Abstract
Signaling via the androgen receptor (AR) plays an important role in human health and disease. All currently available anti-androgens prevent ligand access to the receptor, either by limiting androgen synthesis or by competitive antagonism at the ligand binding domain. It is unknown to what extent various steps of receptor activation may be separable and distinctly targeted by inhibitors. We have previously described the use of fluorescent protein fusions to AR to monitor its subcellular distribution and ligand-induced conformational change by fluorescence resonance energy transfer (FRET). We have now used a microscopy-based screen to identify inhibitors that prevent AR conformational change or nuclear accumulation after ligand activation. Hits were secondarily selected based on their ability to inhibit AR transcription at a PSA-luciferase promoter, and were tested for effects on 3H-DHT binding to AR in cells. We find a strong correlation between compounds that block DHT binding and those that inhibit nuclear accumulation. These compounds are structurally distinct from known antagonists. Additional compounds blocked AR conformational change but did not affect DHT binding or nuclear localization of AR. One compound increased ligand-induced FRET, yet functioned as a potent inhibitor. These results suggest multiple inhibitory conformations of AR are possible, and can be induced by diverse mechanisms. The lead compounds described here may be candidates for the development of novel anti-androgens, and may help identify new therapeutic targets.
Introduction
The androgen receptor (AR) is a member of the nuclear hormone receptor (NR) superfamily, which consists of a large group of ligand-regulated transcription factors (1). AR is expressed in many tissues and influences an enormous range of physiologic processes such as cognition, muscle hypertrophy, bone density, and prostate growth and differentiation (2). AR signaling is directly linked to numerous disorders including benign prostatic hyperplasia (BPH), alopecia, and hirsutism; and it also drives the proliferation of prostate cancer (PCa), even in the setting of therapies that reduce systemic androgen levels. AR is thus the major therapeutic target for this malignancy (3).
AR activation is initiated by binding of testosterone or the more potent dihydrotestosterone (DHT) to its ligand binding domain. However, AR is likely regulated at multiple points subsequent to ligand binding, and can even be activated in the absence of ligand by various cross-talk pathways (4–7). Prior to ligand binding, AR associates with a complex of cytoplasmic factors and molecular chaperones that maintain it in a high-affinity ligand binding conformation (8, 9). Ligand binding induces an intramolecular conformational change that brings the N and C-termini into close proximity, occurs in minutes after DHT treatment (10), and does not occur in cell lysates, suggesting that this process is not protein autonomous, but depends on additional cellular factors (11). After ligand activation, AR accumulates in the nucleus, where it binds DNA as a homodimer at specific androgen response elements (AREs) to regulate gene expression. This requires interactions with positive (coactivator) and negative (corepressor) factors (12). AR is then recycled to the cytoplasm (13). AR degradation is proteasome-dependent, and is mediated in part by an N-terminal proteasome-targeting motif (14). AR activity is also regulated by multiple cross-talk pathways, including HER-2/neu kinase and insulin-like growth factor-1 signaling, which influence AR activity via post-translational modifications such as phosphorylation, sumoylation, and acetylation (12).
All existing approaches to treat AR-associated diseases target ligand binding. This includes direct competition with competitive antagonists such as bicalutamide, reduction of ligand levels with gonadotropin-releasing hormone (GnRH) agonists, blocking testosterone synthesis with CYP17A1 inhibitors, or blocking DHT formation with 5α reductase inhibitors. However, it is clear that AR activity can be inhibited at points distinct from ligand binding (15, 16). Such inhibition could profoundly enhance current anti-androgen therapies. Heat shock proteins, histone deacetylases, and several kinases, such as the HER2/neu kinase are among the targets being explored as ‘indirect’ AR regulators (17–20).
We have previously created a FRET-based conformation reporter system that we exploited in a plate reader assay to identify AR inhibitors (11). This cell-based assay allows identification of inhibitory compounds that directly bind AR, and those that block its activity indirectly, presumably by targeting proteins required for ligand-induced conformational change. However, because it utilizes readings from populations of cells, it cannot simultaneously discriminate multiple aspects of AR activation, such as conformational change and nuclear localization. In this study, we utilized high-content fluorescence microscopy to detect ligand-induced conformational change in the cytoplasm and nucleus of individual cells, and to determine the relative distribution of AR between the cytoplasm and nucleus. By simultaneously monitoring two independent steps in AR signaling, in this screen we defined several new classes of anti-androgens that reflect multiple modes of inhibition.
