Abstract
Many biologically active bacterial natural products contain highly modified deoxysugar residues that are often critical for the activity of the parent compounds. Most of these deoxysugars are secondary metabolites that are biosynthesized in the form of nucleotide diphosphate (NDP) sugars prior to their transfer to natural product aglycones by glycosyltransferases. Over the past decade, many biosynthetic pathways that lead to the formation of these unusual sugars have been unraveled, and the mechanisms of many key enzymatic transformations involved in these pathways have been elucidated. However, obtaining workable quantities of NDP-deoxysugars for in vitro studies is often a difficult task. This limitation has hindered an in-depth investigation of the substrate specificity of deoxysugar biosynthetic enzymes, many of which are promiscuous with respect to their NDP-sugar substrates and are, thus, potentially useful catalysts for natural product glycoengineering. Presented in this review are procedures for the enzymatic synthesis and purification of a variety of NDP-deoxysugars, including some early intermediates in NDP-deoxysugar biosynthetic pathways, and highly modified NDP-deoxysugars that are late intermediates in their respective biosynthetic pathways. The procedures described herein could be used as general guidelines for the development of specific protocols for the synthesis of other NDP-deoxysugars.
Introduction
Glycosylation is important for the biological activity of macrolide, peptide and aminoglycoside antibioticsas well as numerous anticancer, antiparasitic, and antifungal agents of diverse biosynthetic origin (Lamb and Wright, 2005; Mendez and Salas, 2001; Walsh et al., 2003). These sugar residues play crucial biological roles in many natural products and their removal oftentimes results in the loss of biological activity (Mendez and Salas, 2001; Thorson et al., 2001; Weymouth-Wilson, 1997). The most chemically diverse group of carbohydrate moieties found in secondary metabolites are 6-deoxyhexoses, which are produced by a variety of organisms, but are most prevalent in actinomycetes, a group of soil bacteria that are a rich source of biologically active secondary metabolites (Salas and Mendez, 2007). For many of the natural product biosynthetic pathways found in these organisms, a combination of experimental evidence and database comparisons has been used to identify the deoxysugar biosynthetic genes and to delineate the corresponding biosynthetic pathways (reviewed in Thibodeaux et al., 2009).
Altering and/or exchanging the sugar structures and points of aglycone attachment in natural products is a feasible route to enhance or vary the physiological properties of these compounds. Importantly, it has been discovered that many natural product glycosyltransferases (or GTs -enzymes which link activated sugar donors to aglycone acceptors) exhibit relaxed substrate specificity. This discovery has resulted in an explosion in the use of these GTs to engineer natural products with altered glycosylation patterns, and has generated several broad and complementary glycoengineering strategies (Thibodeaux et al., 2007; Thibodeaux et al., 2009). For in vivo glycodiversification, deoxysugar biosynthetic pathways can be altered within a producing bacterial strain using gene disruption and/or heterologous gene expression methods in order to reroute sugar biosynthetic intermedaites to new final products. Alternatively, new biosynthetic pathways can be assembled in hosts that do not normally produce glycosylated natural products. Furthermore, these genetically engineered bacteria can be fed with non-native aglycones or they can be transformed with additional plasmids in order to produce novel compounds in a combinatorial fashion. In vitro, purified wild-type or engineered sugar biosynthetic enzymes can be used to synthesize specific natural product glycoforms, as well as to prepare libraries of novel glycoforms. A detailed understanding of the organization of deoxysugar biosynthetic machinery and of the biochemical properties of the enzymes involved in synthesizing and coupling these sugars to their aglycones is critical to the success of these enzymatic glycoengineering approaches.
A significant limitation to in vitro glycodiversification efforts is the availability of activated sugar donor substrates for the promiscuous glycosyltransferases. There are several methods by which activated deoxysugar donors (i.e., NDP-deoxysugars) are generally obtained. Total chemical synthesis, though feasible, often requires significant technical expertise and can be plagued by low yields. Another approach for obtaining highly modified NDP-deoxysugars is to hydrolyze the desired reducing sugar from the natural product, then to chemically synthesize the NDP sugar from the reducing sugar (Chang et al., 2000; Chen et al., 2002). The recently discovered in vitro reversibility of GT-catalyzed reactions may provide a new synthetic route to some NDP-deoxysugars (Bode and Muller, 2007; Melancon et al., 2006; Minami et al., 2005; Zhang et al., 2006), but it is not yet clear whether this will be a generally applicable method for NDP-deoxysugar synthesis. Thus, we envision that multi-step enzymatic and chemoenzymatic synthesis will continue to play an important role in the production of structurally complex, NDP-activated deoxysugar donors for glycobiology and glycoengineering studies.
Enzymatic synthesis is appealing for several reasons. Enzymes generally catalyze reactions with well-defined regio- and steroeochemical preference and enzymes are a readilly renewable resource. Furthermore, when manipulated in vitro, it is possible to present these enzymes with substrate analogues that contain chemically incorporated non-natural functional groups. Introduction of certain types of functional groups can allow further chemoselective derivitization to enhance the structural diversity of glycoform libraries (Fu et al., 2003). Here, we provide detailed protocols for the enzymatic synthesis and purification of several thymidine diphosphate (TDP)-activated deoxysugar intermediates that are common to many TDP-deoxysugar biosynthetic pathways in bacteria. These methods should be useful for researchers interested in obtaining workable quantities of a desired TDP-deoxysugar. We also highlight several successful examples of in vitro multi-step enzymatic syntheses of highly modified TDP-deoxysugars and discuss how biosynthetic machinery can be manipulated in vivo to generate desired deoxysugar structures. For more information on NDP-deoxysugar synthesis, bacterial deoxysugar biosynthesis, mechanistic studies of deoxysugar biosynthetic enzymes, and the application of these enzymes in glycoengineering efforts, the reader is directed to several recent and comprehensive reviews (He et al., 2000; He and Liu, 2002; Langenhan et al., 2005; Luzhetskyy et al., 2008; Mendez et al., 2008; Rupprath et al., 2005; Salas and Mendez, 2007; Thibodeaux et al., 2009).
1. Enzymatic Synthesis of TDP-α-D-glucose
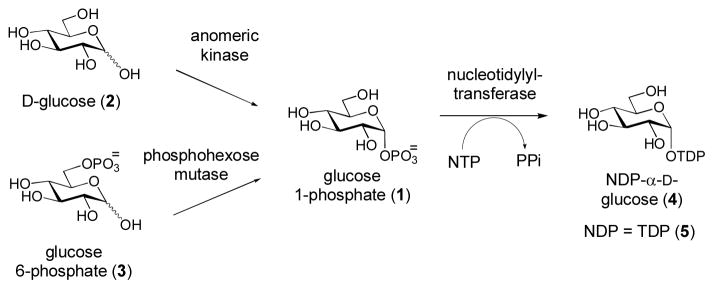
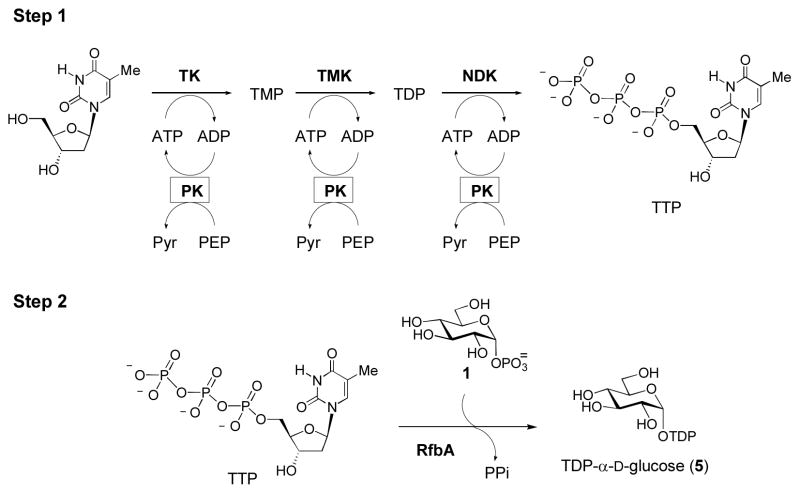
Most of the unusual deoxysugars produced by bacterial biosynthetic pathways are derived from α-D-glucose-1-phosphate (1) which is, in turn, derived from D-glucose (2) by direct anomeric phosphorylation, or from glucose-6-phosphate (3) by a phosphohexose mutase-catalyzed reaction (Scheme 1). A nucleotide monophosphate (NMP) moiety from a nucleotide triphosphate (NTP) is then coupled to 1 by a nucleotidylyltransferase to generate NDP-glucose (4). While some deoxysugars are derived from primary metabolites and can be activated with other NDP groups, the vast majority of bacterial deoxysugars used in glycosylation of secondary metabolites are TDP-sugars that are biosynthetically dervived from TDP-α-D-glucose (5) (Thibodeaux et al., 2007; Thibodeaux et al., 2009). Because TDP-α-D-glucose (5) is rather costly, efficient methods for its preparation are desirable. Towards this end, we have developed an efficient and facile one-pot, two-step enzymatic synthesis for 5 (Scheme 2) using readily available enzymes and inexpensive substrates (Takahashi et al., 2006). In the first step of this reaction, thymidine is converted to thymidine triphosphate (TTP) by the sequential action of three separate ATP-dependent kinases [thymidine kinase (TK), thymidylate kinase (TMK), and nucleotide diphosphate kinase (NDK)]. ATP is continually regenerated by pyruvate kinase at the expense of phosphoenol pyruvate (PEP). In the second step of the synthesis, a TMP moiety from TTP is coupled to α-D-glucose-1-phosphate (1) by RfbA, an α-D-glucose-1-phosphate thymidylyltransferase from Salmonella enterica. The resulting TDP-α-D-glucose (5) can then be purified or used directly for subsequent enzymatic transformation.
