(2D-ATD) | 80–1500 | 0.33 | 1.3 | 5 | 0.13 | 1.2 | 5 |
| NR2D | 190–2700 | 0.35 | 1.6 | 6 | 0.23 | 1.4 | 4 |
| NR2D-ΔATD | 60–580 | 0.28 | 1.7 | 6 | 3.2 | 1.9 | 5 |
| NR2D-(2A-ATD) | 50–240 | 2.4 | 1.8 | 5 | 0.51 | 1.2 | 5 |
| NR2B | 150–3000 | 1.3 | 1.2 | 6 | 0.18 | 2.0 | 5 |
| NR2B-ΔATD | 110–2700 | 0.85 | 1.3 | 6 | 2.0 | 2.2 | 5 |
| NR2C | 500–2400 | 0.80 | 1.4 | 5 | 0.14 | 1.8 | 5 |
| NR2C-ΔATD | 90–1300 | 1.2 | 1.5 | 5 | 3.6 | 1.5 | 5 |
EC50 values were obtained by fitting the composite concentration-response curve with the equation Response = 100%/(1 + (EC50 /[agonist])N) where N is the Hill slope.
Glycine potency was also evaluated in the presence of a maximally effective concentration of glutamate (100 μm). The glycine EC50 increased (i.e., potency decreased) by 10- to 20-fold in the ATD deletion constructs of NR2B, NR2C, and NR2D subunits, whereas the glycine potency was only slightly changed for NR2A-ΔATD (Fig. 2B; Table 1). These data indicate that ATD contributes more significantly to the glycine potency in wild-type NR2B, NR2C and NR2D-containing NMDA receptors than in NR2A-containing NMDA receptors. The chimeric receptors containing NR2A-(2D-ATD) or NR2D-(2A-ATD) showed shifts in the EC50 values toward those of the receptor contributing the ATD, further suggesting ATD can control glycine potency.
The NR2 ATD controls the deactivation time course of current responses
The deactivation time course of NMDA receptors, which contributes to the duration of EPSCs (Lester et al., 1990), varies according to the NR2 subunit incorporated into the receptor. To evaluate the contribution of the ATD to the glutamate deactivation time course, the current response following glutamate removal was measured using a rapid solution exchange system and whole-cell voltage-clamp recordings of wild-type and mutant NMDA receptors expressed in HEK 293 cells. The time course of the response following a rapid jump from glutamate and glycine into glycine alone was fitted with a single exponential function to determine a deactivation time constant (τ). The deletion of the ATD from the NR2A subunit significantly prolonged the deactivation time constant (90 ± 6.6 ms for NR2A-ΔATD; 39 ± 4.2 ms for NR2A; p < 0.01) (Fig. 3A,F; Table 2), while ATD deletion from NR2D significantly accelerated the glutamate deactivation time constant (1200 ± 65 ms for NR2D-ΔATD; 2800 ± 84 ms for NR2D; p < 0.01) (Fig. 3B,F; Table 2). These data indicate that the ATD of NR2A contributes to fast deactivation rate, while the ATD of NR2D contributes to the slow rate. This was further corroborated by experiments using NR2A-NR2D chimeric receptors, which show deactivation rates intermediate to those of wild-type NR2A and NR2D [330 ± 5.2 ms for NR2A-(2D-ATD) and 350 ± 10 ms for NR2D-(2A-ATD)] (Fig. 3C,F; Table 2).
Figure 3.

The NR2 ATD controls the deactivation time course in NMDA receptors. A, Representative whole-cell currents elicited by glycine and glutamate (50 μm each, black bar) are shown in whole-cell voltage-clamp (VHOLD = −60 mV) current recordings from HEK 293 cells expressing recombinant NR1 with NR2A or NR2A-ΔATD. Glycine was present in all solutions. Right panel shows superimposed traces (black: NR2A; gray: NR2A-ΔATD). B, Representative traces of currents from NR1 coexpressed with NR2D or NR2D-ΔATD. C, Representative traces of currents from NR1 coexpressed with NR2A-(2D-ATD) or NR2D-(2A-ATD). D, Representative traces of currents from NR1 coexpressed with NR2B or NR2B-ΔATD. Right panel shows superimposed traces (black: NR2B; gray: NR2B-ΔATD). E, Representative traces of currents from NR1 with NR2C or NR2C-ΔATD. Right panel shows superimposed traces (black: NR2C; gray: NR2C-ΔATD). F, Summary of the mean deactivation time constants. #p < 0.01, one-way ANOVA with Bonferroni's post hoc test, compared with the corresponding wild type. *p < 0.01, unpaired t test, compared with the corresponding wild type.
Table 2.
Summary of deactivation time course data
| Constructs | n | τ (ms) |
|---|---|---|
| NR2Aa | 6 | 39 ± 4.2 |
| NR2A-ΔATD | 7 | 90 ± 6.6# |
| NR2A-(2D-ATD) | 5 | 330 ± 5.2# |
| NR2D | 6 | 2800 ± 84 |
| NR2D-ΔATD | 5 | 1200 ± 65# |
| NR2D-(2A-ATD) | 5 | 350 ± 10# |
| NR2B | 5 | 420 ± 50 |
| NR2B-ΔATD | 5 | 170 ± 8.1* |
| NR2C | 6 | 310 ± 5.3 |
| NR2C-ΔATD | 5 | 190 ± 7.6* |
τ deactivation was measured following removal of glutamate, but not glycine. Values are shown with two significant digits as mean ± SEM, and n is the number of HEK 293 cells used to generate the data.
#p < 0.01, one-way ANOVA with Bonferroni's post hoc test, compared with the corresponding wild type;
*p < 0.01, unpaired t test, compared with the corresponding wild type.
aA minority of the NR2A-transfected cell responses were fit by two components for the deactivation rate with small percentage of slow deactivation rate, which is consistent with other studies (Vicini et al., 1998).
ATD deletion from NR2B and NR2C significantly accelerated the glutamate deactivation rate compared with wild-type NR2B and NR2C (170 ± 8.1 ms for NR2B-ΔATD and 420 ± 50 ms for NR2B, p < 0.01, unpaired t test; 190 ± 7.6 ms for NR2C-ΔATD and 310 ± 5.3 ms for NR2C, p < 0.01, unpaired t test) (Fig. 3D–F; Table 2). These data suggest that the ATDs of NR2B and NR2C, like that of NR2D, contribute to the slow deactivation rates of the current responses.
