Abstract
Benzo[a]pyrene (BaP) is a known genotoxicant that affects both mitochondrial and nuclear DNA (mtDNA, nDNA). Here, we examined mtDNA and nDNA damage in the Atlantic killifish (Fundulus heteroclitus) from a highly contaminated Superfund site (Elizabeth River, VA, USA) and from a reference site (King’s Creek, VA, USA) that were dosed with 10 mg/kg BaP. Using the long amplicon quantitative PCR technique, we observed similar increases in mitochondrial and nuclear DNA damage in King’s Creek fish treated with BaP. Killifish from the Elizabeth River showed high levels of basal nDNA and mtDNA damage compared to fish from the reference site, but the level of damage induced due to BaP treatment was much lower in Elizabeth River killifish compared to King’s Creek fish. Laboratory-reared offspring from both populations showed increased BaP-induced damage in mtDNA, relative to nDNA. Similar to the adult experiment, the Elizabeth River larvae had higher levels of basal DNA damage than those from the reference site, but were less impacted by BaP exposure. Measurements of oxidative DNA damage (8-oxo-deoxyguanine by LC-MS/MS) showed no differences among treatment groups, suggesting that the majority of DNA damage is from covalent binding of BaP metabolites to DNA. This study shows for the first time that mitochondria can be an important target of BaP toxicity in fish, indicating that BaP exposures could have important energetic consequences. Results also suggest that multi-generational exposures in the wild may lead to adaptations that dampen DNA damage arising from BaP exposure.
Keywords: Fundulus heteroclitus, DNA damage, mitochondria, benzo[a]pyrene, Elizabeth River
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) are released into the environment as byproducts of incomplete combustion. These compounds can enter into aquatic ecosystems from anthropogenic sources such as urban runoff, fuel exhaust, and oil shipping and refineries (Latimer and Zheng, 2003). PAHs have been increasing in the aquatic ecosystem in parallel with the rapid urbanization since the 1970s (Van Metre et al., 2000; Van Metre and Mahler, 2005). Thus, the need to understand how these chemicals may affect aquatic systems is gaining in importance.
Several studies in mammalian models indicate that mitochondria are major targets of PAH toxicity within the cell. These studies have shown that PAHs localize to the mitochondria and are correlated with a decrease in ATP production (Zhu et al., 1995), loss of mitochondrial membrane potential and changes in mitochondrial morphology (Li et al., 2003; Xia et al., 2004), and induction of the mitochondria-dependent apoptotic pathway (Ko et al., 2004; Huc et al., 2006).
Mitochondria are responsible for the majority of cellular energy production in eukaryotes. In addition, mitochondria are involved in cellular calcium homeostasis, cell signaling, and apoptosis (Wallace, 1999; Duchen, 2004). As a result, disruption of normal mitochondrial function has been linked with a variety of diseases in mammalian systems, including humans (Chan, 2006). Thousands of mitochondria can exist in each eukaryotic cell, and within each mitochondrion, several copies of the mitochondrial genome are present. These circular chromosomes are believed to be especially prone to damage (Wallace, 1999; Izzotti, 2009). Their proximity to the electron transport chain, where reactive oxygen species are constantly produced, makes mitochondrial DNA (mtDNA) susceptible to oxidative damage. In addition, DNA repair activity in the mitochondria is less efficient than nuclear DNA (nDNA) repair (Shadel and Clayton, 1997). Base excision repair (BER) has been observed in the mitochondria, but nucleotide excision repair (NER) has not been detected in the mitochondria yet (Berneburg et al., 2006; Van Houten et al., 2006; Maynard et al., 2009). Therefore, mtDNA may be more affected by bulky adducts, such as those formed by PAH-metabolites, than nDNA. Finally, mtDNA is considered structurally less protected from damage than the more compact DNA-histone complex formed in the nucleus (Suliman et al., 2004). In mammals, mtDNA is especially susceptible to damage from stable adduct formation by the dihydrodiol-epoxide metabolite of benzo[a]pyrene (BaP), a well-studied PAH (Backer and Weinstein, 1980; Niranjan et al., 1985). Furthermore, Graziewicz et al. (2004) showed that mtDNA replication is hindered by adducts formed from BaP and benzo[c]phenanthrene epoxides in vitro. Almost nothing is known in terms of the role of xenobiotics, including PAHs, in the integrity and function of fish mtDNA. Since fish are generally efficient in metabolizing PAHs (van der Oost et al., 2003), it is plausible that reactive metabolite formation is elevated in these organisms. Therefore, fish may be quite vulnerable to both mtDNA and nDNA damage formed by bulky adducts as well as oxidative adducts, such as 8-hydoroxyguanosine (8-Oxo-dG), generated by PAHs.