Scheme 1. Activation of the Glycolytic Intermediate D-Glucose.
Sugar activation. D-Glucose (2) and glucose-6-phosphate (3) can be converted to α-D-glucose-1-phosphate (1) by anomeric kinase and phosphohexose mutase, respectively. A nucleotide monophosphate (NMP) moiety is then tranferred from the corresponding NTP to 1 by a nucleotidylyltransferase to from NDP- α-D-glucose (4). Most deoxysugars produced in bacterial secondary metabolism are derived from TDP- α-D-glucose (5).
Scheme 2. Enzymatic Synthesis of TDP- agr;-D-glucose (5).
In the first step of TDP- α-D-glucose (5) synthesis, thymidine triphosphate (TTP) is synthesized from thymidine by three successive ATP-dependent phosphorylations catalyzed by thymidine kinase (TK), thymidylate kinase (TMK), and nucleotide diphosphate kinase (NDK). ATP is only needed in catalytic amounts, due to a pyruvate kinase (PK) ATP regeneration system, which transfers a phosphate group from phosphoenol pyruvate (PEP) to ADP, yielding pyruvate (Pyr) and ATP. Removal of the enzymes and addition of α-D-glucose-1-phosphate (1) and RfbA (a thymidylyltransferase from Salmonella enterica) leads to the synthesis of TDP- α-D-glucose (5).
1.1 Preparation of Enzymes Required for in vitro Synthesis of TDP-α-D-glucose (5)
The genes encoding thymidine kinase (TK), thymidylate kinase (TMK), and nucleotide diphosphate kinase (NDK) can be amplified from the genomic DNA of Escherichia coli strain HMS174 and cloned in tandem into a single pET28b (+) plasmid, such that each gene has its own ribosome binding site and a His6-tag. The three enzymes can then be co-expressed from the same plasmid in E. coli BL21(DE3) using standard growth and induction conditions. Typically, we inoculate LB medium (supplemented with 50 μg/mL kanamycin) with a 1:500 dilution from an overnight culture, grow the cells at 37 °C, induce with 0.3 mM isopropyl α-thiogalactoside (IPTG) at OD600 readings of 0.4–0.6, and continue to grow for 18 h at 25 °C. Cells are then harvested by centrifugation, lysed by sonication, and centrifuged to remove cellular debris. The recombinant TK/TMK/NDK enzymes are then purified from the resulting supernatant using Ni-NTA affinity chromatography. We generally use 150 mM NaCl in our lysis, wash, and elution buffers, as opposed to the 300 mM recommended in the QIAexpressionist protocol. The mixture of purified TK/TMK/NDK enzymes is dialyzed against 50 mM potassium phosphate buffer (pH 7.5) containing 15% glycerol.
In parallel, rabbit muscle pyruvate kinase from Sigma (purchased as a 400–800 units/mg ammonium sulfate precipitate) is dissolved in water to a concentration of 2500 units/mL, dialyzed against buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) to remove the ammonium sulfate, dispensed in 500-μL to 1-mL aliquots, flash frozen in liquid nitrogen, and stored at −80 °C. The rfbA gene is PCR-amplified from Salmonella enterica serovar Typhimurium LT2 genomic DNA with an upstream ribosome binding site (RBS)/translational spacer element (TSE) and is inserted into a pUC18 vector. A (His)5-tag is added to the C-terminus of RfbA by PCR amplification of the rfbA insert of plasmid rfbA/pUC18. The newly synthesized gene is then cloned into the same vector, and the resulting plasmid, rfbA-(His)5/pUC18, introduced by transformation into BL21(DE3) for protein expression. Expression and purification conditions are identical to those for the TK/TMK/NDK enzymes, except that 100 μg/mL ampicillin is used for selection. Induction with IPTG is unnecessary. The approximate amount of RfbA purified from an average 6 L culture is 140 mg. Each liter of culture produces enough RfbA to perform one large-scale TDP-α-D-glucose synthesis reaction.
1.2 Enzymatic Synthesis of TDP-α-D-glucose (5)
In the first step of TDP-α-D-glucose synthesis, a 15-mL reaction mixture containing phosphoenol pyruvate (PEP, 85.6 mM), thymidine (27 mM), ATP (1.8 mM), and MgCl2 (30 mM) in 50 mM Tris ·HCl buffer (pH 7.5) is prepared. After the addition of these reagents, the pH of the solution is adjusted to 7.5 using NaOH or HCl prior to the addition of enzymes. Pyruvate kinase (PK, 1000 units) and ~ 30 mg of the TK/TMK/NDK enzyme mixture are added to give final concentrations of ~ 25–30 μM for each of the TK/TMK/NDK enzymes. The reaction mixture is incubated at 37 °C for 4 h, then filtered through an Amicon ultrafiltration cell unit (YM-10 membrane) under nitrogen at 4 °C to remove the enzymes. The flow-through is collected in a 50-mL conical tube and the pH adjusted to 7.5. This flow-through contains TTP and will be used in the next step.
To synthesize TDP-α-D-glucose (5), a mixture containing TTP, α-D-glucose-1-phosphate (1, 4 mM), MgCl2 (30 mM), and recombinant RfbA (47 μM) is prepared. This reaction mixture is incubated at 30–37 °C for 12 to 16 h. It is important to keep the Mg2+ ion concentration similar to that of TTP, as a high molar excess of Mg2+ (over TTP) is known to adversely affect the activity of RfbA (Amann et al., 2001). After the reaction is completed, any precipitate is removed by centrifugation at 5000 × g for 10 min at 4 °C. RfbA is then removed by filtering the supernatant through either a YM-10 Amicon stirred cell unit or a 50 mL conical centrifugal filter unit at 4 °C. TDP-D-glucose (5) present in the flow-through can now be purified or used directly for the following enzymatic reactions.
1.3 Purification of TDP-α-D-glucose (5)
TDP-α-D-glucose (5) can be purified with either anion exchange or size-exclusion chromatography. When using the size-exclusion method, the sample containing 5 should be frozen and lyophilized to reduce the volume to 1–2 mL. It is important to note that many TDP-sugars degrade significantly if lyophilized to dryness without removing salts. For this reason, if the stability of a particular NDP-sugar is not known, it is always best to lyophilize to half-volume, dilute with the appropriate solvent, and lyophilize to half-volume again. This cycle can be repeated as necessary to exchange solvents or to remove volatile substances. When the concentrated TDP-α-D-glucose is thawed, the sample typically appears as a slightly yellowish syrup. If purifying by FPLC using a MonoQ column, the TDP-α-D-glucose sample needs not to be lyophilized/concentrated before purification.
1.3.1 Purification Using Size-Exclusion Chromatography
A P2 biogel (Biorad) column (2.5 × 100 cm) is packed following the manufacturer’s instructions and equilibrated with 1 L of filtered ddH2O at 4 °C. After washing, the solvent above the column bed is removed and the concentrated (1–2 mL) TDP-α-D-glucose-containing reaction mixture is carefully applied to the top of the resin bed. The sample is introduced into the resin by gravity flow, ensuring that the resin bed does not dry. After the sample has been loaded onto the column, water is used to elute the TDP-sugar product. An adjustible pneumatic pump is used to maintain the flow rate at about 6–12 mL per hour. Under these conditions, 5 will generally be eluted from the column around 24–48 h after loading. Fractions eluted from the P2 column can be analyzed by UV-Vis spectroscopy for the presence of 5 (ε267 = 9600 M−1cm−1). Usually, there will be a sharp rise in absorbance in the first fraction containing 5, followed by ~10–12 fractions with high absorbance readings, and then a sharp decrease in absorbance to baseline. The fractions comprising the absorbance plateau are individually frozen and lyophilized until their purity can be verified by HPLC or NMR spectroscopy.