Results and Discussion
Screening for novel anti-androgens using high-throughput microscopy
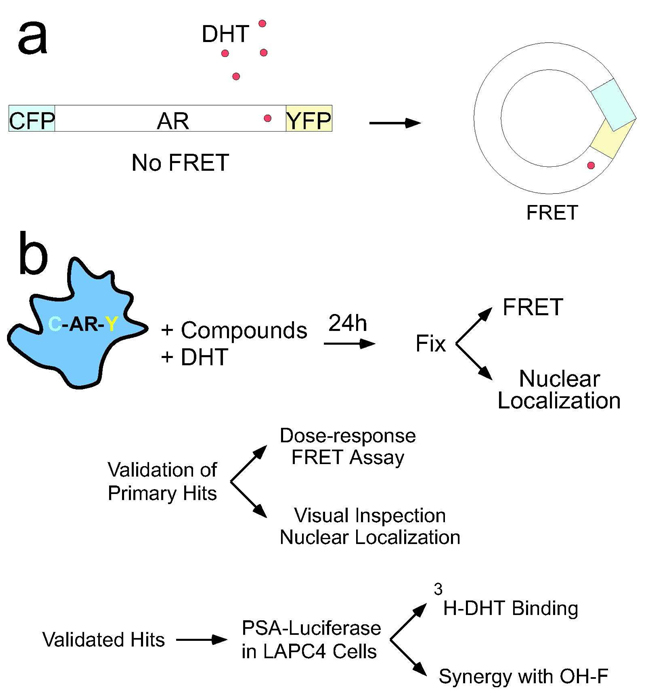
The HEK293/C-AR-Y cell line has been previously described (11). This line stably expresses full-length human AR fused to cyan (CFP) and yellow (YFP) fluorescent proteins at the amino and carboxyl termini, respectively. We developed a high content assay using automated microscopy to simultaneously measure two important steps in AR signaling: ligand induced conformational change and subcellular localization (Figure 1a). HEK293/C-AR-Y cells were stimulated with 10nM DHT, and the inhibitory effect of various compounds was measured after 24h (Figure 1b). In control wells, where cells were treated with DHT and the vehicle DMSO, seventy to eighty percent of cells demonstrated nuclear translocation, as opposed to less than four percent translocation in the absence of DHT (Z’=.72). FRET signal, as measured by FRET:donor ratio, increased 60% in the presence of DHT (Z’=0.24). We used image analysis algorithms to identify cells, delineate cytoplasm from nucleus, and determine the total fluorescence and FRET:donor ratio in each compartment. We excluded from our analysis compounds that reduced the total cell count below 100, and those that altered the total CFP or YFP signal more than two standard deviations from control wells treated with DHT alone. These filters eliminated toxic compounds, non-specific transcription or translation inhibitors, and compounds with inherent fluorescence that would confound analysis. Based on these criteria, ~17% of the compounds were eliminated, which was similar to our previous experience (21).
Figure 1. Screening Strategy.
(a) AR was cloned between CFP (donor) and YFP (acceptor) and stably expressed in cells. DHT binding causes a conformational change in AR that brings the CFP and YFP moieties together to enable FRET. (b) The screening strategy involves treatment of stable HEK293/C-AR-Y cells with DHT and test compounds, followed by microscopy-based analysis for inhibitors of FRET and nuclear localization. Compounds from the primary screen were validated by re-testing with a dose-response for the FRET assay, and by direct visual inspection of cells to confirm inhibition of nuclear localization. Validated hits were also re-tested in the complementary assays. Validated hits were tested next for inhibition of endogenous AR activity by transfecting a PSA-luciferase reporter into LAPC4 cells. All lead compounds were then checked for effects on 3H-DHT binding, and selected compounds were evaluated for synergy with OH-F.
The FRET:donor ratio was quantified as previously described (10). A significant difference existed between the cytoplasmic and nuclear FRET signals in only 0.1% (5 of 4423) of the wells, and in no case could we reproduce this difference on repeated measurements, suggesting that no effects on AR conformation were limited to a particular compartment. Thus, we averaged cytoplasmic and nuclear FRET signals to represent the FRET value from the entire cell. The degree of cytoplasm to nucleus translocation of AR was determined by correlating YFP and Hoechst (nuclear) signals. The maximal conformational change and nuclear accumulation values were derived from cells treated with 10nM DHT alone. Minimal FRET values were derived from cells treated with vehicle control (DMSO). Using these values, we calculated the percent inhibition of conformational change and nuclear accumulation. Our prior work with the HEK293/C-AR-Y reporter cell line indicated that a four standard deviaton (SD) FRET cutoff would limit a screen to about 1–5% of all compounds, of which a high percentage would be validated in secondary assays(11). A 50% inhibition of nuclear translocation or FRET signal (which represented at least a four SD reduction from the maximal value) was used to select compounds for secondary analysis.
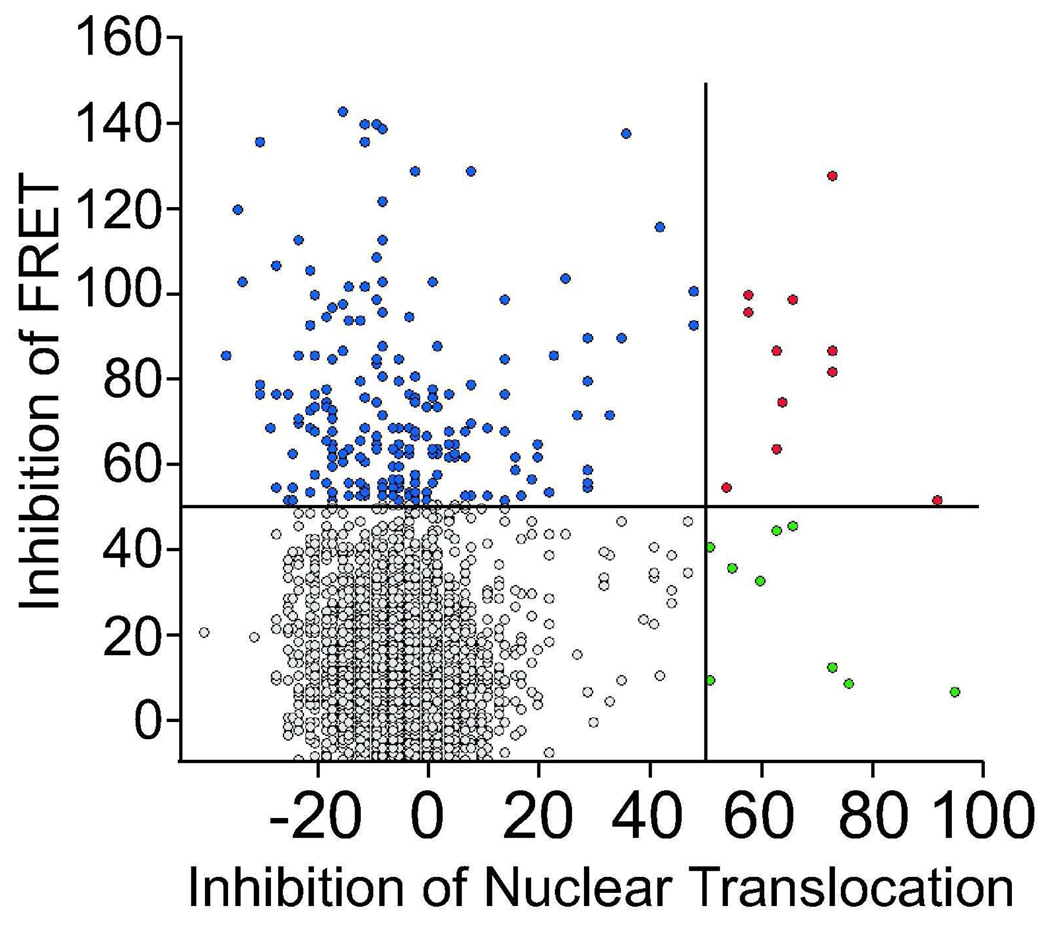