The NR2 ATD controls open probability as measured by the degree of MTSEA potentiation
Di-heteromeric NR1/NR2 NMDA receptors also have distinct open probabilities that vary >50-fold depending on the identity of the NR2 subunit in the receptor assembly (Wyllie et al., 1998; Erreger et al., 2005a; Yuan et al., 2005; Dravid et al., 2008). As a first step toward assessing the role of the ATD in controlling open probability, the NR2 ATD deletion and NR2A-D chimeric constructs were coexpressed with NR1-A652C (alanine to cysteine mutation in the seventh amino acid of a conserved 9 amino acid motif in M3 of NR1; SYTANLAAF) in oocytes (Jones et al., 2002; Yuan et al., 2005). This mutation allowed us to evaluate the agonist-induced accessibility changes for the sulfhydryl-modifying reagent MTSEA, which reacts with the cysteine residue in the mutant subunit NR1-A652C. Application of MTSEA to agonist-bound NR1-A652C-containing channels irreversibly locks open the pore following covalent modification (Jones et al., 2002). We have assumed that the modified receptors have an open probability (POPEN) of ∼1.0, which renders the degree of MTSEA potentiation of the maximal agonist response (corrected for effects of MTSEA on conductance) inversely related to the open probability of the unmodified mutant channel. The POPEN can be estimated using the following equation: POPEN = (γMTSEA/γCONTROL) × (1/Potentiation), where Potentiation is equal to IMTSEA/ICONTROL, γMTSEA is the conductance of the MTSEA-modified channel, γCONTROL is the conductance of wild-type channels, ICONTROL is the current response induced by application of saturating concentrations of both glutamate and glycine, and IMTSEA is the current response following application of MTSEA.
The effect of MTSEA on unitary conductance was evaluated in outside-out patch recordings from NR1-A652C/NR2A-transfected HEK 293 cells, which showed γCONTROL is 65 pS and γMTSEA is 45 pS for NR2A (n = 3). These values are virtually identical to those previously determined for MTSEA—modification of NR1/NR2A-A650C (γCONTROL 64 pS; γMTSEA 44 pS; Yuan et al., 2005), suggesting similar effects of MTSEA-modification of NR1 or NR2 on unitary conductance. The conductance change by MTSEA was also investigated on NR2B subunits coexpressed with NR1-A652C, which showed γCONTROL is 75 pS and γMTSEA is 49 pS (n = 3). From these and other conductance measurements (data not shown), we determined γMTSEA/γCONTROL values of 0.69 for NR1-A652C/NR2A, 0.66 for NR1-A652C/NR2B, 0.69 for NR1-A652C/NR2C, and 0.69 for NR1-A652C/NR2D. Whereas estimates of open probability obtained by MTSEA-potentiation of mutant NR1/NR2 channels are useful for initial comparisons of the properties of various NR2 constructs, these estimates should not be compared with open probability of wild-type NR1/NR2 channels measured from single-channel openings in outside-out patches from HEK 293 cells. Estimates of POPEN from MTSEA-modified oocyte currents do not provide a quantitative measure of the open probability of an individual wild-type NMDA receptor.
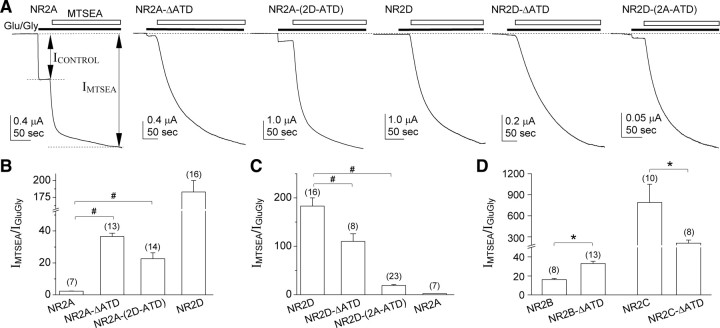
Coapplication of 200 μm MTSEA with glutamate and glycine potentiated the agonist-evoked NMDA receptor currents for all NR2 constructs coexpressed with NR1-A652C (Fig. 4). MTSEA potentiation of NR2A-(2D-ATD) suggested that POPEN was 0.04 (Fig. 4A,B; Table 3), significantly lower than wild-type NR2A-containing receptors (POPEN 0.31). The degree of MTSEA potentiation suggested a POPEN value of 0.04 for NR2D-(2A-ATD), higher than that estimated for wild-type NR2D (POPEN < 0.01) (Fig. 4A,C; Table 3). This result is consistent with the idea that the ATD of NR2A can enhance open probability of receptors containing the full-length NR2 subunit.
Figure 4.
NR2 ATD control of open probability evaluated by the degree of MTSEA potentiation. A, Representative two-electrode voltage-clamp current recordings obtained from oocytes coexpressing recombinant NR1-A652C with NR2A, NR2A-ΔATD, NR2A-(2D-ATD), NR2D, NR2D-ΔATD, or NR2D-(2A-ATD) to evaluate the degree of potentiation by 0.2 mm MTSEA after the receptors are activated by glutamate and glycine (100 μm). Application of MTSEA to agonist-bound NR1-A652C-containing channels irreversibly locks open the pore following covalent modification. MTSEA-modified receptors have an open probability (POPEN) of ∼1.0, which renders the degree of MTSEA potentiation of the maximal agonist response inversely related to the open probability of the channel. B–D, Summary of the degree of MTSEA potentiation. Open probability estimated from these data are given in Table 3; #p < 0.01, one-way ANOVA with Bonferroni's post hoc test, compared with the corresponding wild type. *p < 0.01, unpaired t test, compared with the corresponding wild type.
Table 3.
Summary of degree of MTSEA potentiation and calculated open probability
| Constructs | n | Degree of MTSEA potentiation | Calculated POPEN |
|---|---|---|---|
| NR2A | 7 | 2.3 ± 0.1 | 0.31 ± 0.02 |
| NR2A-ΔATD | 13 | 37 ± 2.1# | 0.02 ± 0.002 |
| NR2A-(2D-ATD) | 14 | 23 ± 3.7# | 0.04 ± 0.01 |
| NR2D | 16 | 180 ± 17 | <0.01 |
| NR2D-ΔATD | 8 | 110 ± 16# | <0.01 |
| NR2D-(2A-ATD) | 23 | 19 ± 1.9# | 0.04 ± 0.03 |
| NR2B | 8 | 16 ± 1.2 | 0.04 ± 0.003 |
| NR2B-ΔATD | 13 | 33 ± 2.4* | 0.02 ± 0.002 |
| NR2C | 10 | 790 ± 260 | <0.01 |
| NR2C-ΔATD | 8 | 210 ± 44* | <0.01 |
Values are shown with two significant digits as mean ± SEM, and n is the number of oocytes used to generate the data. POPEN values less than 0.01 reflect exceptionally low-current amplitudes prior to MTSEA application and are not reported.
#p < 0.01, one-way ANOVA with Bonferroni's post hoc test, compared with the corresponding wild type;
*p < 0.01, unpaired t test, compared with the corresponding wild type.