To verify the purity of the fractions from the P2 size-exclusion column, HPLC analysis is performed using a Dionex Carbopac PA1 column and a 20-μL sample injection volume. With water as solvent A and 500 mM NH4OAc (adjusted to pH 7.0 with aqueous NH3) as solvent B, the following gradient elution is typically used: 5–20% B over 15 min, 20–60% B over 20 min, 60–100% B over 2 min, 3 min wash at 100% B, 100-5% B over 5 min, and re-equilibration at 5% B for 15 min. The flow rate is 1 mL/min and the detector is set at 267 nm. Under these conditions, the retention times for compounds are as follows: TDP-α-D-glucose (31–33 min), TMP (25 min), TDP (41–42 min). In samples of high concentration, TDP-α-D-glucose (5) may be eluted anywhere from 30 to 34 min, but will still appear as a very sharp, well-defined peak in the HPLC trace. Fractions are pooled based on purity either before or after lyophilization. The final concentration of 5 in pooled fractions can be determined spectrophotometrically (ε267 = 9600 M−1cm−1). The theoretical yield for this reaction is 228 mg (based on a 15 mL reaction and the substrate concentrations described in 1.2), but the actual yields range from 150 to 200 mg, depending on one’s experience with the methodology. Most commonly, one will find that 10% of the product will be greater than 90% pure, 40% will be 90–80% pure, 40% will be 80-70% pure, and the remaining 10% will be less than 70% pure. The most abundant contaminants using P2 size-exclusion chromatography as the method of purification are TMP and TDP, both of which can be removed by further FPLC purification. However, we have found that further purification is usually unnecessary if 5 is to be used in enzymatic reactions with high yield (i.e., > 90% conversion).
1.3.2 FPLC Purification of TDP-α-D-glucose (5)
As mentioned above, P2 chromatography will separate the majority of unreacted α-D-glucose-1-phosphate (1) from the desired TDP-α-D-glucose product (5). However, only about 10% of 5 will be > 90% pure, with the remainder being contaminated by TDP and TMP. The majority of TDP and TMP in these samples can be removed if the P2 fractions are subjected to anion exchange chromatography. It should be noted that, in many cases, the enzymatically synthesized TDP-sugars can be purified directly by FPLC, bypassing the P2 purification step. Purification of 5 can be achieved by FPLC using a Mono Q 10/10 or 16/10 column with a gradient, where HPLC-grade water is buffer A and 400 mM NH4HCO3 in water (pH, 8.2) is buffer B. Flow rates of 1 or 4 mL/min are used for the 10/10 and 16/10 columns, respectively. To elute 5, the following gradient is used: from 0 to 10% B over 0.5 column volumes, from 10 to 40% B over four column volumes (which will elute the TDP-sugar), and from 40 to 100% B over 0.5 column volumes. The column is then washed with 100% B for two column volumes, followed by reduction to 0% B over 0.5 column volumes, and reequilibration at 0% B over three column volumes. TDP-α-D-glucose (5) will be eluted at approximately 30 min. Other important peaks include TMP and TDP, which have retention times of 29 min and 34 min, respectively.
Fractions containing the major peak from each injection should be lyophilized individually, redissolved in water, and lyophilized again to remove NH4HCO3. Alternatively, fractions can be desalted using a G-10 column (see Section 1.3.3), lyophilized, redissolved in water, and lyophilized again. The purities of these fractions should be analyzed by 1H and 31P NMR spectroscopy and typically range from 50 to 90%. The most common residual contaminant is TMP. The resolution of the Mono-Q column is not sufficient to completely separate TMP from TDP-α-D-glucose (5) in cases of large injection volumes or high concentrations of samples. From this method, one can typically obtain an average of 25 mg of 90% pure TDP-α-D-glucose.
1.3.3 Desalting FPLC Fractions
To desalt the FPLC fractions, a Sephadex G-10 desalting column (25 mm × 50 cm) is prepared according to manufacturer’s instructions. After loading the FPLC fraction, the column is washed with 1 L HPLC-grade water using gravity flow (~24 mL/h) at 4 °C. Fractions of 2 mL are collected over a period of 12–15 h. TDP-α-D-glucose is typically eluted over 5–10 fractions within 8 h. Fractions displaying significant absorbance at 267 nm are combined, lyophilized to near dryness, resuspended in HPLC-grade water, and lyophilized again to remove the remaining NH4HCO3 from the TDP-sugar. After desalting, TDP-α-D-glucose is quite stable and can be stored for months at −80 °C, although multiple freeze-thaw cycles should be avoided.
For the majority of enzymatic reactions, we have found that the presence of small amounts of comtaminanting TMP or TDP do not significantly affect subsequent enzymatic reactions. Thus, TDP-α-D-glucose of >85% purity obtained after P2 size-exclusion chromatography or FPLC purification is generally sufficient. For applications that require a much higher substrate purity, such as kinetic analysis or in situ 1H NMR spectroscopic assays to determine the activity of sugar biosynthetic enzymes, the complete purification sequence of P2, FPLC and G-10 chromatography should be followed to achieve >95% purity.
2.1 Generation of TDP-4-keto-6-deoxy-α-D-glucose (6)
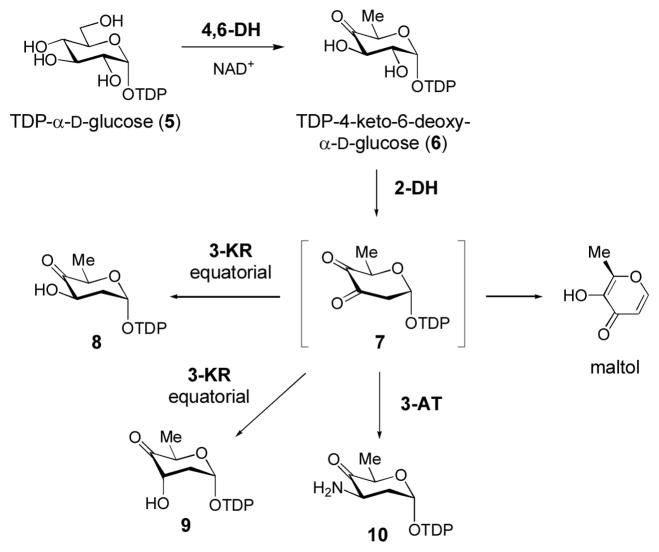
TDP-4-keto-6-deoxy-α-D-glucose (6) is produced from TDP-α-D-glucose (5) by the action of TDP-glucose-4,6-dehydratase (4,6-DH, Scheme 3). TDP-4-keto-6-deoxy-α-D-glucose is a common intermediate in many sugar biosynthetic pathways. The 4-keto group provides a chemically versatile handle that can be manipulated by deoxysugar biosynthetic enzymes to produce structurally diverse products. Due to such versatility, the ready availability of this sugar is important for the investigation of many interesting deoxysugar biosynthetic enzymes. Not surprisingly, several useful methods for the synthesis of 6 have been developed (Elling et al., 2005; Oh et al., 2003; Rupprath et al., 2005; Stein et al., 1998).
Scheme 3. Early TDP-deoxysugar Biosynthetic Intermediates.
The key intermediate in TDP-deoxysugar biosynthesis is TDP-4-keto-6-deoxy- α-D-glucose (6), which is synthesized from 5 by a TDP-glucose-4,6-dehydratase enzyme (4,6-DH). This intermediate is a branching point for the biosynthetic pathways of TDP-deoxysugars. For 2,6-dideoxyhexoses, 6 is converted to the unstable intermediate 7 by a 2-dehydratse (2-DH). This intermediate can then be reduced by a 3-ketoreductase (3-KR) to give 8 or 9, or it can be transaminated by a 3-aminotransferase (3-AT) to give 10.
To enzymatically synthesize this sugar, we cloned rfbB (the 4,6-DH from Salmonella enterica serovar Typhimurium LT2) into the pUC18 vector and expressed the recombinant RfbB-(His)5 protein in E. coli BL21(DE3) in a manner similar to that described in Section 1.1 for the RfbA-(His)5 construct (Melancon et al., 2007; Takahashi et al., 2006). Recombinant RfbB was purified by affinity chromatography using Ni-NTA resin (Qiagen) according to the manufacturer’s instructions, with the exception that 10% glycerol was included in the lysis, wash, and elution buffers. RfbB was then dialyzed against 50 mM NaH2PO4 buffer, 300 mM NaCl, 15% glycerol (pH 8.0), concentrated, flash frozen in liquid nitrogen, and stored at −80 °C until use. The yield of RfbB is generally ~ 80 mg per liter of growth culture.