We screened 4423 compounds from an in-house small molecule collection at the Broad Institute. This was compiled from known bioactive molecules, including many FDA approved drugs that are commercially available from several vendors (Figure 2). 308 compounds (~7%) inhibited the FRET signal by >50%. 20 compounds (~0.5%) inhibited nuclear accumulation by >50%. 11 compounds (~0.3%) inhibited both conformational change and nuclear accumulation by >50%. To reduce subsequent analyses, when multiple hits with similar structures were identified, only one was validated in secondary assays. For example, of gambogic acid, gambogic acid amide, and dihydrogambogic acid, only gambogic acid was analyzed further. We also excluded known competitive antagonists (e.g. nilutamide), as their mechanisms of action are already known. Based on these considerations, potency in the primary assays, and the availability of compounds, we selected 121 compounds that inhibited FRET > 50% and 9 compounds that inhibited nuclear accumulation >50% in the primary assays. These represented more than 70% of non-redundant primary hits from both the conformational change and nuclear accumulation screens. An example of different cellular responses to hits is shown in Figure 3.
Figure 2. Distribution of hits from the primary screen.
After filtering the data, compounds were ranked for their ability to inhibit AR conformational change (blue), nuclear accumulation (green), or both (red). A 50% inhibition, or approximately 4 standard deviations below DHT alone, was used as a cutoff to determine which compounds to analyze in secondary assays.
Figure 3. Examples of cellular responses in the primary screen.
20x images of HEK293/C-AR-Y cells from the cell imager were collected. The YFP and Hoechst channels represent primary image acquisition; the FRET channel represents the relative FRET intensity. The first row represents untreated cells, with predominant localization of C-AR-Y in the cytoplasm; the second row represents 10nM DHT stimulation with mainly nuclear localization; the third row illustrates cells treated with DHT and diflorasone, a steroid that blocked AR conformational change, but not nuclear localization; the fourth row illustrates cells treated with DHT and Chembridge 5107769, which blocked nuclear import, but did not affect FRET.
We validated primary hits in the FRET assay by re-testing each compound in a dose titration in quadruplicate. 38/121 compounds (31%) scored as true positives using this approach, consistent with our prior study (11). Many primary hits did not exhibit a dose response, often because their toxic concentrations were similar to their effective concentrations in our assay. Other hits failed validation because their fluorescence profiles affected the FRET readings. To validate nuclear accumulation inhibitors, HEK293/C-AR-Y cells were pre-treated with each compound for 1hr, and then treated with 1nM DHT. Cells were fixed at 2hrs and 24hrs post-DHT exposure and examined by visual inspection using fluorescence microscopy. All putative nuclear accumulation inhibitors scored as true positives in this assay, reflecting the power of the microscopy-based primary screen. Two validated compounds initially scored positive as both conformation and nuclear accumulation inhibitors.
We cross-examined hits from one part of the screen for activity in the other. Four nuclear accumulation inhibitors that had not scored positive in the conformational change screen actually did inhibit conformational change. None of the original conformational change inhibitors from the primary assay blocked nuclear translocation upon subsequent analysis. Thus, while some inhibitors block all aspects of AR function, ligand-induced conformational change and nuclear accumulation are not necessarily linked, and are separable targets for AR inhibition.
Next we tested for inhibition of endogenous AR transcriptional activity. LAPC4 cells, which are derived from prostate cancer and express wild-type AR (22), were transfected with an androgen-dependent PSA promoter-firefly luciferase reporter plasmid and an androgen-independent renilla luciferase control. Validated hits were tested in a dose-response. After 24hrs, AR-dependent transcription was measured using renilla-normalized firefly luciferase activity. Every validated inhibitor of both conformational change and nuclear accumulation also inhibited the transcriptional activity of endogenous AR, indicating the very strong predictive power of a multi-modal readout. Some compounds had nanomolar potency (Table 1, column 4).
Table 1.