We interpret the potentiation of NR2A-ΔATD (37-fold) to reflect an estimated open probability (POPEN) of 0.02, considerably lower than the open probability estimated for wild-type NR2A-containing receptors of 0.31 (Table 3). ATD deletion in the NR2B subunit significantly decreased the estimated POPEN from 0.04 for wild-type receptors to 0.02 (Fig. 4D; Table 3). POPEN values estimated using MTSEA potentiation were all <0.01 for wild-type NR2C, wild-type NR2D, NR2C-ΔATD, and NR2D-ΔATD receptors (Fig. 4A,C,D; Table 3), which reflected low current responses in the absence of MTSEA. These results suggest that the presence of ATD increases POPEN, for NR2A and NR2B receptors, compared with receptors containing NR2 subunits that lack ATD.
The NR2 ATD controls single-channel open probability of NMDA receptors
To understand more clearly the effects of the ATD on open probability, we studied single-channel recordings in outside-out patches excised from HEK 293 cells that were transiently transfected with NR1 and wild-type or mutated NR2A or NR2D. For all experiments reported here, analyses were restricted to only excised patches that contained a single active channel (see Materials and Methods), which allows unambiguous determination of the open probability and interpretation of closed time intervals. Steady-state single-channel unitary currents were recorded from NR2A and NR2A-ΔATD in response to maximally effective concentrations of glutamate and glycine. Figure 5Aa1 shows typical wild-type NR1/NR2A single-channel currents that have a complex bursting behavior, long desensitized periods of inactivity, one predominant conductance state (69 pS) in the presence of a relatively low extracellular Ca2+ concentration (0.5 mm) (Erreger et al., 2005a), and a high open probability (e.g., 0.41 in this patch). Open period and shut duration histograms were fitted with two and five exponential components, respectively (Fig. 5Aa2). A parallel set of recordings for NR1 coexpressed with the ATD deletion construct (NR2A-ΔATD) (Fig. 5Bb1) showed significantly fewer open events with prolonged shut durations, resulting in a very low open probability (0.002 in this patch). The area percentages of the exponential components of the longest shut durations were increased in NR2A-ΔATD compared with wild-type NR2A, whereas the major conductance level remained unchanged (66 pS) (Fig. 5Bb2). Table 4 summarizes the pooled data describing single-channel properties from 4 patches of each NR2A and NR2A-ΔATD constructs. The mean open probability significantly decreased from 0.48 for NR2A to 0.006 for NR2A-ΔATD (p < 0.01, unpaired t test). The mean shut duration increased from 1.9 ms for NR2A to 360 ms for NR2A-ΔATD, consistent with a profound decrease in opening frequency. The mean open duration (1.7 ± 0.2 ms for NR2A, n = 4; 1.6 ± 0.3 ms for NR2A-ΔATD, n = 4, p > 0.05) and major-conductance level (69 pS for NR2A; 66 pS for NR2A-ΔATD) remained unchanged. These data support the conclusion from the MTSEA potentiation experiments that the removal of the ATD from the NR2A subunit significantly reduces the open probability.
Figure 5.
ATD deletion from NR2A decreases the single-channel open probabilities in outside-out patch-clamp recordings from transiently transfected HEK 293 cells. Aa1, Bb1, Steady-state recordings of NR1/NR2A (Aa1) and NR1/NR2A-ΔATD (Bb1) unitary currents in an outside-out patch that contained one active channel activated by 50 μm glycine and 1 mm glutamate are shown at different time scales. Aa2 and Bb2 show the open period histogram, shut duration histogram, amplitude histogram, and the stability plot of the amplitude for the channel in this patch.
Table 4.
Summary of single channel properties
| Parameters | NR2A | NR2A-ΔATD | NR2A-(2D-ATD) | NR2D | NR2D-ΔATD | NR2D-(2A-ATD) |
|---|---|---|---|---|---|---|
| N | 4 | 4 | 5 | 6 | 3 | 4 |
| POPEN | 0.48 ± 0.05 | 0.006 ± 0.003a | 0.035 ± 0.017a | 0.012 ± 0.002 | 0.065 ± 0.015a | 0.059 ± 0.011a |
| Open duration (ms) | 1.7 ± 0.20 | 1.6 ± 0.33 | 1.1 ± 0.08a | 0.61 ± 0.04 | 0.86 ± 0.08a | 0.99 ± 0.04a |
| τ1, ms (%) | 0.1 (9%) | 0.25 (38%) | 0.1 (9%) | 0.1 (21%) | 0.12 (1%) | 0.10 (1%) |
| τ2, ms (%) | 1.7 (91%) | 2.8 (62%) | 1.1 (91%) | 0.61 (79%) | 0.86 (99%) | 0.99 (99%) |
| Shut duration (ms) | 1.9 ± 0.45 | 360 ± 86a | 69 ± 21a | 60 ± 13 | 14 ± 2.6a | 17 ± 4.0a |
| τ0, ms (%) | 0.02 (49%) | 0.02 (53%) | 0.02 (41%) | 0.027 (30%) | 0.029 (33%) | 0.023 (48%) |
| τ1, ms (%) | 0.20 (16%) | 0.36 (26%) | 0.25 (15%) | 0.28 (14%) | 0.20 (11%) | 0.26 (23%) |
| τ2, ms (%) | 0.93 (27%) | 2.9 (7%) | 1.3 (13%) | 13 (20%) | 1.3 (11%) | 1.9 (9%) |
| τ3, ms (%) | 2.8 (8%) | 17 (7%) | 13 (26%) | 66 (31%) | 12 (35%) | 14 (13%) |
| τ4, ms (%) | 540 (<1%) | 61 (3%) | 85 (3%) | 260 (5%) | 46 (10%) | 76 (7%) |
| τ5, ms (%) | – | 4400 (4%) | 3200 (2%) | 16000 (<1%) | 500 (<1%) | 1300 (<1%) |
| Amplitude 1 (pA) | 5.5 ± 0.04 | 5.3 ± 0.06 | 5.5 ± 0.11 | 4.3 ± 0.6 | 4.3 ± 0.26 | 4.7 ± 0.21 |
| Amplitude 2 (pA) | – | – | 4.4 ± 0.07 | 2.5 ± 0.3 | 2.6 ± 0.11 | 2.9 ± 0.12 |
| γ1, ps (%) | 69 (100%) | 66 (100%) | 69 (88%) | 53 (70%) | 54 (70%) | 58 (66%) |
| γ2, ps (%) | –b | –b | 55 (12%) | 31 (30%) | 33 (30%) | 36 (34%) |
| H→ L (%) | – | – | – | 64 | 60 | 58 |
| L→H (%) | – | – | – | 36 | 40 | 42 |
Values are shown with two significant digits as mean or mean ± SEM; n is the number of single channel patches from HEK 293 cells used to generate the data. Component values of open periods and shut durations and the transitions between high (H) and low (L) conductance levels are from the averages of the fittings from each of the single channel patches.
aFor mean open probability, open duration, shut duration, and unitary current amplitude, p < 0.01, one-way ANOVA with Bonferroni's post hoc test, compared with the corresponding wild type.
bA minority of patches from NR2A- and NR2A-ΔATD-transfected cells show a subconductance level with percentage less than 5%.