A typical RfbB reaction contains 5 (25 mg) and RfbB (5 mg) in 2 mL of 50 mM Tris·HCl buffer (pH 7.5). Incubation is carried out at 30 °C for ~ 3–4 h. The reaction is slightly slower at 30 °C, but less degradation of 5 occurs at this temperature. Some published procedures (Amann et al., 2001; Stein et al., 1998) use alkaline phosphatase (2 units/mL) to suppress any competitive inhibition by TDP, but we have found its inclusion to be unnecessary. The progress of the reaction can be monitored by HPLC using a Carbopac-PA1 column (Dionex). To assess the progress of the reaction, 20- to 30-μL reaction aliquots are removed at appropriate time intervals and subjected to a prewashed YM-10 microcon centrifugal filter unit to remove RfbB. The flow-through is then injected into HPLC and the TDP-sugars (5 and 6) are eluted using the conditions described in Section 1.3.1. The retention times for 5 and 6 are 31–33 min and 33.5–35 min, respectively. The peak for 6 is slightly broader than that for 5. For maximum yields, it is best to use fresh RfbB enzyme that has not been subjected to multiple freeze-thaw cycles. Using this method, we have achieved a reproducible yield of 95% (24 mg of 6) before purification. After the conversion of 5 to 6 is completed, RfbB can be removed by filtration though either an Amicon stirred cell unit equipped with a YM-10 membrane or a larger centrifugal filter unit. The filtrate, which contains 6, can be directly used in subsequent biosynthetic reactions or divided into smaller aliquots and frozen at −80 °C for future use. Desalting is necessary if the sample is lyophilized and stored in dry form.
2.2 Generation of TDP-2,6-dideoxysugars
Another useful TDP-deoxysugar is TDP-4-keto-2,6-dideoxy-α-D-glucose (8), which is produced from TDP-4-keto-6-deoxy-α-D-glucose (6) at an early stage in the biosynthesis of 2,6-dideoxy sugars (Scheme 3). The 2-deoxygenation reaction is catalyzed by a group of enzymes called 2-dehydratases (2-DH) that employ a metal ion to convert 6 into a 3,4-diketo sugar intermediate 7 (Chen et al., 1999; Draeger et al., 1999). The 3-keto group of 7 has one of two fates in different biosynthetic pathways: it can be reduced by a 3-ketoreductase (3-KR) to a hydroxyl group with either equatorial (8) or axial (9) stereochemistry, or it can be transaminated by a 3-aminotransferase (3-AT) to give a 3-aminosugar (10). Our group employs TylX3 (a 2-DH) and TylC1 (an axial 3-KR from the tylosin biosynthetic pathway of Streptomyces fradiae) to synthesize 9 from 6 (Chen et al., 1999; Takahashi et al., 2006), and TylX3 and KijD10 (an equatorial 3-KR from the kijanimicin biosynthetic gene cluster of Actinomadura kijaniata) to synthesize 8 from 6 (Zhang et al., 2007). In our experience, the generation of the equatorial 3-OH product (8) from 6 is straightforward, while synthesis of the axial 3-OH sugar (9) from 6 is more problematic. In both cases, excess reductase (2.5 molar excess of either TylC1 or KijD10 as compared to TylX3) is used to drive the reaction forward.
Cloning, expression and purification of TylX3, TylC1, and KijD10 have been described (Chen et al., 1999; Takahashi et al., 2006; Zhang et al., 2007). In general, these proteins are purified following the protocols given above for RfbA and RfbB. Importantly, 20% glycerol should be included in all purification buffers for TylX3, and 10% glycerol should be present in all reactions performed with TylX3. Due to the instability of the TylX3 product (Scheme 3, compound 7), TylX3 should be the last component added to initiate the coupled reactions. Both TylC1 and KijD10 are NADPH-dependent, but many other 3-KR enzymes utilize NADH.
2.2.1 Synthesis of TDP-4-keto-2,6-dideoxy Sugars with Equatorial C3-OH Stereochemistry
To synthesize the TDP-4-keto-2,6-dideoxy sugar (8) with equatorial C3-OH stereochemistry, a 3 mL reaction containing 6 (21.8 mM), NADPH (53 mM), and 10% glycerol in 50 mM Tris ·HCl buffer (pH 7.5) is prepared. KijD10 (360 nM) and TylX3 (160 nM) are added to initiate the reaction. The reaction mixture is incubated at 25 °C and the progress of the reaction is monitored by HPLC using the conditions described in section 1.3.1 (the retention times of 6 and 8 are 33–34 min and 32–33 min, respectivevly). A degradation product of 7 (maltol, see Scheme 3) eluted at 1.7–1.8 min is always observed. When freshly-purified TylX3 and KijD10 are used, this reaction is complete within 2–3 h (after repeated freeze-thaw cycles, KijD10 loses activity). The TylX3/KijD10 coupled reaction is quite efficient and, on average, affords approximately 85% conversion to product (or ~ 20 mg of 8 from 24 mg of 6). With freshly prepared enzyme, we have observed nearly quantitative conversion within 6 h.
Prior to its use in any subsequent enzymatic reactions, compound 8 should be purified due to the presence of NADP+ and NADPH in the reaction. The sample (containing 8) should be filtered to remove enzymes and lyophilized to a yellow syrup. Importantly, the frozen sample must be removed from the lyophilizer as soon as it begins to thaw. If lyophilization is allowed to proceed to dryness, there is often substantial (> 50%) degradation of the product. To purify 8 using P2 size-exclusion chromatography, the procedure provided in Section 1.3.1 should be followed, and 25 mM NH4HCO3 instead of ddH2O should be used to wash and equilibrate the column, and to elute the sample. A typical flow rate is 6–8 mL/h. Samples with absorbance at 267 nm begin to elute after 24–30 h, and continue for the next 12–20 h. As described for TDP-α-D-glucose (5), a similar absorbance plateau is commonly observed. However, 8 should be eluted towards the end of the plateau region (just prior to the reduction of A267 back to baseline) over only 2–4 fractions. The fractions at the beginning of the plateau region contain NADP+ and NADPH. Generally, two of the four fractions are greater than 85% pure (as determined by HPLC), while one or two are of lower purity. The desired fractions are combined and lyophilized. Care should be taken not to lyophilize to dryness, as this will cause significant degradation, as observed by 1H-NMR spectroscopy. Usually, approximately 10 mg of 8 is recovered from this purification process (Zhang et al., 2007).
2.2.2 Synthesis of TDP-4-keto-2,6-dideoxy Sugars with Axial C3-OH Stereochemistry
To generate the TDP-4-keto-2,6-dideoxy sugar 9 with axial C3-OH stereochemistry, we use TylC1 in place of KijD10. A typical 3-mL reaction mixture contains 6 (21.8 mM), NADPH (53 mM), and 10% glycerol in 50 mM Tris ·HCl buffer (pH 7.5). TylC1 (40 μM) and TylX3 (8 μM) are added to initiate the reaction. The incubation is carried out at 25 °C and product formation is monitored by HPLC. Under the HPLC conditions described in Section 1.3.1, the retention times of substrate (6) and product (9) are 33.6–34 min and 32.3–33 min, respectively. Generation of 9 is not as facile as the C-3 equatorial product (8). We obtain approximately 50% conversion of 6 → 9, even after overnight incubation. Gel filtration using a P2 column to remove NADP+ and NADPH can be carried out by the method described in Section 2.2.1 for compound 8, and typical yields are about 5 mg of purified sugar. Compound 9 can often be generated in situ in sufficient quantities to be processed by downstream biosynthetic enzymes.
3.1 In vitro Reconstitution of Entire Deoxysugar Biosynthetic Pathways
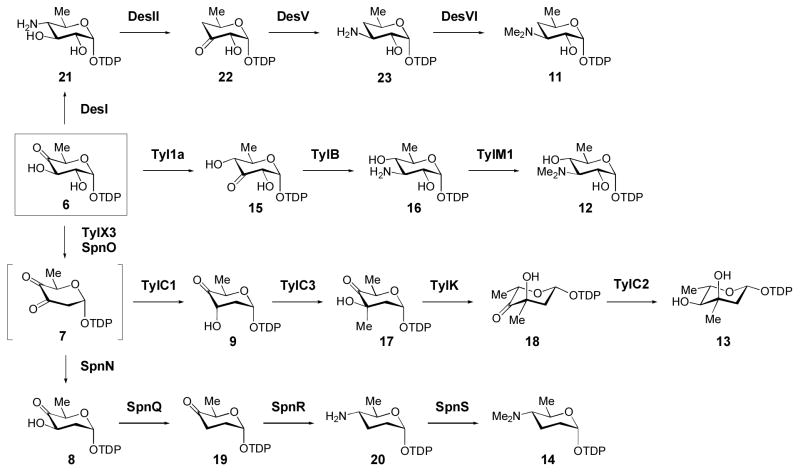
Up to this point, we have summarized methodology to synthesize and purify several common (early) intermediates (5, 6, 8, and 9, Scheme 3) produced in deoxysugar biosynthetic pathways. These intermediates can be used as starting materials for more elaborate enzymatic syntheses. To date, only a handful of TDP-deoxysugar biosynthetic pathways have been fully reconstituted in vitro using native pathway enzymes. Some of these highly modified TDP-sugars (Scheme 4) that have been enzymatically prepared include TDP-α-D-desosamine (11), TDP-α-D-mycaminose (12) (Melancon et al., 2007), TDP-β-L-mycarose (13) (Takahashi et al., 2006), TDP-α-D-forosamine (14) (Hong et al., 2008), TDP-β-L-epivancosamine (Chen et al., 2000), and TDP-β-L-digitoxose (Zhang et al., 2007). Due to the variation in the reactivity and stability of different TDP-sugar intermediates, and to the differences in catalytic efficiencies of the various biosynthetic enzymes involved, there is no general protocol that can be used to synthesize all TDP-sugars. Instead, syntheses must be optimized on a case-by-case basis. The fact that few in vitro sugar biosyntheses are known reflects the dearth of biochemical information available for deoxysugar biosynthetic enzymes, and clearly suggests that more thorough kinetic investigations of these important enzymes are warranted. Here, we present procedures for the enzymatic synthesis of several TDP-deoxysugars (11–14) that have been investigated in our laboratory.