The Elizabeth River, located in Portsmouth, Virginia, is a tributary of the James River in the Chesapeake Bay watershed. The sediment, groundwater, and soil at the Atlantic Wood Industries Superfund site on the southern branch of the Elizabeth River are heavily contaminated with PAHs (EPA, 2007; Hartwell and Hameedi, 2007). This site has been listed as a National Priorities Superfund site since 1990 (EPA, 1995). Total PAH concentrations at the site can be as high as 500 µg/g dry sediment, with BaP being one of the most prevalent PAHs, accounting for about 11 % of total PAHs (Vogelbein et al., 2008). These concentrations suggest that the Atlantic Wood Industries Superfund site is comparable to some of the most PAH-contaminated rivers/estuaries in the world (Walker et al., 2004).
Atlantic killifish (Fundulus heteroclitus) at this site show increased liver tumor formation (Vogelbein et al., 1990; Vogelbein et al., 1999). They also show refractory cytochrome P450 1A (CYP1A) expression and activity (Van Veld and Westbrook, 1995; Meyer et al., 2003b). Compared to fish from reference sites, these fish show resistance to developmental toxicity from the sediment collected at the site (Ownby et al., 2002; Meyer et al., 2003b; Wassenberg and Di Giulio, 2004). Interestingly, this population shows greater sensitivity to hypoxia (Meyer and Di Giulio, 2003), upregulated antioxidant defense (Meyer et al., 2003a; Bacanskas et al., 2004), and higher expression levels of enzymes involved in glycolytic energy metabolism (Meyer et al., 2005). Such reports indicate that there may be alterations in mitochondrial structure and/or function in this population. This could have important ecological implications. Fish mitochondria are already exposed to higher levels of oxidative stress, most likely due to higher conversion of oxygen to H2O2 and episodes of environmental and physiological hypoxia (Abele and Puntarulo, 2004). However fish generally have lower mitochondrial antioxidant activity than terrestrial vertebrates (Wilhelm Filho, 2007). Added oxidative stress may result in reduced bioenergetic fitness and subsequently, may result in reduced survival of these fish.
In this study, we hypothesized that treatment with BaP would induce damage in killifish mtDNA to a greater extent than in nDNA. We also hypothesized that Superfund site fish dosed with BaP would show little increase in DNA damage due to adaptation to PAHs. To test our hypotheses we compared the basal DNA damage levels, as well as changes in DNA damage levels in response to BaP for both mtDNA and nDNA of adult killifish collected from Elizabeth River and a reference site, and laboratory-reared progeny from these populations. In addition, we evaluated the amount of oxidative DNA damage in the two populations of fish with or without BaP treatment.
2. Materials and Methods
2.1. Fish care
Adult killifish were captured using baited minnow traps from the Atlantic Wood Superfund site at the Elizabeth River in Portsmouth, VA (36°48’27.4” N, 76°17’36.1” W) on July 18, 2006. Fish were caught on the same day from King’s Creek, a tributary of the Severn River in Gloucester County, VA (37°17’52.4”N, 76°25’31.4”W). After capture, fish were transported to the Duke University Ecotoxicology Laboratory and maintained as described previously (Matson et al., 2008). Fish were depurated in the laboratory for at least four weeks before adult and larval experiments were conducted.
Fish were manually spawned and eggs were incubated on plates with wet filter paper at 27°C. After 14 days, fish were hatched by the addition of 25 ppt artificial sea water (ASW, Instant Ocean®, Aquarium Systems, Forster & Smith, Rhinelander, WI, USA) and gentle shaking for about 30 minutes. Larvae were kept in 2-liter beakers in 25 ppt ASW and fed brine shrimp daily until initiation of the experiment.
2.2. BaP treatment and DNA isolation
For the adult exposure experiment, male fish from both populations were moved to individual aerated tanks with 3 L of 25 ppt artificial seawater (Instant Ocean®, Aquarium Systems, Forster & Smith, Rhinelander, WI, USA) 24 hours before treatment. Ten fish from each population were injected intraperitoneally with BaP dissolved in corn oil. 10 mg/kg of the chemical was injected with an injection volume of 5 µL/g wet weight. This dose was selected as it is high enough to increase the level of proteins that are involved in the metabolism of BaP, but low enough not to show any signs of cellular or organismal level of toxicity. Additionally, ten male fish from each population were injected with 5 µL/g wet weight of corn oil as an experimental control. The fish were fed Tetramin® Tropical Fish Food (Tetra Systems, Blacksburg, VA, USA) everyday and sacrificed 72 hours post-treatment.