Single-channel currents also were recorded from NR1/NR2D and/NR2D-ΔATD (Fig. 6). Figure 6Aa1 shows typical NR1/NR2D single-channel currents that have characteristic short open periods and long closed durations, a very low open probability (0.012), and two conductance levels (54 pS and 33 pS) in the presence of 0.5 mm extracellular Ca2+ (Wyllie et al., 1998). Open duration and shut duration histograms were described by two and six exponential components, respectively (Fig. 6Aa2). A parallel set of recordings for the ATD deletion construct NR2D-ΔATD (Fig. 6Bb1) showed significantly more open events with increased mean open period and decreased long-lasting shut durations, resulting in a fivefold higher open probability (0.075 for the NR1/NR2D-ΔATD channel shown in Fig. 6). The percentage area of exponential components of long shut durations decreased compared with the wild type, consistent with an increase in opening frequency. The major- and subconductance levels remain unchanged (49 pS and 31 pS) (Fig. 6Bb2). Table 4 summarizes the single-channel properties determined from analysis of 6 and 3 patches of NR2D and NR2D-ΔATD, respectively. Mean open probability significantly increased from 0.012 for NR2D to 0.065 for NR2D-ΔATD (p < 0.01, unpaired t test), mean open duration increased from 0.61 ms for NR2D to 0.86 ms for NR2D-ΔATD (p < 0.01, unpaired t test), and mean shut duration decreased from 60 ms for NR2D to 14 ms for NR2D-ΔATD (p < 0.01, unpaired t test). Major- and subconductance levels and their transitions remain comparable between NR2D and NR2D-ΔATD. These data clearly show that removal of ATD from NR2D significantly increases the open probabilities and mean open durations. Thus, the ATD on NR2A increases open probability, whereas the ATD on NR2D decreases open probability.
Figure 6.
ATD deletion from NR2D increases the single-channel open probability and mean open duration. Aa1, Bb1, Steady-state recordings of NR1 with NR2D (Aa1) or NR2D-ΔATD (Bb1) unitary currents in an outside-out patch that contained one active channel are shown at different time scales. Aa2 and Bb2 show the open period histogram, shut duration histogram, amplitude histogram, and the stability plot of the amplitude for the channel in this patch.
To evaluate whether the function of the ATD from NR2A can control the single-channel properties of the NR2D receptor (and vice versa), single-channel currents were recorded from NR1 coexpressed with either the NR2A-(2D-ATD) (Fig. 7) or NR2D-(2A-ATD) (Fig. 8). Figure 7A shows that single-channel currents from NR2A-(2D-ATD) have shorter open durations with a greatly reduced open probability (0.06 in this patch) compared with the wild-type NR2A. That is, chimeric channels with the NR2D ATD within the NR2A subunit have channel properties more similar to NR2D channels. Table 4 summarizes the pooled data describing single-channel properties from 5 patches of NR2A-(2D-ATD). The major-conductance level of NR2A-(2D-ATD) (69 pS) remains unchanged compared with the NR2A, although interestingly there is a minor new subconductance level apparent (55 pS) in some patches (Fig. 7B, right panel).
Figure 7.
Chimeric NR2A-(2D-ATD) shows lower open probability and briefer open periods than wild-type NR2A. A, Steady-state recordings of NR1/NR2A-(2D-ATD) unitary currents in an outside-out patch that contained one active channel are shown at different time scales. B shows the open period histogram and amplitude histogram.
Figure 8.
Chimeric NR2D-(2A-ATD) shows an increased open probability and longer open periods than NR2D. A, Steady-state recordings of NR1/NR2D-(2A-ATD) unitary currents in an outside-out patch that contained one active channel are shown at different time scales. B shows the open period histogram and amplitude histogram.
A parallel set of recordings for NR1/NR2D-(2A-ATD) (Fig. 8A) showed longer open durations and decreased long-lasting shut durations, resulting in a higher open probability (0.052 in this patch) for NR1/NR2D-(2A-ATD) channels compared with the wild-type NR1/NR2D (0.012). The major- and subconductance levels remain unchanged (58 pS and 37 pS in this patch) (Fig. 8B, right panel). Table 4 summarizes the pooled data describing single-channel properties from 4 patches of NR1/NR2D-(2A-ATD). Together, these data suggest that removal of the NR2A-ATD or replacement of the NR2A ATD with the NR2D ATD decreases open probability, whereas removal of the NR2D ATD or replacement of the NR2D ATD with the NR2A ATD increases open probability. This result supports the idea that the ATD from NR2A in general contributes to the high open probability, whereas the ATD from NR2D contributes to the low open probability of NMDA receptors. We also note that POPEN for NR2A-ATD deletion is lower than that for NR2A-(2D-ATD). However, we do not believe this comparison is informative, because it compares two very different mutant receptors. Rather, we restricted our conclusions to comparison of properties of deletion/chimeric receptors to the properties of wild-type receptors.
The NR2 ATD differentially controls specific shut duration components
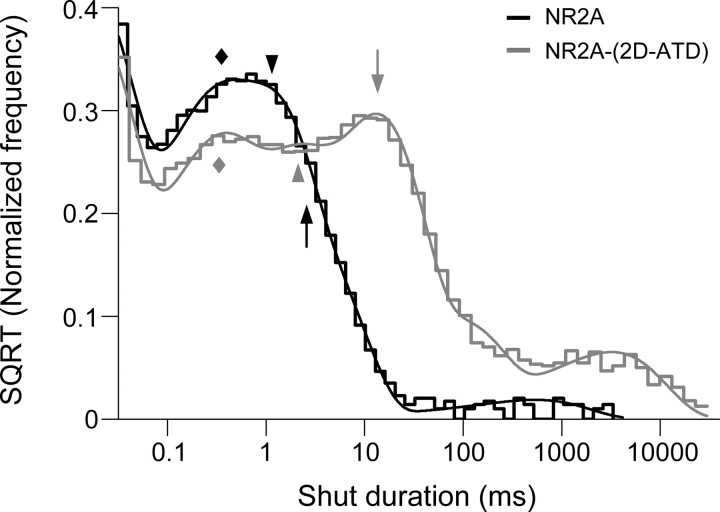
Analysis of single-channel recordings of patches with only a single NR2A-NR2D chimeric receptor allows us to interpret shut duration components, which in this situation contain information related to rate-limiting pregating steps. We found that a minimum of 5 exponential components were required to describe the histogram of shut durations for NR1/NR2A, and 6 components were necessary for the remaining receptors. Focusing on NR1/NR2A-(2D-ATD), we find an ultrafast ∼20 μs shut duration component that was not resolved in our previous studies of NR2A function (Erreger et al., 2005a,b; Erreger and Traynelis, 2008), but observed in other studies that similarly used time course fitting methods (Schorge et al., 2005). Omitting this component, the subsequent three intraburst shut duration components (τ1, τ2, τ3) matched reasonably well with values that we have previously reported. Figure 9 compares the composite pooled shut duration histograms (normalized to the number of events) for wild-type NR2A (n = 4) and NR2A-(2D-ATD) (n = 5). What is clear from the superimposed histograms is that the major effect of the NR2D ATD on NR2A function is to markedly slow the third shut duration component, with more modest effects on faster time constants. The low area (e.g., <1% for NR2A) and variability of τ4 and τ5 preclude meaningful comparisons, and thus these components were not evaluated. We have observed in a number of previous NR2A publications perturbations that reduce NR1/NR2A receptor function (such as partial agonists, increased proton concentrations, and Zn2+ binding to the ATD) shift this same shut duration component to a greater extent than the other components, suggesting that this time constant is strongly influenced by events occurring within the NR2A subunit (Table 5).