Scheme 4. In vitro enzymatic synthesis of highly-modified TDP-deoxysugars.
Biosynthetic pathways for several highly modified TDP-deoxysugars (11–14) that have been synthesized in vitro using native pathway enzymes. See text for details.
3.2.1 One-pot Synthesis of TDP-α-D-mycaminose (12)
The synthesis of TDP-α-D-mycaminose (TDP-3-N, N-dimethylamino-3,6-dideoxy-α-D-glucose, 12, Scheme 4) from TDP-4-keto-6-deoxy-α-D-glucose (6) can be carried out in a single reaction using three enzymes (Tyl1a, TylB, and TylM1) from the tylosin biosynthetic pathway of S. fradiae (Scheme 4). Tyl1a and TylB catalyze the conversion of 6 to TDP-3-amino-6-deoxy glucose (16) through a TDP-3-keto-6-deoxy sugar intermediate (15) (Melancon et al., 2007). TylM1 is an N, N-dimethyltransferase that converts 16 to TDP-α-D-mycaminose (12) (Chen et al., 1998; Chen et al., 2002). Genes encoding Tyl1a, TylB, and TylM1 can be cloned into either pET24b or pET28b, expressed in E. coli as the His6-tagged fusion proteins, and purified using standard protocols.
For the synthesis of TDP-α-D-mycaminose (12), compound 6, prepared as described in Section 2.1, is lyophilized and resuspended in 50 mM potassium phosphate buffer, pH 7.5. The reaction mixture contains 6 (1 mM), L-glutamate (30 mM), pyridoxal 5′-phosphate (PLP, 150 μM), S-adenosyl-L-methionine (SAM, 2 mM), TylB (30 μM), and TylM1(60 μM) in a total volume of 2 mL of 50 mM potassium phosphate buffer (pH 7.5). Tyl1a (3 μM) is added to initiate the reaction. It is best to use newly prepared enzymes and a high level of glycerol in the reaction mixture should be avoided. After ~ 12 h at 25 °C, 40–60% conversion of 6 to 12 should be observed by HPLC. Using the HPLC protocol described in Section 1.3.1, retention times for TDP-sugars 12, 16, 6, and 15 are 7–8 min, 13 min, 35–36 min, and 39.0 min, respectively. In addition to the TDP-sugar peaks, peaks for TDP (41.9 min), (2R,3R)-2-methyl-3,5-dihydroxy-4-keto-2,3-dihydropyran, a degradation product of 15 (1.8 min), SAM and its degradation products (3.2 min, 21 min and 23 min), and S-adenosyl-L-homocysteine (2.1 min) are also visible. An Adsorbosphere SAX column can also be employed to purify 12 (Chen et al., 2002). This column is especially useful for isolating TDP-aminosugars, which are significantly more polar than most other TDP-deoxysugars (see below).
After the TDP-α-D-mycaminose (12) peak is collected, the sample can be readily desalted using HPLC for small-scale reactions, or by FPLC for large-scale preparations. When using HPLC, the sample is applied to an analytical C18 column and H2O is used to elute 12 (retention time is 5–10 min). For larger-scale preparations, FPLC purification can be carried out using the buffers and column described in Section 1.3.2, applying a linear gradient from 0 to 100% B over 25 min. Under these conditions, TDP-α-D-mycaminose (12) will elute at approximately 12.5 min. During FPLC purification, most of the unused substrate, TDP-4-keto-6-deoxy-α-D-glucose (6, retetion time 15 min), can be recovered and desalted using the G-10 column. The average yield from a 2-mL reaction mixtue for the enzymatic synthesis of TDP-α-D-mycaminose is 11% (~ 0.11 mg of 12 from 1 mg of 6), which is a 3-fold improvement over the chemical preparation method (Chen et al., 2002).
3.2.2 Two-stage One-pot Synthesis of TDP-β-L-mycarose (13)
The functions of each of the enzymes in TDP-β-L-mycarose (13) biosynthesis in the tylosin producer, S. fradiae, have been verified in vitro and are shown in Scheme 4 (Chen et al., 1999; Chen et al., 2001; Takahashi et al., 2005). From TDP-4-keto-6-deoxy-α-D-glucose (6), TylX3 and TylC1 catalyze the formation of 9. Following SAM-dependent C3 methylation of 9 by TylC3, compound 17 is converted to TDP-β-L-mycarose (13) by the sequential action of TylK, a 5-epimerase, and TylC2, an NAD+-dependent 4-ketoreductase. Two features make a one-pot synthesis of 13 attractive. First, as noted above, the instability of the TylX3 product (7) requires the presence of a 3-ketoreducatse (TylC1) to drive the reaction forward and to avoid the decomposition of 7 into TDP and maltol (Chen et al., 1999). Second, the presence of the 4-ketoreductase (TylC2) is required in order to drive the formation of 13 from the epimerized TylK product (18), which is in equilibrium with 17 (Takahashi et al., 2005).
Thus, a two-stage, one-pot synthesis of TDP-β-L-mycarose (13) was developed, wherein compound 6 is first generated by procedures similar to those outlined in Sections 1.2 and 2.1 with slight modifications (Takahashi et al., 2006). Following the completion of TTP synthesis from thymidine and ATP, the TM/TMK/NDK/PK enzymes are removed by ultrafiltration. Next, α-D-glucose-1-phosphate (1, 3 mM) and RfbA (57 μM) are added to the reaction mixture to convert 1 to 5. After a 30-min incubation period at 30 °C, RfbB (28 μM) is added and the mixture is allowed to incubate for 1 h at 37 °C to convert 5 to 6. At this point, it is not necessary to remove either RfbA or RfbB from the incubation mixture. In the next stage of the reaction, the five TDP-β-L-mycarose (13) biosynthetic enzymes (TylX3/C1/C3/K/C2, 30 μM each) are added along with 6 mM NADPH and 3 mM SAM. The reaction is allowed to proceed for 1 h at room temperature to convert 6 to 13. The cloning, expression, and purification of the mycarose biosynthetic enzymes have been previously described (Chen et al., 1999; Chen et al., 2001; Takahashi et al., 2005). TDP-β-L-mycarose (13) is purified from the reaction mixture by FPLC using a MonoQ 10/10 column that is eluted with H2O over 2-column volumes, followed by a linear gradient from 0–280 mM NH4HCO3 buffer (pH 7.0) over 2-column volumes at a flow rate of 1 mL/min. Following FPLC purification of 13, desalting is carried out using a Sephadex G-10 column (see Section 1.3.3). The identity of 13 is confirmed by 1H NMR spectroscopy and high-resolution MS analysis (Takahashi et al., 2005). The final yield of TDP-β-L-mycarose (13) is 16% from glucose-1-phosphate (1) (Takahashi et al., 2006).
3.2.3 Multi-step Enzymatic Synthesis of TDP-α-D-forosamine (14)
TDP-α-D-forosamine is a highly modified tetradeoxy sugar produced during spinosyn biosynthesis in Saccharopolyspora spinosa (Scheme 4). Unlike the preparation of TDP-mycarose (13) from 6, a one-pot synthesis of 14 from 6 is impractical. This is mainly because the SpnQ-catalyzed reaction (8 → 19) is sensitive to O2 and, therefore, must be conducted using deoxygenated buffers under anaerobic conditions or in the presence of sodium dithionite. We have recently demonstrated the activities for each of the TDP-α-D-forosamine biosynthetic enzymes in vitro (Hong et al., 2008). The activities of SpnO and SpnN (6 → 7 → 8) were established in a coupled assay by following the consumption (loss in absorbance at 340 nm) of NADPH by SpnN. Formation of the SpnN product (8) was verified by HPLC analysis as described in Hong et al., 2008. However, the yield of compound 8 is not as high as that obtained in the TylX3/KijD10 reaction described in Section 2.2.1. Thus, for preparative purposes, we recommend using the optimized TylX3/KijD10 system to synthesize 8.