In the larval experiment, 7 day post hatch (dph) larvae (10–12 from each population per treatment) were exposed to BaP individually in 20-mL scintillation vials containing 10 mL artificial sea water. The water was dosed with equal volumes of DMSO (dimethyl sulfoxide, Sigma-Aldrich, St. Louis, MO, USA) containing BaP (Sigma-Aldrich) to final concentrations of 0, 50, 100, and 200 µg/L BaP. The solvent concentrations did not exceed 0.03 % for all treatments. Larvae were sacrificed 5 days post-treatment.
After sacrifice, individual tissues (brain, liver, muscle) from adults and whole individual larvae were flash frozen in 20% glycerol and stored in −80°C until further analysis. Tissues were later ground in liquid nitrogen, and total DNA was extracted with Genomic-tip 20/G (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s protocol.
2.3. Long amplicon quantitative PCR
Long amplicon quantitative PCR (LA-QPCR) is a method that indirectly measures general structural DNA damage. When extremely long PCR targets (~ 10 kB) are amplified in control and experimental groups, DNA templates with lesions and/or structural alterations hinder DNA polymerase activity. Quantification of amplification product allows one to calculate lesion frequency in the target genome. With different primers, one can simultaneously measure both mtDNA and nDNA in the same sample.
LA-QPCR was performed as described previously (Santos et al., 2006; Jung et al., 2009). Briefly, 10 ng DNA was amplified with rTth polymerase (Applied Biosystems, Foster City, CA, USA) using primers for long mitochondrial and nuclear targets as described by Jung et al. (2009). Small mtDNA and nDNA targets were amplified simultaneously for normalization. PicoGreen dye (Invitrogen, Carlsbad, CA, USA) was used to quantify the template and PCR product, and relative amplifications were converted to relative lesion frequency per 10 kB DNA.
2.4. 8-oxo-dG analysis
For DNA adduct analysis, additional adult male killifish were treated with either BaP or corn oil (vehicle control) as described above. At 72 hours post-treatment, fish livers were harvested, flash frozen in liquid nitrogen, and kept at −80°C until further analysis. For liver samples weighing less than 100 mg, livers from two individuals were pooled to ensure that enough DNA could be isolated. For isolation of DNA, 2,2,6,6-tetramethyl-piperidinoxyl (TEMPO, 20 mM final concentration, Sigma-Aldrich) was added to all solutions. Frozen tissues were thawed at 4°C and homogenized in PBS with a Polytron® (Kinematica, Lucerne, Switzerland). After centrifugation at 1700×g for 10 min, the nuclear pellets were incubated in lysis buffer (Qiagen. Valencia, CA, USA) overnight at 4°C with proteinase K (Applied Biosystems, Foster City, CA, USA). Protein was precipitated with protein precipitation solution and centrifugation at 2000×g for 10 min. The DNA and RNA in the supernatant were precipitated by mixing with isopropanol for subsequent centrifugation at 2000×g for 5 min at 4°C. The DNA/RNA pellet was washed with 70% ice cold ethanol and centrifuged at 2000×g for 3 min at 4°C. After air drying, it was resuspended in cell lysis solution and incubated with Ribonuclease A (Qiagen) for 30 min at 37°C. The enzyme was precipitated with protein precipitation solution (Qiagen) followed by DNA precipitation by propanol and washing with 70% ethanol as described above. The washed and dried DNA was resuspended in sterilized double distilled water. DNA concentration was measured using a NanoDrop (Thermo Scientific, Wilmington, DE, USA) and DNA was stored at −80°C until assayed.
For enzymatic hydrolysis, 50 µg DNA was mixed with 50 µl of 80 mM Tris-HCl, 20 mM MgCl2 buffer (pH 7) and appropriate standards ([15N5]8-Oxo-dG internal standard (Cambridge Isotope Laboratories, Andover, MA, USA) for all samples and 8-Oxo-dG analyte standard (Sigma-Aldrich) for positive controls). The hydrolysis was started by addition of 32 U DNAse I (Sigma-Aldrich) and incubation at 37°C for 15 min. 3.4 mU phosphodiesterase I (Sigma-Aldrich) and 2.5 U alkaline phosphatase (Sigma-Aldrich) were then added and the samples were incubated again at 37°C for 60 min. The final volume of each sample containing all reagents and internal standard was 300 µl. The samples went through an enzyme removal process by centrifugal filtration using pre-washed Centricon YM-10 microcentrifuge filters (Millipore, Bedford, MA, USA).