| Compound | Conformational Change |
Nuclear Accumulation |
Transcription IC50 (nM) |
DHT Binding IC50 (nM) |
|---|---|---|---|---|
| Chembridge 5107769 | X | 341 | 182 | |
| sanguinarine sulfate | X | 500 | 779 | |
| dihydrocelastrol | X* | X | 52 | 57 |
| gambogic acid | X* | X | 269 | 36 |
| thimerosal | X* | X | 347 | no effect |
| helenine | X* | X | 1010 | 6360 |
| radicicol | X | X | 3.4 | 10971 |
| Chembridge 5128773 | X | X | 834 | no effect |
| actinomycin D | X | 1.1 | no effect | |
| 17-AAG | X | 1.7 | no effect | |
| cucurbitacin I | X | 1.00 | 256 | |
| puromycin HCl | X | 19 | no effect | |
| AG 592 | X | 215 | no effect | |
| oxindole I | X | 224 | no effect | |
| xanthohumol | X | 276 | no effect | |
| sertaconazole nitrate | X | 554 | 1153 | |
| acrisorcin | X | 915 | 718 | |
| bromoconduritol | X | 1000 | 4600 | |
| cadmium acetate | X | 1102 | no effect | |
| 2,5-dihydroxycinnamic acid | X | 1142 | 10811 | |
| epigallocatechin-3-monogallate | X | 1432 | no effect | |
| oxiconazole nitrate | X | 1491 | 1534 | |
| mechlorethamine | x | 1560 | 7670 | |
| luffariellolide | x | 1563 | no effect | |
| madecassic acid | x | 1989 | no effect | |
| EGFR/ErbB-2 Inhibitor | x | 2426 | no effect | |
| Chembridge 5404078 | x | 2480 | no effect | |
| GSK-3b Inhibitor III | x | 2555 | no effect | |
| thapsigargicin | x | 2776 | no effect | |
| WR 216174 | x | 3108 | 12750 | |
| myoseverin | x | 4048 | no effect | |
| MDL-12,330A, HCl | x | 4412 | no effect | |
| xanthyletin | x | 5000 | no effect | |
| retusoquinone | x | 5616 | no effect | |
| mebendazole | x | 8473 | no effect | |
| glutethimide | x | 9795 | no effect | |
| catechin | x | 10000 | no effect | |
| Chembridge 5255637 | x | 10000 | no effect | |
| blasticidine S | x | 10000 | no effect | |
| epoxomicin | x | 10000 | no effect | |
| chlorpromazine HCl | x | 11950 | no effect | |
| ikarugamycin | x | 14304 | no effect | |
| heudelottin c | x | 16632 | no effect | |
| Chembridge 5213395 | x | 17000 | no effect |
indicates that positive result was obtained by cross-assay validation, as opposed to a result from the primary screen. Structures of all non-competitive inhibitors are shown in Supplementary Figure 1.
Novel antagonists of DHT binding to AR
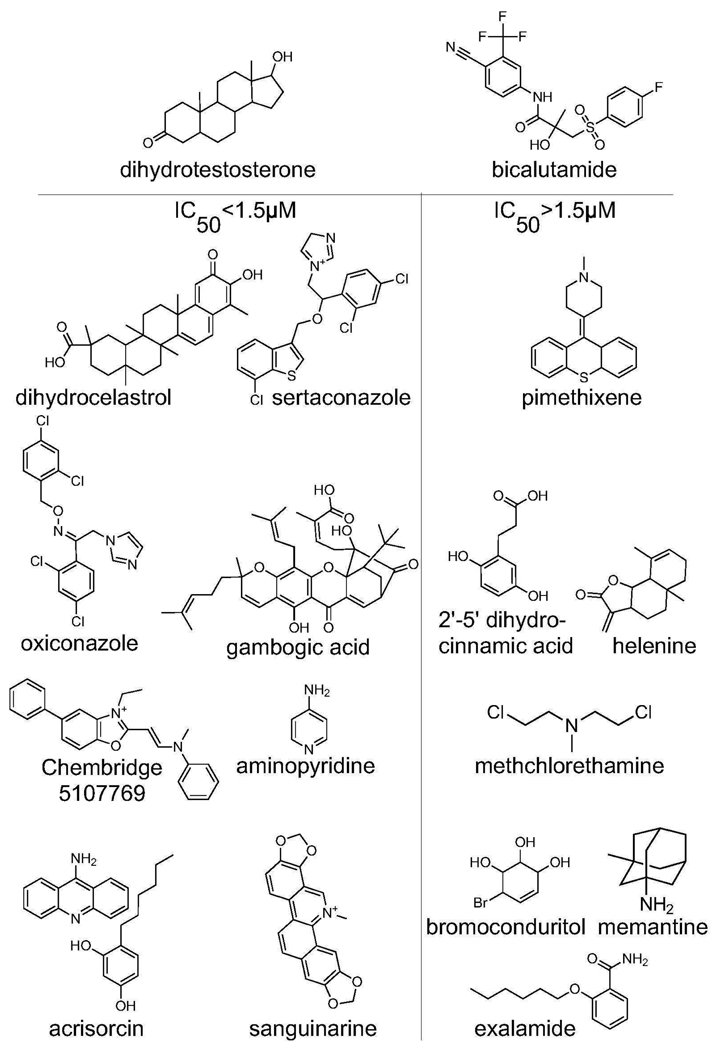
We employed a whole cell assay to test whether any validated compounds would inhibit ligand binding to AR. HEK293/C-AR-Y cells were incubated for 1hr with 1nM 3H-DHT and various doses of test compounds. Binding of 3H-DHT to AR was quantified via scintillation counter. We calculated the concentration at which each compound inhibited DHT binding by 50% (Table I, column 5). 6/8 nuclear accumulation inhibitors prevented DHT binding to AR. 12/42 conformation change inhibitors (including the nuclear accumulation inhibitors that subsequently scored in the conformation change assay) also prevented DHT binding. None of these compounds has a structure similar to known steroidal or non-steroidal competitive antagonists (Figure 4). These leads thus may represent new types of ligand binding inhibitors.
Figure 4. Structures of putative competitive antagonists.
Structures of compounds that inhibit DHT binding to AR are shown, with more potent compounds on the left. The commercial compound known as acrisorcin is a mixture of the two indicated chemicals.
The whole cell DHT binding assay does not exclusively report competitive antagonists as any compound that disrupts the conformation of the ligand binding pocket of AR could also block ligand binding activity. The electrophilic nature of a number of the compounds suggests that they could covalently modify AR, or AR accessory proteins. It is possible that these electrophilic compounds bind the newly recognized BF-3 site on the AR ligand binding domain, similar to previously identified anti-androgens with electrophilic characteristics (23). Further studies are required to determine the exact binding sites for these compounds, however.