SpnQ catalyzes the 3-deoxygenation of 8 to give 19. SpnQ is a [2Fe-2S] cluster containing, pyridoxamine 5′-phosphate (PMP)-dependent enzyme, that is a homologue of E1 - a mechanistically well-characterized 3-dehydrase from Yersinia pseudotuberculosis (reviewed in He et al., 2000). These 3-dehydrase enzymes also require a reductase component to complete their catalytic cycles, which involve single electron transfer radical chemistry. Because of the sensitivity of the [2Fe-2S] center in SpnQ to O2, the reaction must be carried out anaerobically. A typical SpnQ reaction contains compound 8 (0.7 mM), PMP (250 μM), SpnQ (30 μM), NADPH (0.7 mM), and a physiological reductase system comprised of either flavodoxin/flavodoxin reductase (30 μM each) or ferredoxin/ferredoxin reductase (30 μM each) in 50 mM potassium phosphate buffer (pH 7.5). Sodium dithionite (0.6 mM) can also be used as the reductant. Production of 19 is monitored by HPLC (see below).
The next step in the pathway is catalyzed by a PLP-dependent 4-aminotransferase, SpnR. The substrate of this enzyme, 19, can be generated in situ by SpnQ using the conditions described above. Following a 3-h incubation period at 24 °C to convert 8 → 19, SpnQ, ferridoxin, and ferridoxin reductase are removed by ultrafiltration through a YM-10 membrane. SpnR (37 μM), PLP (305 μM), L-glutamate (12.2 mM), and MgCl2 (1.2 mM) are then added and the reaction mixture is incubated at 24 °C for an additional 2 h. The formation of the SpnR product (20) can be monitored by HPLC analysis (see below). The final N-methylation reaction to afford TDP-α-D-forosamine (14), catalyzed by the methyltransferase SpnS, is accomplished by the incubation of the SpnR product (20) with SpnS (10 μM), SAM (2.0 mM), MgCl2 (2.0 mM), and DTT (2.0 mM) in 50 mM potassium phosphate (pH 7.5) at 37 °C. Both monomethylated and dimethylated products are observed.
The HPLC conditions used to resolve the TDP-α-D-forosamine biosynthetic intermediates are as follows. For the SpnO/N, SpnQ, and SpnQ/R reactions, a Dionex CarboPac PA1 analytical column (4 ×250 mm) equipped with a CarboPac PA1 guard column (4 × 250 mm) is employed, and the elution conditions are identical to those given in Section 1.3.1. Using a 1 mL/min flow rate and detection at 267 nm, the HPLC retention times for TDP-sugars 8, 19, and 20 are 33.4, 36.2, and 9.2 min, respectively. The above HPLC conditions cannot resolve the mono- and di-methylated SpnS-catalyzed reaction products from the other assay components. Thus, to purify these sugars, an Adsorbosphere SAX column (5 μm, 4.6 × 250 mm) is used. Here, a linear gradient from 0 to 20% buffer B (500 mM KH2PO4 buffer, pH 3.5) in buffer A (50 mM KH2PO4 buffer, pH 3.5) over 20 min is applied.
3.2.4 TDP-α-D-desosamine (11)
TDP-α-D-desosamine is a 4,6-dideoxysugar found in several macrolide antibiotics, including erythromycin, oleandomycin, mycinamicin, methymycin/pikromycin, and megalomicin. The biosynthetic pathway for 11 (6 → 21 → 22 → 23 → 11, Scheme 4) has been fully established through biochemical studies of the pathway enzymes. DesI is a PLP-dependent 4-aminotransferase that converts 6 to 21 (Zhao et al., 2001). DesII, a member of the radical SAM enzyme superfamily, catalyzes the oxidative deamination of 21 to produce 22 (Szu et al., 2005). The PLP-dependent 3-aminotransferase, DesV, transaminates 22 to generate 23 (Szu et al., 2005; Zhao, 2000). The final step, N, N-dimethylation, is catalyzed by DesVI (Chang et al., 2000; Chen et al., 2002) to complete the biosynthesis of 11.
Starting from 6, the DesV product (23) can be prepared in a one-pot, two-step reaction from 6 (Szu et al., 2005). In the first step of this synthesis, 6 is converted into 22 by the combined action of DesI and DesII. Due to the sensitivity of the [4Fe-4S] cluster of DesII to O2, and to the fact that this cluster must be reduced for activity, this coupled reaction must be performed anaerobically. Prior to the enzymatic synthesis, the inactive [4Fe-4S]2+ cluster of DesII (190 μM) must be reduced under anaerobic conditions to the [4Fe-4S]1+ state by sodium dithionite (1.2 mM) in 100 mM Tris ·HCl buffer (pH 8.0) for 40 min. The reduction of the [4Fe-4S]2+ cluster can be monitored by following the decrease in absorbance at 420 nm. A 1-mL reaction mixture containing compound 6 (0.6 mM), L-glutamate (0.5 mM), PLP (0.14 mM), DesI (26 μM), the above-reduced DesII (100 μM), SAM (0.1 mM), and DTT (2 mM) in 100 mM Tris·HCl buffer (pH 8.0) is carried out in an anaerobic chamber using degassed buffers. The reaction mixture is incubated at 25 °C for 3 h. At this point, DesV (100 μM), along with additional L-glutamate (10 mM) and PLP (0.8 mM), are added to the reaction mixture and incubated at 25 °C for another 30 min. The DesI, DesII, and DesV enzymes are removed by ultrafiltration through a YM10 membrane, and the reaction progress is analyzed by HPLC using a Dionex anion exchange column (4 × 250 mm). Using a flow rate of 0.6 mL/min, detection at 267 nm, and a linear gradient from 20 to 35% of eluent B (1 M NH4OAc, pH 7.0) in eluent A (H2O) over 30 min, the retention times for compound 6 (the DesI substrate), compound 22 (the DesII product), and compound 23 (the DesV product) are 13.8, 28.2, and 3.7 min, respectively. It should also be noted that a preparative-scale enzymatic synthesis of the DesI product (compound 21) has been reported (Zhao et al., 2001).
For the final step of TDP-α-D-desosamine (11) synthesis, a small-scale reaction (50 μL) containing 23 (1.2 mM), DTT (2 mM), SAM (10 mM) and N, N-dimethyltransferase DesVI can be carried out for 3 h at 25 °C in 50 mM potassium phosphate buffer (pH 7.5). These conditions yield roughly 80% conversion of 23 → 11 (Chen et al., 2002). The reaction products can be purified using an Adsorbosphere SAX column and the elution conditions given in Section 3.2.3. Under these conditions, the retention times for 23 and 11 are 6.5 and 17 min, respectively. A small amount of an N-monomethylated DesVI reaction intermediate may also be present (retention time 9.3 min). A preparative-scale synthesis of 11 from 23 (Chen et al., 2002) can be performed by incubating 23 (5.7 mM), SAM (30 mM), DTT (2 mM), and DesVI (1.8 mg) in 1 mL of 50 mM potassium phosphate buffer (15% glycerol, pH 7.5) for 3 h at 25 °C. After removal of DesVI by ultrafiltration (YM-10 membrane), TDP-α-D-desosamine (11) is isolated by size-exclusion chromatography using a P2 column (2 × 100 cm) with a 0.5 M NH4HCO3 solution as the eluent. Fractions containing 11 (identified by absorbance at 267 nm) are concentrated and further purified with an FPLC MonoQ HR (10/10) column using a flow rate of 3 mL/min and a linear gradient of 0–0.15 M NH4HCO3 over 15 min to elute compound 11 (retention time 8 min).
4.1 Synthesis of Deoxysugars in vivo by Metabolic Pathway Engineering
While using purified biosynthetic enzymes to synthesize TDP-deoxysugars in vitro is desirable, this approach may not always be feasible if the sugar biosynthetic enzymes required for a particular synthesis cannot be expressed and purified in suitable quantities, or are not stable. In these cases, an in vivo biosynthetic approach may be useful, wherein a glycosylated natural product is isolated from cultures of a producing bacterial strain, and the deoxysugar of interest is then recovered from the glycosylated product either through reverse GT catalysis or by hydrolysis and chemical derivitization. One method is to heterologously express the deosysugar biosynthetic gene(s) of interest in a host that naturally produces glycosylated natural products with similar structures. In this case, it may be advantageous to disrupt some of the host’s deoxysugar biosynthetic genes to increase the intracellular concentration of the substrate(s) for the heterologously expressed biosynthetic enzyme(s). Another approach involves the expression of entire biosynthetic pathways in a non-producing, but tolerant strain such as Streptomyces lividans or Streptomyces albus. In these organisms, the heterologously expressed sugar biosynthetic enzymes will not have to compete with endogenous enzymes for their substrate, and the glycosylated products will be excreted from the host cells, limiting their toxicity. It should be noted that synthesis of TDP-deoxysugars in large quantities is not the primary focus of many of the in vivo pathway engineering studies reported in the literature, but the biological systems used in these studies could potentially serve as good sources for TDP-deoxysugar production. Below we highlight some of our recent in vivo studies on deoxysugar biosynthesis and pathway engineering in Streptomyces venezuelae (Thibodeaux and Liu, 2007), that produces several macrolide derivatives bearing D-desosamine moieties.