Figure 9.
NR2D-ATD controls τ3 from the exponential components fitted to the shut duration histogram. Overlay of the normalized composite fitted histograms of shut durations from wild-type NR2A (black line) and NR2A-(2D-ATD) (gray line). Each histogram was normalized by dividing the frequency of each dwell time (i.e., each bin) with the total number of dwell times used to generate the histogram. τ3 (arrow) of NR2A-(2D-ATD) was right-shifted 4.6-fold compared with the wild-type NR2A, while τ1 (diamond) and τ2 (arrowhead) show either no shift and shifted to right side only by 1.4-fold, respectively.
Table 5.
NR2D-ATD alters τ3 describing the shut duration histogram
| Control | NR1/NR2A experiment | τ2 (ms) control | τ2 (ms) exp | Con/exp (%) | τ3 (ms) control | τ3 (ms) exp | Con/exp (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Glutamate | Homoquinolinate | 0.49 | 0.46 | 110% | 4.7 | 6.2 | 130% | Erreger et al., 2005b |
| pH 7.3 | pH 6.7 | 0.79 | 0.71 | 90% | 4.3 | 7.4 | 170% | Unpublisheda |
| EDTA | 300 nm Zn2+ | 1 | 0.63 | 63% | 4 | 8 | 200% | Erreger and Traynelis, 2008 |
| NR2A | NR2A-(2D-ATD) | 0.91 | 1.3 | 140% | 2.8 | 13 | 460% | Current study |
Two intraburst shut duration constants (excluding τ0, the ultrafast 0.02 ms shut duration resolved using time course fitting for NR2A/D chimera in this study, and τ1) are compared for recombinant NR1/NR2A receptor, evaluated in outside out patches in control and the indicated experimental conditions. All experiments were performed in outside out patches at pH 7.3 in response to 100 μm glutamate 30 μm glycine in the presence of 10 μm EDTA to chelate contaminant Zn2+, except where indicated and for NR2A-NR2D ATD chimera, pH 8.0. Paired recordings were made from the same patch in control and experimental conditions. Fitted time constants are given for control recordings and the indicated experimental manipulations, and the percent change is shown. Data for homoquniolinic acid and 300 nm Zn2+ were from Erreger et al. (2005b) and Erreger and Traynelis (2008) and are included for comparison to data from this report.
aComposite shut duration histograms were constructed from recordings of NR1/NR2A in outside out patches (n = 7), and fitted as described in Materials and Methods (K. Erreger and S. F. Traynelis, unpublished data).
The linker between the ATD and the agonist-binding domain influences receptor open probability
The ATD of the NR2 subunit shows weak homology with bacterial periplasmic amino acid binding proteins and mGluR1, whose atomic structures have been determined by crystallographic methods (Paoletti et al., 2000; Madden, 2002; Malherbe et al., 2003; Marinelli et al., 2007). The periplasmic amino acid binding proteins as a group are shorter than the amino-terminal domain of the NR2 subunit, raising the possibility that a distinct linker region exists between a folded semi-autonomous ATD and the S1 region of the ligand-binding domain. Because our ATD deletion and chimeric constructs included regions beyond that for which homology models exist, we reasoned that a distinct linker region might control some of the effects that we had attributed to the ATD. To assess whether this region of the receptor is important in the results we found, we evaluated two additional chimeric receptors [NR2A-(2D-linker) and NR2D-(2A-linker)] constructed by interchanging the 16 residues in NR2A and NR2D between the end of the homology of the ATD with periplasmic binding proteins and the start of the S1 region of the ligand-binding domain (Fig. 10A). The EC50 values for glutamate and glycine of NR2A-(2D-linker) and NR2D-(2A-linker) expressed with NR1 in oocytes are comparable to the corresponding wild-type receptors [glutamate EC50: NR2A-(2D-linker) 2.6 μm; NR2D-(2A-linker) 0.77 μm; glycine EC50: NR2A-(2D-linker) 1.0 μm; NR2D-(2A-linker) 0.52 μm]. To evaluate the open probability, these chimeric NR2A or NR2D subunits were coexpressed with NR1-A652C in Xenopus oocytes, and the degree of MTSEA potentiation on agonist-evoked currents was compared with the potentiation of corresponding wild-type NR2A- or NR2D-containing receptors, or NR2A-(2D-ATD)- or NR2D-(2A-ATD)-containing receptors. The degree of potentiation induced by 200 μm MTSEA with glutamate and glycine was 11-fold for NR2A-(2D-linker) (Fig. 10B, left panel), which corresponds to an estimated POPEN of 0.08. This value was closer to that for the chimeric NR2A subunit in which the entire ATD was exchanged, NR2A-(2D-ATD) (POPEN 0.04), than to wild-type NR2A-containing receptors (POPEN 0.31). The degree of MTSEA potentiation on NR2D-(2A-linker) was 97-fold (Fig. 10B, right panel), which is less than that of the wild-type NR2D, although these data cannot be interpreted conclusively because agonist-evoked currents for NR2D-containing receptors are low in the absence of MTSEA. The deactivation time courses of NR2A-(2D-linker) and NR2D-(2A-linker) were evaluated by whole-cell voltage-clamp recordings of mutant NMDA receptors expressed in HEK 293 cells. The chimera NR2A-(2D-linker) has a more prolonged deactivation time constant (94 ± 7 ms; p < 0.01; one-way ANOVA with Bonferroni's post hoc test; compared with the wild-type NR2A) (Fig. 10C, left panel), while the chimera NR2D-(2A-linker) has an accelerated deactivation time constant (1980 ± 79 ms; p < 0.01; one-way ANOVA with Bonferroni's post hoc test; compared with the wild-type NR2D) (Fig. 10C, right panel). Both the degree of MTSEA potentiation and the deactivation rate of NR2A-(2D-linker) and NR2D-(2A-linker) lie intermediately between the corresponding wild-type NMDA receptors and NR2A-NR2D ATD chimeric receptors. These data indicate that the linker region between the ATD and the start of the ordered structure of the S1 region in the NR2 subunits contributes to the effect of the ATD on the open probability and the deactivation time course of NMDA receptors. Evaluation of additional chimeric receptors suggests that the boundaries defining the functional effects of this linker region may extend into the S1 region of the ligand-binding domain, since addition of 13 residues in NR2D-S1 adjacent to the NR2D linker region further decreases open probability in chimeric NR2A receptors (see supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
Figure 10.