Compound Synergy
To gain further insight into mechanism, we tested whether combinations of the most potent compounds would act in an additive vs. synergistic manner with the competitive antagonist hydroxyflutamide (OH-F) to inhibit AR activity in the LAPC4 luciferase reporter assay. Two competitive antagonists in combination should inhibit AR activity in an additive manner. Conversely, a compound with a different mechanism of action may have an additive, antagonistic, or synergistic effect with a competitive antagonist. Cells were treated with an increasing concentration of compound, OH-F, or their combination at a constant ratio, and the relative luciferase activities were measured. After creating a mean-effect plot for each combination and determining the expected additive IC50 vs. the actual IC50, we used the combination index (CI) to evaluate the relationship between the compounds (Table 2), where a CI<1 indicates synergy, a CI~1 indicates additivity, and a CI>1 indicates antagonism (24). As expected, the combination of gambogic acid or CB5107769, two putative competitive antagonists, with OH-F resulted in a CI50 of ~1, indicating an additive effect. Other compounds exhibited synergy with OH-F (Table 2).
Table 2.
| Ratio of inhibitor treatments |
Actual IC50 (nM) | Expected IC50 (nM) | CI at IC50 |
|---|---|---|---|
| actinomycin:OH-F 1:10 | 0.78 | 1.4 | 0.3 |
| radicicol:OH-F 1:10 | 0.66 | 5.5 | 0.08 |
| cucurbitacin:OH-F 1:100 | 0.35 | 0.55 | 0.4 |
| oxindole:OH-F 3:1 | 31 | 223 | 0.1 |
| gambogic acid:OH-F 1:1 | 72 | 81 | 1 |
| CB5107769:OH-F 3:1 | 276 | 340 | 0.8 |
Compounds that interfere with ligand binding
One compound, sanguinarine, a natural product, has previously been shown to compete with 10nM dexamethasone for binding to the glucocorticoid receptor (GR) with an IC50 of ~10µM (25). We observed competition for 1nM DHT with an IC50 of less than 1µM (Table 1, column 5), suggesting a greater affinity for AR than GR. It is likely that sanguinarine binds a conserved surface on the NRs, probably within the ligand binding pocket, and could serve as a scaffold for the design of new antagonists for AR and GR, and possibly for other related NRs.
Ketoconazole binds and inhibits cytochrome P-450 dependent steroidogenic enzymes with high affinity, thus inhibiting testosterone synthesis, but it can also bind to AR with a much lower affinity (~60µM) (26). We found that sertaconazole and oxiconazole, two derivatives of ketoconazole, competed with DHT at ~1µM. Similarly, ketoconazole and miconazole, another derivative, have been shown to competitively antagonize dexamethasone binding to the glucocorticoid receptor (GR) (27). Ketoconazole also directly inhibits pregnane X receptor activity by disrupting its association with the steroid receptor coactivator-1 (28). Ketoconazole and related compounds have been used to treat androgen dependent diseases by inhibiting DHT synthesis, but sertaconazole and oxiconazole could also competitively antagonize AR, and might be therapeutic leads in this regard.
We found an isomer of dihydrocinnamic acid, a known competitive antagonist of 5α reductase (29), to have apparent affinity for AR as well (Table 1). It has previously been suggested that dihydrocinnamic acid could be used to treat BPH and PCa (29). Our results suggest that it may directly inhibit AR, in addition to blocking 5α reductase. Two other natural products, gambogic acid and celastrol, have been observed to inhibit the growth of prostate cancer cells in xenograft mouse models (30, 31). The mechanism of celastrol has been attributed to proteasome inhibition and gambogic acid to VEGF receptor 2 inhibition, but we found that these compounds prevented >50% of DHT binding at 58nM and 36nM respectively, suggesting that they could inhibit prostate cancer growth primarily by preventing ligand binding to AR. It remains to be seen whether any of the putative competitive antagonists identified in our screen associate with the AR ligand binding pocket in the same orientation as other known AR ligands or competitive antagonists. If they do, they could provide new scaffolds for the design of antagonists.
Novel, non-competitive AR inhibitors
We identified multiple, novel non-competitive, or indirect, AR inhibitors, some with low nanomolar potencies (Table 1 and Supplementary Figure 1). Two Hsp90 inhibitors, 17-AAG and radicicol, inhibited AR-dependent transcription in LAPC4 cells with potencies of 1–3nM (Table 1, column 4). The interaction between Hsp90 and AR is well-documented, and Hsp90 is required for proper AR function (9). However, 17-AAG did not compete for DHT binding and radicicol inhibited DHT binding to AR only at concentrations of >1000x its potency as a transcription inhibitor (Table 1, column 5). Thus each appears to influence AR activity by a mechanism distinct from blocking DHT binding. 17-AAG is a widely used Hsp90 inhibitor and has previously been shown to inhibit AR activity and reduce prostate tumor growth in a xenograft model (32). Radicicol, which was identified in both the conformational change and nuclear accumulation screens, has previously been shown to inhibit AR nuclear accumulation (33), corroborating our results. Because Hsp90 inhibitors work by a different mechanism than competitive antagonists, we hypothesized that they would synergize. We treated LAPC4 cells transfected with PSA-luciferase with dose titrations of OH-F, radicicol, or a combination of the compounds, and measured the resultant luciferase activities (Table 2). A 1:10 combination of radicicol and OH-F synergistically inhibited AR activity with picomolar efficacy.
Cucurbitacin I, a natural product, inhibited AR transcription with a potency of approximately 1nM, and inhibited DHT binding at approximately 250nM, which may account for some, but not all of its activity. In a synergy analysis, a 1:100 combination of cucurbitacin I and OH-F had a CI50 of 0.4 (Table 2), a borderline synergistic effect, suggesting that both competitive and non-competitive mechanisms of AR inhibition may be involved. Cucurbitacin I has been identified as a potent and selective inhibitor of JAK/STAT3 signaling (34), suggesting that this cross-talk pathway might contribute to the regulation of AR conformational change and downstream activity. We also found that actinomycin D, a nonspecific transcriptional inhibitor, blocked AR transcriptional activity with an IC50 of approximately 1nM. At this concentration the drug had no effect on the activity of the control renilla luciferase reporter, consistent with a more specific effect on AR conformation. Actinomycin D also synergized with OH-F (Table 2), suggesting that these two compounds inhibit AR activity by different mechanisms. Actinomycin D has been used as a general cytotoxic agent to treat various cancers, including PCa, but to our knowledge it has not been used specifically as an anti-androgen.