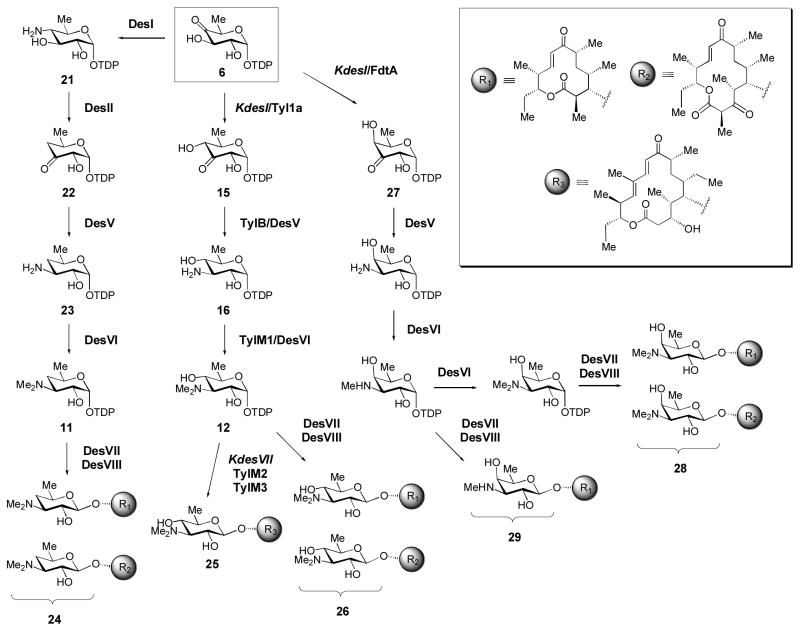
The native biosynthetic pathways for production of D-desosaminylated 12- and 14-membered ring macrolactones in S. venezuelae are shown in Scheme 5 (6 → 21 → 22 → 23 → 11 → 24). In an attempt to generate TDP-α-D-mycaminose (12) in S. venezuelae, we constructed an S. venezuelae double knockout mutant (termed KdesI/desVII) lacking the 4-aminotransferase (DesI) that catalyzes 6 → 21, and the endogenous desosaminyltransferase (DesVII) that catalyzes 11 → 24 (Borisova et al., 1999; Melancon et al., 2005). This mutant was designed to enable the accumulation of 6, which could then be processed by heterologously expressed genes from the D-mycaminose pathway of S, fradiae (Scheme 5, 6 → 15 → 16 → 12) in order to make 12 – a deoxysugar that is not normally produced by S. venezuelae. The tyl1a, tylB, and tylM1 genes were heterologously expressed in this S. venezuelae mutant along with the genes rerquired for mycaminosyltransfer (tylM2 and tylM3). When this strain was fed with tylactone (the native aglycone substrate of TylM2/M3), D-mycaminosylated tylactone (25) was generated (Melancon et al., 2005), indicating that the heterologously expressed tyl genes indeed converted 6 into 12.
Scheme 5. Manipulating deoxysugar structures by metabolic pathway engineering.
A combination of gene knockout, heterologous gene expression, and precursor feeding experiments allowed the production of several novel compounds in Streptomyces venezuelae. See text for details.
Interestingly, when the tyl1a gene alone was heterologously expressed in a separate S. venezuelae mutant that lacked only the desI gene (Scheme 5), novel D-mycaminosylated macrolide derivatives (26) were obtained (Borisova et al., 1999; Melancon et al., 2005). Thus, heterologous expression of a single S. fradiae gene (tyl1a) was sufficient to convert the native D-desosamine biosynthetic pathway into a D-mycaminose pathway in S. venezuelae. From this experiment, it is clear that several of the desosamine biosynthetic enzymes (DesV, DesVI, and DesVII/DesVIII) are capable of processing alternative TDP-deoxysugar substrates (each containing an equatorial 4-OH group that is not present in the natural substrates for DesV-VIII). The above example not only illustrates how deoxysugar biosynthesis can be manipulated in a producing strain by metabolic pathway engineering, but it also reveals the inherent relaxed substrate specificity of many deoxysugar biosynthetic enzymes – a property that should be useful for the synthesis or engineering of deoxysugar structures in other pathways.
Indeed, we have exploited the relaxed substrate specificity of the desosamine biosynthetic enzymes to synthesize deoxysugars that have not yet been identified from natural sources (Melancon and Liu, 2007). For example, we expressed fdtA (from the Gram-negative Aneurinibacillus thermoaerophilus), a 3,4-ketoisomerase homologue of tyl1a that produces 27, the C4 epimer of compound 15 (Davis et al., 2007), in the S. venezuelae KdesI mutant. The mutant cultures were found to produce macrolide derivatives bearing either 4-epi-D-mycaminose (28) or 3-N-monomethyl-3-deoxy-D-fucose (29) – two unnatural deoxysugars.
Summary
The deoxysugar moieties of many glycosylated bacterial secondary metabolites are often essential for the biological activity of these compounds. Changing the structure of these sugar moieties has the potential to generate new glycoforms with altered or improved biological activity. Thus, convenient methods for synthesizing highly modified deoxysugars with defined structures are desirable. Chemical synthesis of deoxysugars is feasible, but is often tedious, technically demanding, and typically suffers from low overall yields. On the other hand, in vitro and in vivo (chemo)enzymatic synthesis of activated nucleotide sugars exploits Nature’s biosynthetic machinery and has several benefits over chemical synthesis methods. First, the enzymes used in these methods are a readilly available, relatively cheap, and renewable resource. Second, enzymes also typically enable more stringent control over the regio- and stereochemical outcome of a reaction. Third, these enzymatic reactions are performed under mild, biological conditions and nucleotide sugar purification usually requires only a few simple and familiar chromatographic steps. Here, we have outlined detailed synthesis and purification procedures for several common TDP-sugar intermediates as well as for several specific highly-modified TDP-deoxysugars. It is hoped that the procedures outlined here will provide useful guidelines for the development of synthetic protocols for other unusual sugars. This, in turn, should facilitate the development of workable quantities of glycoforms with defined (and potentially novel) sugar strucutres.
Acknowledgments
We thank the National Institutes of Health (GM35906, GM54346) and the Welch Foundation (F-1511) for their generous support of our research work, and Prof. Christian P. Whitman from the College of Pharmacy, University of Texas, Austin, for his critical reading of the manuscript.