The linker between the ATD and the agonist binding domain influences receptor open probability. A, Amino acid alignment of the linker region between the ATD and the agonist binding domain of NR2A and NR2D. The chimeras NR2A-(2D-linker) and NR2D-(2A-linker) were generated by interchanging the linker between the ATD and S1 (16 aa) from NR2A (V389-N404) and NR2D (L413-Q427). B, Summary of the degree of MTSEA (200 μm) potentiation of responses to glutamate and glycine (100 μm) from oocytes coexpressing NR1-A652C with NR2A-(2D-linker) or NR2D-(2A-linker). C, Summary of the mean deactivation time constants of responses to 50 μm glutamate obtained from whole-cell voltage-clamp (VHOLD = −60 mV) current recordings from HEK 293 cells transfected with NR1 and NR2A-(2D-linker) or NR2D-(2A-linker); 50 μm glycine was present in all solutions. The number of oocytes and HEK 293 cells is indicated in parenthesis above each bar. #p < 0.01, one-way ANOVA with Bonferroni's post hoc test, compared with the corresponding wild type.
Discussion
The most important conclusion of this study is that the ATD of the NR2 subunits controls biophysical and pharmacological properties of the NMDA receptor, including agonist potency, deactivation time course, open probability (POPEN), and mean open/shut duration. These properties are uniquely controlled by different NR2 subunits in wild-type NMDA receptors and fine-tune the physiological roles of native NMDA receptor subtypes. Because the ATD governs open probability and glutamate deactivation time course, this domain will be an important determinant of the signaling properties of synaptic NMDA receptors, and thus of critical importance to understand as a potential therapeutic target.
The ATD controls agonist potency of NMDA receptor subtypes
Our experiments show that there is a ninefold difference in glutamate EC50 between NR2A- and NR2D-containing receptors, which is consistent with other studies (Kutsuwada et al., 1992; Monyer et al., 1992; Erreger et al., 2007). When the NR2A ATD is replaced with the NR2D ATD, the glutamate EC50 value is reduced tenfold, becoming similar to that of the wild-type NR2D. Similarly, when the NR2D ATD is replaced by the NR2A ATD, the glutamate EC50 value is increased sevenfold, becoming similar to that of NR2A. However, we found potentially conflicting results between our studies of NR2A-NR2D chimeric subunits and our ATD deletion studies. Whereas glutamate potency in chimeric receptors reflects that of the subunit contributing the ATD, removal of the ATD from all four NR2 subunits has virtually no effect on the glutamate EC50 value. We interpret these data to suggest that in full-length NR2 subunits, the ATD influences the potency of glutamate, presumably through actions on gating and possibly stability of the glutamate-bound agonist-binding domain, which can on its own influence glutamate potency (Erreger et al., 2007). However, the effect of ATD deletion on glutamate potency is more difficult to interpret—possibly due to different effects of removing both intra- and inter-subunit allosteric interactions of the ATD. Surprisingly, removal of the NR2 ATD appears to markedly influence the EC50 values for glycine activation for NR2B-, NR2C-, and NR2D-containing receptors, with little effect on NR2A-containing receptors. This is consistent with previous data suggesting that the S2 region of the NR2A ligand binding may control glycine potency (Chen et al., 2008), and also suggests that the structural determinants of inter-subunit interactions are not identical for receptors containing the NR2A subunit compared with other NR2 subunits.
These data suggest that the identity of the ATD is one important factor driving the difference in glutamate and glycine potency among NR2A- and NR2D-containing NMDA receptors. This result further implies that the unique configurations of the NR2A-ATD and NR2D-ATD may have distinct allosteric interactions within the protein complex. The apparent effects of ATD deletion or substitution on glycine EC50 further suggests that the sensitivity of the receptor to glycine (especially at NR2B, NR2C, and NR2D) is influenced by the presence of the ATD, which might interact in some manner with the ATD of NR1.
The ATD controls glutamate deactivation of NMDA receptor subtypes
The deactivation time course of NMDA receptor current responses following removal of glutamate determines the duration of EPSCs (Lester et al., 1990), and is relatively fast in NR2A-containing receptors, but slow in NR2D-containing receptors (Vicini et al., 1998; Wyllie et al., 1998). Thus, because the ATD can influence the time course of deactivation, this domain sets the temporal features of synaptic NMDA receptor signaling. Our study shows that removal of the ATD from NR2A slows deactivation, while removal of the ATD from NR2B, NR2C, and NR2D accelerates deactivation. These effects are not correlated with glutamate EC50 values, which are largely unchanged by removal of the NR2 ATD. The ability of the ATD to influence deactivation likely reflects ATD control of receptor gating. Interestingly, replacement of the NR2A ATD with the NR2D ATD or replacement of the NR2D ATD with the NR2A ATD results in intermediate deactivation time constants, which are tenfold higher than wild-type NR2A and tenfold lower than wild-type NR2D. Thus, the inherent actions of the NR2A ATD can be transferred to NR2D, and the inherent actions of the NR2D ATD can be transferred to NR2A. This suggests that these semi-autonomous domains can drive similar long-range actions on NMDA receptor channel function regardless of the NR2 identity, suggesting that the downstream pathway and mechanism by which ATD acts is conserved within the NR2 subunit family.
Structural mechanism for the actions of the ATD on receptor function
The presence of the NR2A ATD on the NR2A subunit allows receptors to open with high probability, whereas the NR2D ATD present on either NR2D or NR2A impedes channel opening compared with wild-type receptors and is associated with a low open probability for agonist-bound receptors. That is, the stability and the frequency of reaching the active conformation are controlled by the structural nature of the ATD in receptors containing full-length NR2. Single-channel data confirm that removal or replacement of the ATD has no effect on the major conductance levels, suggesting that the conformation of the pore of the mutant receptor is similar to that of the wild-type receptor. The ATD of the NR2 subunits is a bilobed structure similar to the bacterial periplasmic amino acid binding proteins and the glutamate binding domain of mGluR1 (Paoletti et al., 2000; Malherbe et al., 2003; Marinelli et al., 2007). Recently, it has been proposed that Zn2+ binding to the NR2A ATD induces some degree of closure around a central cleft in the bilobed ATD, which induces intra-protein rearrangements that inhibit NMDA receptor function, perhaps involving a rearrangement of the interface of an NR1-NR2 ATD dimer and enhancement of proton sensitivity (Paoletti et al., 2000; Erreger and Traynelis, 2008; Gielen et al., 2008). A similar hypothesis has been advanced for ifenprodil binding to the NR2B ATD (Perin-Dureau et al., 2002). If this idea is correct, one might speculate that the cleft of the NR2C and NR2D ATDs naturally adopt a more closed conformation relative to the ATD of NR2A and (to a lesser extent) NR2B. Interestingly, our data estimating open probability support this subdivision of the effects of the NR2 ATD, with NR2A/B ATD enhancing open probability and NR2C/D ATD decreasing open probability. This latter observation reinforces the idea that NR2A/B and NR2C/D each share operational principles that reflect in part the actions of the ATDs.