A novel conformational path to AR inhibition
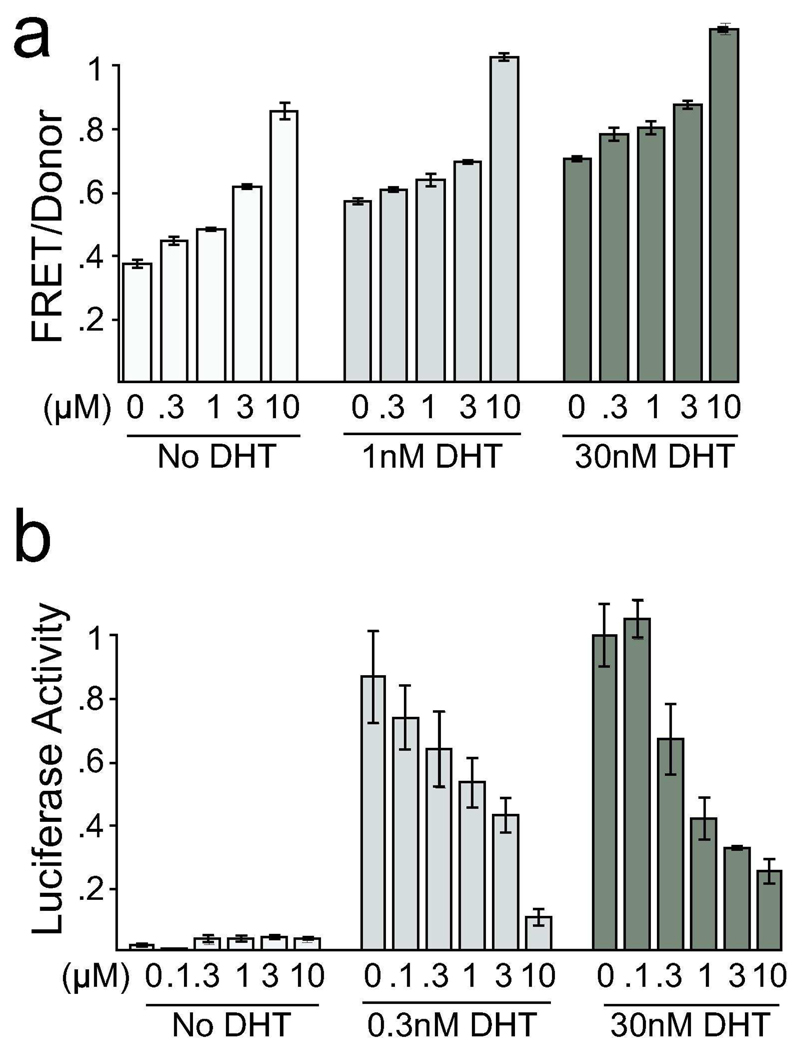
One compound, oxindole I, increased the FRET signal in HEK293/C-AR-Y cells (Figure 5A), without affecting absolute fluorescence values. This suggests that oxindole I may lead to a more “compact” AR conformation, in which the N and C termini are brought closer together. Oxindole I blocked AR-dependent transcription in LAPC4 cells with an IC50 of 224nM (Figure 5B). It did not compete for DHT binding in the whole cell radiolabel assay, and a combination of oxindole I and OH-F synergistically inhibited AR transcription with a CI50 of 0.1 (Table 1 and Table 2). In the absence of DHT, oxindole I induced a conformational change in AR, without inducing transcriptional activity, though not to the extent of DHT. The compound also increased the FRET:donor ratio, even at saturating levels of DHT (30nM), though it still inhibited AR transcriptional activity at these high DHT levels (Figure 5B). Oxindole I inhibits the VEGF receptor tyrosine kinase, fetal liver kinase (Flk-1) with an IC50 of 390nM, possibly by binding its ATP-binding pocket (35). It is unclear at this point how Flk-1 might alter AR conformation, but these results indicate multiple, distinct effects on AR conformation can be produced by various inhibitors.
Figure 5. Oxindole 1 increases AR conformational change but inhibits AR-dependent transcription.
(a) HEK293/C-AR-Y cells were treated with 0nM, 1nM, or 30nM DHT and increasing amounts of oxindole 1. FRET was recorded on a fluorescence plate reader. Oxindole 1 increased FRET with and without DHT present (b) LAPC4 cells transfected with luciferase reporters were treated with 0nM, 0.3nM, and 30nM DHT and increasing amounts of oxindole 1, which decreased AR-dependent transcription.
Conclusion
The development of new types of AR inhibitors might play an important role in the future treatment of human disease. This study illustrates how a multifaceted screen based on high-throughput microscopy increases detection power, and corroborates prior efforts (36). The combination of nuclear localization with conformational change as a readout predicted bona fide AR inhibitors with 100% specificity. While nuclear accumulation and ligand binding appear to be tightly linked, conformational change relies on many factors in addition to ligand binding, since compounds that prevented conformational change did not necessarily prevent DHT binding to AR. This cell-based assay thus has the power to identify compounds that inhibit AR activity by directly binding AR, and also those that inhibit AR activity indirectly, presumably by targeting accessory or regulatory factors. The identification of separate inhibitors of conformational change and nuclear accumulation that block transcriptional activity of AR highlights how each step in the AR signaling pathway contributes to downstream activity, and may be targeted pharmacologically. The spectrum of potential AR antagonists is thus quite large.