References
- Amann S, Drager G, Rupprath G, Kirschning A, Ellinga L. Chemoenzymatic synthesis of dTDP-activated 2,6-dideoxysugars as building blocks of polyketide antibiotics. Carbohydr Res. 2001;335:23–32. doi: 10.1016/s0008-6215(01)00195-1. [DOI] [PubMed] [Google Scholar]
- Bode HB, Muller R. Reversible sugar transfer by glycosyltransferases as a tool for natural product biosynthesis. Angew Chem Int Ed. 2007;46:2147–2150. doi: 10.1002/anie.200604671. [DOI] [PubMed] [Google Scholar]
- Borisova SA, Zhao L, Sherman DH, Liu Hw. Biosynthesis of desosamine: construction of a new macrolide carrying a genetically designed sugar moiety. Org Lett. 1999;1:133–136. doi: 10.1021/ol9906007. [DOI] [PubMed] [Google Scholar]
- Chang C, Zhao L, Yamase H, Liu Hw. DesVI: A new member of the sugar N, N-dimethyltransferase family involved in the biosynthesis of desosamine. Angew Chem Int Ed. 2000;39:2160–2163. doi: 10.1002/1521-3773(20000616)39:12<2160::aid-anie2160>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Chen H, Agnihotri G, Guo Z, Que NLS, Chen X, Liu Hw. Biosynthesis of mycarose: isolation and characterization of enzymes involved in the C-2 deoxygenation. J Am Chem Soc. 1999;121:8124–8125. [Google Scholar]
- Chen H, Guo Z, Liu Hw. Expression, purification, and characterization of TylM1, an N, N-dimethyltransferase involved in the biosynthesis of mycaminose. J Am Chem Soc. 1998;120:9951–9952. [Google Scholar]
- Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Deoxysugars in glycopeptide antibiotics: enzymatic synthesis of TDP-L-epivancosamine in chloroeremomycin biosynthesis. Proc Natl Acad Sci USA. 2000;97:11942–7. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yamase H, Murakami K, Chang CW, Zhao L, Zhao Z, Liu Hw. Expression, purification, and characterization of two N, N-dimethyltransferases, tylM1 and desVI, involved in the biosynthesis of mycaminose and desosamine. Biochemistry. 2002;41:9165–9183. doi: 10.1021/bi020245j. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhao Z, Hallis TM, Guo Z, Liu Hw. Insights into the branched-chain formation of mycarose: methylation catalyzed by an (S)-adenosylmethionine-dependent methyltransferase. Angew Chem Int Ed. 2001;40:607–610. doi: 10.1002/1521-3773(20010202)40:3<607::AID-ANIE607>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Davis ML, Thoden JB, Holden HM. The X-ray structure of dTDP-4-keto-6-deoxy-D-glucose-3,4-ketoisomerase. J Biol Chem. 2007;282:19227–19236. doi: 10.1074/jbc.M702529200. [DOI] [PubMed] [Google Scholar]
- Draeger G, Park SH, Floss HG. Mechanism of the 2-deoxygenation step in the biosynthesis of the deoxyhexose loieties of the antibiotics granaticin and oleandomycin. J Am Chem Soc. 1999;121:2611–2612. [Google Scholar]
- Elling L, Rupprath C, Gunther N, Romer U, Verseck S, Weingarten P, Drager G, Kirschning A, Piepersberg W. An enzyme module system for the synthesis of dTDP-activated deoxysugars from dTMP and sucrose. Chembiochem. 2005;6:1423–1430. doi: 10.1002/cbic.200500037. [DOI] [PubMed] [Google Scholar]
- Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Antibiotic optimization via in vitro glycorandomization. Nat Biotechnol. 2003;21:1467–1469. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- He X, Agnihotri G, Liu Hw. Novel enzymatic mechanisms in carbohydrate metabolism. Chem Rev. 2000;100:4615–4662. doi: 10.1021/cr9902998. [DOI] [PubMed] [Google Scholar]
- He X, Liu Hw. Mechanisms of enzymatic C-O bond cleavages in deoxyhexose biosynthesis. Curr Opin Chem Biol. 2002;6:590–597. doi: 10.1016/s1367-5931(02)00367-8. [DOI] [PubMed] [Google Scholar]
- Hong L, Zhao Z, Melancon CE, 3rd, Zhang H, Liu H-w. In vitro characterization of the enzymes involved in TDP-D-forosamine biosynthesis in the spinosyn pathway of Saccharopolyspora spinosa. J Am Chem Soc. 2008;130:4954–4967. doi: 10.1021/ja0771383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb S, Wright G. Accessorizing natural products: Adding to nature’s toolbox. Proc Natl Acad Sci USA. 2005;102:519–520. doi: 10.1073/pnas.0408858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan JM, Griffith BR, Thorson JS. Neoglycorandomization and chemoenzymatic glycorandomization: Two complementary tools for natural product diversification. J Nat Prod. 2005;68:1696–1711. doi: 10.1021/np0502084. [DOI] [PubMed] [Google Scholar]
- Luzhetskyy A, Mendez C, Salas JA, Bechthold A. Glycosyltransferases, important tools for drug design. Curr Topics Med Chem. 2008;8:680–709. doi: 10.2174/156802608784221514. [DOI] [PubMed] [Google Scholar]
- Melancon CE, 3rd, Hong L, White JA, Liu Y-n, Liu Hw. Characterization of TDP-4-keto-6-deoxy-D-glucose-3,4-ketoisomerase from the D-mycaminose biosynthetic pathway of Streptomyces fradiae: in vitro activity and substrate specificity studies. Biochemistry. 2007;46:577–590. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon CE, 3rd, Liu Hw. Engineered biosynthesis of macrolide derivatives bearing the non-natural deoxysugars 4-epi-D-mycaminose and 3-N-monomethyl-3-deoxy-D-fucose. J Am Chem Soc. 2007;129:4896–4897. doi: 10.1021/ja068254t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon CE, 3rd, Thibodeaux CJ, Liu Hw. Glyco-stripping and glyco-swapping. ACS Chem Biol. 2006;1:499–504. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]
- Melancon CE, 3rd, Yu W-L, Liu Hw. TDP-mycaminose biosynthetic pathway revised and conversion of desosamine pathway to mycaminose pathway with one gene. J Am Chem Soc. 2005;127:12240–12241. doi: 10.1021/ja053835o. [DOI] [PubMed] [Google Scholar]
- Mendez C, Luzhetskyy A, Bechthold A, Salas JA. Deoxysugars in bioactive natural products: development of novel derivatives by altering the sugar pattern. Curr Topics Med Chem. 2008;8:710–724. doi: 10.2174/156802608784221532. [DOI] [PubMed] [Google Scholar]
- Mendez C, Salas JA. Altering the glycosylation pattern of bioactive compounds. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- Minami A, Kakinuma K, Eguchi T. Aglycon switch approach toward unnatural glycosides from natural glycoside with glycosyltransferase VinC. Tetrahedron Lett. 2005;46:6187–6190. [Google Scholar]
- Oh J, Lee SG, Kim BG, Sohng JK, Liou K, Lee HC. One-pot enzymatic production of dTDP-4-keto-6-deoxy-D-glucose from dTMP and glucose-1-phosphate. Biotechnol Bioeng. 2003;84:452–458. doi: 10.1002/bit.10789. [DOI] [PubMed] [Google Scholar]
- Rupprath C, Schumacher T, Elling L. Nucleotide deoxysugars: Essential tools for the glycosylation engineering of novel bioactive compounds. Curr Med Chem. 2005;12:1637–1675. doi: 10.2174/0929867054367167. [DOI] [PubMed] [Google Scholar]
- Salas JA, Mendez C. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Stein A, Kula MR, Elling L. Combined preparative enzymatic synthesis of dTDP-6-deoxy-4-keto-D-glucose from dTDP and sucrose. Glycoconj J. 1998;15:139–145. doi: 10.1023/a:1006912121278. [DOI] [PubMed] [Google Scholar]
- Szu PH, He X, Zhao L, Liu Hw. Biosynthesis of TDP-D-desosamine: identification of a strategy for C4 deoxygenation. Angew Chem Int Ed. 2005;44:6742–6746. doi: 10.1002/anie.200501998. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Liu Y-n, Chen H, Liu Hw. Biosynthesis of TDP-L-mycarose: the specificity of a single enzyme governs the outcome of the pathway. J Am Chem Soc. 2005;127:9340–9341. doi: 10.1021/ja051409x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Liu Y-n, Liu Hw. A two-stage one-pot enzymatic synthesis of TDP-L-mycarose from thymidine and glucose-1-phosphate. J Am Chem Soc. 2006;128:1432–1433. doi: 10.1021/ja0562144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeaux CJ, Liu Hw. Manipulating nature’s sugar biosynthetic machineries for glycodiversification of macrolides: recent advances and future prospects. Pure Appl Chem. 2007;79:785–799. [Google Scholar]
- Thibodeaux CJ, Melancon CE, 3rd, Liu Hw. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- Thibodeaux CJ, Melancon CE, 3rd, Liu H-w. Natural product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed. 2008;47 doi: 10.1002/anie.200801204. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorson JS, Hosted TJ, Jr, Jiang J, Biggins JB, Ahlert J. Nature’s carbohydrate chemists: the enzymatic glycosylation of bioactive bacterial metabolites. Curr Org Chem. 2001;5:139–167. [Google Scholar]
- Walsh CT, Freel Meyers CL, Losey HC. Antibiotic glycosyltransferases: antibiotic maturation and prospects for reprogramming. J Med Chem. 2003;46:3425–3426. doi: 10.1021/jm030257i. [DOI] [PubMed] [Google Scholar]
- Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- Zhang H, White-Phillip JA, Melancon CE, 3rd, Kwon H-j, Yu W-l, Liu H-w. Elucidation of the kijanimicin gene cluster: Insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J Am Chem Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. Ph.D. Thesis. Vol. 233. University of Minnesota; Minneapolis: 2000. Biosynthetic studies of D-desosamine and engineered biosynthesis of methymycin/pikromycin analogues carrying modified deoxysugars. [Google Scholar]
- Zhao L, Borisova S, Yeung SM, Liu Hw. Study of C-4 deoxygenation in the biosynthesis of desosamine: evidence implicating a novel mechanism. J Am Chem Soc. 2001;123:7909–7910. doi: 10.1021/ja010587x. [DOI] [PubMed] [Google Scholar]