Samples of hydrolyzed DNA were chromatographed on an Agilent 1200 HPLC system with an automated fraction collector to separate 8-oxo-dG from matrix. Separation was performed on an Ultrasphere ODS C18 4.6 × 250 mm 5 µm column (Beckman, Fullerton, CA) using a gradient of 10 mM ammonium formate in water (adjusted to pH 4.3 with formic acid) and methanol. Methanol composition was held at 7% from 0 to 22 min, then increased linearly to 80% in 1 min, was held at 80% for 6 min to elute TEMPO, decreased to 7% in 1 min, and held at 7% for 6 min for column re-equilibration. A 275-µL aliquot of sample was injected, and the flow rate was 1 mL per min. The column oven, autosampler tray, and fraction collector chamber temperatures were maintained at 30°C, 4°C and 4°C, respectively. The retention time of 8-oxo-dG, which was determined by using 2’-deoxyguanosine (dG) as a retention time marker and multiplying its retention time by 1.5, was ~18 min. Fractions containing 8-oxo-dG were automatically collected from 1.5 min before until 1.5 min after its predicted retention time. The fraction collection tubes were placed in a SpeedVac concentrator (ThermoFinnigan, San Jose, CA) and evaporated to dryness. Sample residue was transferred to autosampler vials via 2×130 µL washings with 50:50::water:methanol, evaporated to dryness in a SpeedVac concentrator, and finally redissolved in 20 µL HPLC grade water for subsequent analysis by LC-MS/MS. The dG amount in each sample was determined during fraction collection by comparison with dG calibration standards using UV detection at 264 nm.
The quantitative analysis of 8-oxo-dG was performed with an Acquity UPLC (Waters, Milford, MA) coupled to a TSQ-Quantum Ultra triple-quadrupole mass analyzer (ThermoFinnigan) using heat assisted electrospray ionization (HESI) in positive mode. Separation was performed on a 2.1 × 100 mm HSS T3 C18, 1.8 µm column (Waters, Milford MA) with gradient elution at a flow rate of 200 µL per min using 0.1% acetic acid in water and methanol. Methanol composition started at 1% and increased linearly to 5% B in 2 min, was held at 5% for 8 min, increased linearly to 80% in 2 min, held at 80% for 2 min, decreased to 1% in 1 min, then held at 1% for 4 min for column re-equilibration. The retention time of 8-oxo-dG was 9 min, and the total run time was 20 min. The analyte and internal standard were detected in selected reaction monitoring mode (SRM), monitoring the transitions of m/z 284.1 to 168.05 and m/z 289.1 to 173.05 for 8-oxo-dG and [15N5]8-Oxo-dG, respectively. The electrospray conditions were as follows: positive mode, spray voltage of 3000 V, vaporizer (HESI) temperature of 250°C, sheath gas flow rate 35 (arbitrary units), auxiliary gas flow rate 30 (arbitrary units), capillary temperature of 285°C, and collision energy of 12 eV.
2.5. Statistics
Statistical analyses were performed with SPSS, version 15 for Windows (SPSS Inc., Chicago, IL, USA). Normality was checked for all data sets using the Shapiro-Wilk test. Student’s t-test and analysis of variance (ANOVA) with Fisher’s Protected Least-Significant Differences (LSD) were used where appropriate (α = 0.05).