The crystallographic structure of the heterodimer between the isolated agonist binding domains of NR1 and NR2A show several important atomic contacts between NR1 and NR2A (Furukawa et al., 2005). In AMPA and kainate receptors, as well as GluRδ2 receptors, it is well know that the dimer interface between the agonist binding domains is an important site for functional regulation (Partin et al., 1996; Sun et al., 2002; Jin et al., 2005; Mayer, 2006; Weston et al., 2006; Hansen et al., 2007, 2009; Plested and Mayer, 2007; Plested et al., 2008). Consistent with this important precedent, it has been previously proposed (Kunishima et al., 2000; Huggins and Grant, 2005; Gielen et al., 2008) and recently shown that the AMPA and kainate receptor ATDs form a dimer similar to the dimers of the agonist binding domain (Jin et al., 2009; Kumar et al., 2009). If this hypothesis transfers to NMDA receptors, it is possible that changes in agonist potency, deactivation time course, and single-channel properties in the NR2 ATD deletion constructs and ATD chimeras of NMDA receptors may be the result of changes in the conformation of the dimer interface between agonist-binding domains caused by disruption or altered configuration of the ATD (Gielen et al., 2008). Alternatively, such changes could reflect differences in the ATD dimer interface for the various NR2 subunits. Future studies are needed to clarify whether similar perturbations of an ATD dimer interface could explain how the NR2 ATD can control the NMDA receptor properties.
Conclusion
The present study establishes the ATD within the NR2 subunits as a modulatory domain of NMDA receptors that can be deleted or swapped between different NR2 subunits with a predictive effect on receptor function. The effect of the modulatory ATD on receptor function is highly subunit-dependent, suggesting that the ATD plays an important role in fine-tuning the functional properties of the individual NMDA receptor subtypes. These findings provide new insight into functional and structural differences of the individual NR2 subunits that make up the NMDA receptor subtypes.
Footnotes
This work was supported by the National Institutes of Health (Grant NS036654; to S.F.T.), the Michael J. Fox Foundation (S.F.T.), the Lundbeck Foundation (K.B.H.), and the Villum Kann Rasmussen Foundation (K.B.H.). We thank Phuong Le and Kimberly Vellano for excellent technical assistance. We thank Dr. Chian-Ming Low for providing us with the cDNAs of NR2B-(ΔS28-M394) and Dr. Kevin Erreger for sharing unpublished data describing the effect of protons on the shut duration histogram for NR1/NR2A receptors (Table 5).
References
- Béhé P, Colquhoun D, Wyllie DJ. Activation of Single AMPA- and NMDA-Type Glutamate-Receptor Channels. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS. Springer; 1999. pp. 175–218. [Google Scholar]
- Chen PE, Geballe MT, Katz E, Erreger K, Livesey MR, O'Toole KK, Le P, Lee CJ, Snyder JP, Traynelis SF, Wyllie DJ. Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol. 2008;586:227–245. doi: 10.1113/jphysiol.2007.143172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chen HV, Lipton SA. Three pairs of cysteine residues mediate both redox and Zn2+ modulation of the NMDA receptor. J Neurosci. 2001;21:392–400. doi: 10.1523/JNEUROSCI.21-02-00392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YB, Lipton SA. Identification and mechanism of action of two histidine residues underlying high-affinity Zn2+ inhibition of the NMDA receptor. Neuron. 1999;23:171–180. doi: 10.1016/s0896-6273(00)80763-1. [DOI] [PubMed] [Google Scholar]
- Colquhoun D, Hawkes AG. Stochastic properties of ion channel openings and bursts in a membrane patch that contains two channels: evidence concerning the number of channels present when a record containing only single openings is observed. Proc R Soc Lond B Biol Sci. 1990;240:453–477. doi: 10.1098/rspb.1990.0048. [DOI] [PubMed] [Google Scholar]
- Cowan CM, Raymond LA. Selective neuronal degeneration in Huntington's disease. Curr Top Dev Biol. 2006;75:25–71. doi: 10.1016/S0070-2153(06)75002-5. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Dravid SM, Prakash A, Traynelis SF. Activation of recombinant NR1/NR2C NMDA receptors. J Physiol. 2008;586:4425–4439. doi: 10.1113/jphysiol.2008.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF. Zinc inhibition of rat NR1/NR2A N-methyl-d-aspartate receptors. J Physiol. 2008;586:763–778. doi: 10.1113/jphysiol.2007.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Chen PE, Wyllie DJ, Traynelis SF. Glutamate receptor gating. Crit Rev Neurobiol. 2004;16:187–224. doi: 10.1615/critrevneurobiol.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J Physiol. 2005a;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Dravid SM, Snyder JP, Wyllie DJ, Traynelis SF. Mechanism of partial agonism at NMDA receptors for a conformationally restricted glutamate analog. J Neurosci. 2005b;25:7858–7866. doi: 10.1523/JNEUROSCI.1613-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Kristensen A, Chen PE, Hansen KB, Lee CJ, Yuan H, Le P, Lyuboslavsky PN, Micale N, Jørgensen L, Clausen RP, Wyllie DJ, Snyder JP, Traynelis SF. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-d-aspartate glutamate receptors. Mol Pharmacol. 2007;72:907–920. doi: 10.1124/mol.107.037333. [DOI] [PubMed] [Google Scholar]
- Farrant M, Feldmeyer D, Takahashi T, Cull-Candy SG. NMDA-receptor channel diversity in the developing cerebellum. Nature. 1994;368:335–339. doi: 10.1038/368335a0. [DOI] [PubMed] [Google Scholar]
- Fayyazuddin A, Villarroel A, Le Goff A, Lerma J, Neyton J. Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron. 2000;25:683–694. doi: 10.1016/s0896-6273(00)81070-3. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett PJ, Standaert DG. Rationale for and use of NMDA receptor antagonists in Parkinson's disease. Pharmacol Ther. 2004;102:155–174. doi: 10.1016/j.pharmthera.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Hansen KB, Yuan H, Traynelis SF. Structural aspects of AMPA receptor activation, desensitization and deactivation. Curr Opin Neurobiol. 2007;17:281–288. doi: 10.1016/j.conb.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Hansen KB, Naur P, Kurtkaya NL, Kristensen AS, Gajhede M, Kastrup JS, Traynelis SF. Modulation of the dimer interface at ionotropic glutamate-like receptor delta2 by d-serine and extracellular calcium. J Neurosci. 2009;29:907–917. doi: 10.1523/JNEUROSCI.4081-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins DJ, Grant GH. The function of the amino terminal domain in NMDA receptor modulation. J Mol Graph Model. 2005;23:381–388. doi: 10.1016/j.jmgm.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int. 2004;45:583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N, Masu M. Molecular characterization of the family of the N-methyl-d-aspartate receptor subunits. J Biol Chem. 1993;268:2836–2843. [PubMed] [Google Scholar]
- Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behavior of the GluR2 amino terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, VanDongen HM, VanDongen AM. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci. 2002;22:2044–2053. doi: 10.1523/JNEUROSCI.22-06-02044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T, Arakawa M. Molecular diversity of the NMDA receptor channel. Nature. 1992;358:36–41. doi: 10.1038/358036a0. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Bartke I, Schoepfer R, Naujoks K, Seeburg PH. Regional, developmental and interspecies expression of the four NMDAR2 subunits, examined using monoclonal antibodies. Brain Res Mol Brain Res. 1997;51:23–32. doi: 10.1016/s0169-328x(97)00206-4. [DOI] [PubMed] [Google Scholar]
- Lester RA, Clements JD, Westbrook GL, Jahr CE. Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature. 1990;346:565–567. doi: 10.1038/346565a0. [DOI] [PubMed] [Google Scholar]
- Low CM, Zheng F, Lyuboslavsky P, Traynelis SF. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-d-aspartate NR1/NR2A receptors. Proc Natl Acad Sci U S A. 2000;97:11062–11067. doi: 10.1073/pnas.180307497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DR. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci. 2002;3:91–101. doi: 10.1038/nrn725. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Mutel V, Broger C, Perin-Dureau F, Kemp JA, Neyton J, Paoletti P, Kew JN. Identification of critical residues in the amino terminal domain of the human NR2B subunit involved in the RO 25–6981 binding pocket. J Pharmacol Exp Ther. 2003;307:897–905. doi: 10.1124/jpet.103.056291. [DOI] [PubMed] [Google Scholar]
- Marinelli L, Cosconati S, Steinbrecher T, Limongelli V, Bertamino A, Novellino E, Case DA. Homology modeling of NR2B modulatory domain of NMDA receptor and analysis of ifenprodil binding. ChemMedChem. 2007;2:1498–1510. doi: 10.1002/cmdc.200700091. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- Mehta SL, Manhas N, Raghubir R. Molecular targets in cerebral ischemia for developing novel therapeutics. Brain Res Rev. 2007;54:34–66. doi: 10.1016/j.brainresrev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Ng FM, Geballe MT, Snyder JP, Traynelis SF, Low CM. Structural insights into phenylethanolamines high-affinity binding site in NR2B from binding and molecular modeling studies. Mol Brain. 2008;1:16. doi: 10.1186/1756-6606-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Perin-Dureau F, Fayyazuddin A, Le Goff A, Callebaut I, Neyton J. Molecular organization of a zinc binding n-terminal modulatory domain in a NMDA receptor subunit. Neuron. 2000;28:911–925. doi: 10.1016/s0896-6273(00)00163-x. [DOI] [PubMed] [Google Scholar]
- Partin KM, Fleck MW, Mayer ML. AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J Neurosci. 1996;16:6634–6647. doi: 10.1523/JNEUROSCI.16-21-06634.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin-Dureau F, Rachline J, Neyton J, Paoletti P. Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors. J Neurosci. 2002;22:5955–5965. doi: 10.1523/JNEUROSCI.22-14-05955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RX, Reid IC, Stewart CA. The N-methyl-d-aspartate receptor, synaptic plasticity, and depressive disorder. A critical review. Pharmacol Ther. 2000;87:11–25. doi: 10.1016/s0163-7258(00)00063-2. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel G, Platt B, Micheau J. Glutamate receptor function in learning and memory. Behav Brain Res. 2003;140:1–47. doi: 10.1016/s0166-4328(02)00272-3. [DOI] [PubMed] [Google Scholar]
- Schorge S, Elenes S, Colquhoun D. Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J Physiol. 2005;569:395–418. doi: 10.1113/jphysiol.2005.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern P, Béhé P, Schoepfer R, Colquhoun D. Single-channel conductances of NMDA receptors expressed from cloned cDNAs: comparison with native receptors. Proc Biol Sci. 1992;250:271–277. doi: 10.1098/rspb.1992.0159. [DOI] [PubMed] [Google Scholar]
- Sun L, Shipley MT, Lidow MS. Expression of NR1, NR2A-D, and NR3 subunits of the NMDA receptor in the cerebral cortex and olfactory bulb of adult rat. Synapse. 2000;35:212–221. doi: 10.1002/(SICI)1098-2396(20000301)35:3<212::AID-SYN6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci. 1998;18:6163–6175. doi: 10.1523/JNEUROSCI.18-16-06163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai G, Coyle JT. Glutamatergic mechanisms in schizophrenia. Annu Rev Pharmacol Toxicol. 2002;42:165–179. doi: 10.1146/annurev.pharmtox.42.082701.160735. [DOI] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. Functional and pharmacological differences between recombinant N-methyl-d-aspartate receptors. J Neurophysiol. 1998;79:555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nat Struct Mol Biol. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- Wong E, Ng FM, Yu CY, Lim P, Lim LH, Traynelis SF, Low CM. Expression and characterization of soluble amino-terminal domain of NR2B subunit of N-methyl-d-aspartate receptor. Protein Sci. 2005;14:2275–2283. doi: 10.1110/ps.051509905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie DJ, Béhé P, Colquhoun D. Single-channel activations and concentration jumps: comparison of recombinant NR1a/NR2A and NR1a/NR2D NMDA receptors. J Physiol. 1998;510:1–18. doi: 10.1111/j.1469-7793.1998.001bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Erreger K, Dravid SM, Traynelis SF. Conserved structural and functional control of N-methyl-d-aspartate receptor gating by transmembrane domain M3. J Biol Chem. 2005;280:29708–29716. doi: 10.1074/jbc.M414215200. [DOI] [PubMed] [Google Scholar]
- Yuan H, Vance KM, Junge CE, Geballe MT, Snyder JP, Hepler JR, Yepes M, Low CM, Traynelis SF. The serine protease plasmin cleaves the amino-terminal domain of the NR2A subunit to relieve zinc inhibition of the N-Methyl-d-aspartate receptors. J Biol Chem. 2009;284:12862–12873. doi: 10.1074/jbc.M805123200. [DOI] [PMC free article] [PubMed] [Google Scholar]