Materials and Methods
Cell culture
HEK293 and HEK293/C-AR-Y cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with antibiotics and 5% fetal bovine serum (FBS). LAPC4 cells were maintained in phenol-red free RPMI 1640 media supplemented with antibiotics and 10% FBS. Cells were transferred to media containing 5% charcoal-stripped FBS 48hr prior to FRET or transcription assays.
High-throughput screening
HEK293/C-AR-Y cells were dispensed by Multidrop Combi (Thermo Scientific) to 384-well plates in the presence of 10nM DHT and library compounds. Twenty four hours later, cells were fixed for 30 minutes in 4% formaldehyde/PBS and stained with 0.5 ug/ml Hoechst for 30 minutes before the cells were washed once in PBS. In all liquid exchange steps, dispense was performed by Wellmate (Matrix Technologies) and aspiration by ELX405HT (Bio-Tek). Images were acquired by automated microscopy (ImageXpress micro, MDS Analytical Technologies) with plates being fed to the microscope by a CRS robot (Thermo Scientific). The images were acquired with 20x objective for CFP, YFP, and FRET channels. HEK293 cells not expressing the C-AR-Y reporter were included as a control for background fluorescence. HEK293 cells transfected with respective CFP-, YFP-, and CFP-YFP-expressing plasmids were used to calibrate the bleed through between channels. Images were analyzed using MetaXpress (MDS Analytical Technologies) to determine degree of AR nuclear translocation and the total fluorescence and FRET:donor ratio in cytoplasm and nucleus.
Transcription assays
For all transfections, pools of cells were transfected using Lipofectamine Plus (Invitrogen) with pRL-SV40 (Promega) and PSA-luciferase as previously described (21). This region has been shown to induce expression of a similar luciferase reporter gene upon treatment with androgen (37)). The following day, cells and drugs were distributed to 96 well plates. 24hrs later, luciferase activity was measured (Dual luciferase assay kit, Promega). Mean-effect plots (log[compound] vs log[fractional effect]) were generated to determine the IC50 values for each compound or combinations of compounds at constant ratios. Microsoft Excel was used to calculate the statistics for a line using the “least squares” method. The F statistic was used to determine whether the observed relationship between the dependent and independent variables occurred by chance. Only data with an r2 value greater than 0.95 and an F value that was greater than that indicated by the F table for alpha=0.05 were used for analysis. The methods of Chou and Talalay were used to determine whether two compounds had antagonistic, additive, or synergistic reactions toward each other (24). Briefly, a combination index (CI) was established for a range of fractional effects, where a CI~1 indicates additivity, CI > 1 indicates antagonism, and a CI < 1 indicates synergy. The CI’s were based upon an exclusive or non-exclusive assumption, as determined by the slope of the line of the combination of drugs from the mean-effect plot.
Radioligand competition binding assay
5×105 HEK293/C-AR-Y cells were seeded in 24-well plates in phenol-red free media containing 5% charcoal-stripped FBS. After 3 days, media was replaced with serum-free media containing 3nM 3H-DHT in the absence or presence of 0.1–1000 fold molar excess of unlabeled competitor ligands for 90min at 37°C. Cells were washed with phosphate buffer, bound ligand was extracted in ethanol for 30min at RT, and detected using a scintillation counter.
Supplementary Material
Acknowledgments
This work was supported by NIH-5F32CA123750 (JOJ), the NIH/NCI 1R01CA131226 (MID), the Prostate Cancer Foundation (MID), and the Sandler Family Supporting Foundation (MID). The project has been funded in whole or in part with Federal funds from the National Cancer Institute's Initiative for Chemical Genetics, National Institutes of Health, under Contract No. N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Service, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
References
- 1.Katzenellenbogen JA, Katzenellenbogen BS. Nuclear hormone receptors: ligand-activated regulators of transcription and diverse cell responses. Chem Biol. 1996;3:529–536. doi: 10.1016/s1074-5521(96)90143-x. [DOI] [PubMed] [Google Scholar]
- 2.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat Med. 1999;5:280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 5.Klocker H, Culig Z, Hobisch A, Cato AC, Bartsch G. Androgen receptor alterations in prostatic carcinoma. Prostate. 1994;25:266–273. doi: 10.1002/pros.2990250506. [DOI] [PubMed] [Google Scholar]
- 6.Yeh S, Lin HK, Kang HY, Thin TH, Lin MF, Chang C. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc Natl Acad Sci U S A. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueda T, Bruchovsky N, Sadar MD. Activation of the androgen receptor N-terminal domain by interleukin-6 via MAPK and STAT3 signal transduction pathways. J Biol Chem. 2002;277:7076–7085. doi: 10.1074/jbc.M108255200. [DOI] [PubMed] [Google Scholar]
- 8.Cardozo CP, Michaud C, Ost MC, Fliss AE, Yang E, Patterson C, Hall SJ, Caplan AJ. C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch Biochem Biophys. 2003;410:134–140. doi: 10.1016/s0003-9861(02)00680-x. [DOI] [PubMed] [Google Scholar]
- 9.Georget V, Terouanne B, Nicolas JC, Sultan C. Mechanism of antiandrogen action: key role of hsp90 in conformational change and transcriptional activity of the androgen receptor. Biochemistry. 2002;41:11824–11831. doi: 10.1021/bi0259150. [DOI] [PubMed] [Google Scholar]
- 10.Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AA, Miner JN, Diamond MI. The structural basis of androgen receptor activation: intramolecular and intermolecular amino-carboxy interactions. Proc Natl Acad Sci U S A. 2005;102:9802–9807. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones JO, Diamond MI. A cellular conformation-based screen for androgen receptor inhibitors. ACS Chem Biol. 2008;3:412–418. doi: 10.1021/cb800054w. [DOI] [PubMed] [Google Scholar]
- 12.Poletti A. The polyglutamine tract of androgen receptor: from functions to dysfunctions in motor neurons. Front Neuroendocrinol. 2004;25:1–26. doi: 10.1016/j.yfrne.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Tyagi RK, Lavrovsky Y, Ahn SC, Song CS, Chatterjee B, Roy AK. Dynamics of intracellular movement and nucleocytoplasmic recycling of the ligand-activated androgen receptor in living cells. Mol Endocrinol. 2000;14:1162–1174. doi: 10.1210/mend.14.8.0497. [DOI] [PubMed] [Google Scholar]
- 14.Chandra S, Shao J, Li JX, Li M, Longo FM, Diamond MI. A common motif targets huntingtin and the androgen receptor to the proteasome. J Biol Chem. 2008;283:23950–23955. doi: 10.1074/jbc.M800467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao J, Wang S, Qiao R, Vivanco I, Watson PA, Sawyers CL, Wu H. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007;67:6083–6091. doi: 10.1158/0008-5472.CAN-06-4202. [DOI] [PubMed] [Google Scholar]
- 16.Jones JO, Bolton EricC, Huang Yong, Feau Clementine, Guy RKiplin, Yamamoto KeithR, Hann Byron, Diamond MarcI. Non-competitive Androgen Receptor Inhibition In Vitro and In Vivo. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0807282106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobosy JR, Roberts JL, Fu VX, Jarrard DF. The expanding role of epigenetics in the development, diagnosis and treatment of prostate cancer and benign prostatic hyperplasia. J Urol. 2007;177:822–831. doi: 10.1016/j.juro.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 18.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Mellinghoff IK, Vivanco I, Kwon A, Tran C, Wongvipat J, Sawyers CL. HER2/neu kinase-dependent modulation of androgen receptor function through effects on DNA binding and stability. Cancer Cell. 2004;6:517–527. doi: 10.1016/j.ccr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 20.Solit DB, Scher HI, Rosen N. Hsp90 as a therapeutic target in prostate cancer. Semin Oncol. 2003;30:709–716. doi: 10.1016/s0093-7754(03)00346-4. [DOI] [PubMed] [Google Scholar]
- 21.Jones JO, Diamond MI. A Cellular Conformation-Based Screen for Androgen Receptor Inhibitors. ACS Chem Biol. 2008;3:412–418. doi: 10.1021/cb800054w. [DOI] [PubMed] [Google Scholar]
- 22.Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, Lamb DJ, Marcelli M, Belldegrun A, Witte ON, Sawyers CL. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 23.Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A. 2007;104:16074–16079. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 25.Dvorak Z, Vrzal R, Maurel P, Ulrichova J. Differential effects of selected natural compounds with anti-inflammatory activity on the glucocorticoid receptor and NF-kappaB in HeLa cells. Chem Biol Interact. 2006;159:117–128. doi: 10.1016/j.cbi.2005.10.105. [DOI] [PubMed] [Google Scholar]
- 26.Eil C. Ketoconazole binds to the human androgen receptor. Horm Metab Res. 1992;24:367–370. doi: 10.1055/s-2007-1003337. [DOI] [PubMed] [Google Scholar]
- 27.Duret C, Daujat-Chavanieu M, Pascussi JM, Pichard-Garcia L, Balaguer P, Fabre JM, Vilarem MJ, Maurel P, Gerbal-Chaloin S. Ketoconazole and miconazole are antagonists of the human glucocorticoid receptor: consequences on the expression and function of the constitutive androstane receptor and the pregnane X receptor. Mol Pharmacol. 2006;70:329–339. doi: 10.1124/mol.105.022046. [DOI] [PubMed] [Google Scholar]
- 28.Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26:258–268. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 29.Hiipakka RA, Zhang HZ, Dai W, Dai Q, Liao S. Structure-activity relationships for inhibition of human 5alpha-reductases by polyphenols. Biochem Pharmacol. 2002;63:1165–1176. doi: 10.1016/s0006-2952(02)00848-1. [DOI] [PubMed] [Google Scholar]
- 30.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Celastrol, a triterpene extracted from the Chinese "Thunder of God Vine," is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66:4758–4765. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 31.Yi T, Yi Z, Cho SG, Luo J, Pandey MK, Aggarwal BB, Liu M. Gambogic acid inhibits angiogenesis and prostate tumor growth by suppressing vascular endothelial growth factor receptor 2 signaling. Cancer Res. 2008;68:1843–1850. doi: 10.1158/0008-5472.CAN-07-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams CR, Tabios R, Linehan WM, Neckers L. Intratumor injection of the Hsp90 inhibitor 17AAG decreases tumor growth and induces apoptosis in a prostate cancer xenograft model. J Urol. 2007;178:1528–1532. doi: 10.1016/j.juro.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 33.Thomas M, Dadgar N, Aphale A, Harrell JM, Kunkel R, Pratt WB, Lieberman AP. Androgen receptor acetylation site mutations cause trafficking defects, misfolding, and aggregation similar to expanded glutamine tracts. J Biol Chem. 2004;279:8389–8395. doi: 10.1074/jbc.M311761200. [DOI] [PubMed] [Google Scholar]
- 34.Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270–1279. [PubMed] [Google Scholar]
- 35.Sun L, Tran N, Tang F, App H, Hirth P, McMahon G, Tang C. Synthesis and biological evaluations of 3-substituted indolin-2-ones: a novel class of tyrosine kinase inhibitors that exhibit selectivity toward particular receptor tyrosine kinases. J Med Chem. 1998;41:2588–2603. doi: 10.1021/jm980123i. [DOI] [PubMed] [Google Scholar]
- 36.Marcelli M, Stenoien DL, Szafran AT, Simeoni S, Agoulnik IU, Weigel NL, Moran T, Mikic I, Price JH, Mancini MA. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J Cell Biochem. 2006;98:770–788. doi: 10.1002/jcb.20593. [DOI] [PubMed] [Google Scholar]
- 37.Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.