3. Results
3.1. Basal DNA damage in King’s Creek and Elizabeth River population
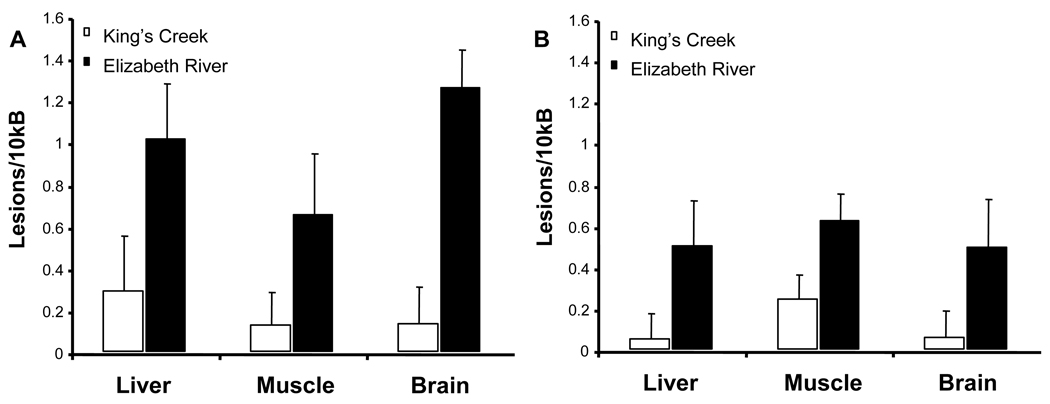
There was a significant overall difference in the basal lesion frequency levels in the mtDNA (Figure 1A) and nDNA (Figure 1B) of liver, brain, and muscle between King’s Creek and Elizabeth River fish (p = 0.003). Three-way ANOVA was performed to test the effect of population, genome type, and tissue type. Significant differences were seen between the populations (p < 0.001). Highest lesion frequencies were seen in the liver and brain mtDNA of Elizabeth River population. Within the Elizabeth River population, higher lesion frequencies were observed in the mitochondria than in the nucleus, but the difference was not significant (p = 0.071). Additionally, there was no difference among tissue types (p = 0.860). Among the tissues, the liver and brain mitochondria basal lesion frequencies were significantly different between the populations (p = 0.009 and p = 0.005 respectively) according to Student’s t-test.
Figure 1. Basal DNA damage in adult killifish from King’s Creek and Elizabeth River Superfund Site.
Comparison of the basal level of DNA damage in both the mitochondria (A) and the nucleus (B) between the two populations show that Atlantic Wood fish have higher levels of DNA damage than fish from King’s Creek (p < 0.001). Three-way ANOVA on population, genome type, and tissue type showed that only population was a significant factor (p < 0.001). Error bars represent standard error of means.
3.2 DNA damage in response to BaP in adults
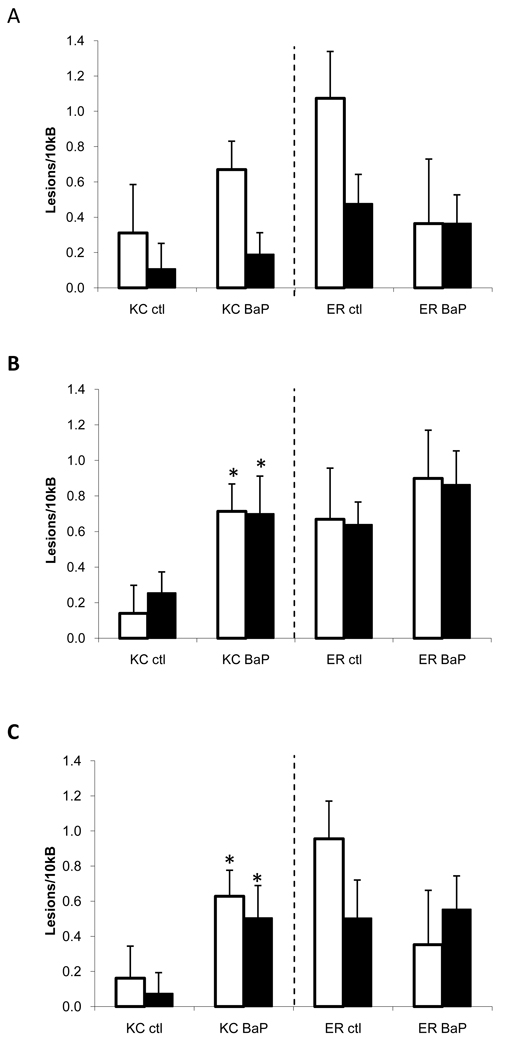
When treated with BaP, there was also an overall significance among treatment groups (p = 0.014) (Figure 2). Population was a significant factor influencing the differences (p = 0.002), and the two populations had significantly different responses to BaP treatment (p = 0.002). BaP-treated King’s Creek fish showed significant increases in DNA damage in both genome types in muscle and brain compared to the control fish, but there were no differences in DNA damage in Elizabeth River killifish in any of the tissues. Specifically, King’s Creek fish demonstrated two to seven fold increases in DNA damage when treated with BaP. However, Elizabeth River fish showed less than a two-fold increase in damage in the BaP treated group compared to the control group.
Figure 2. DNA damage in response to BaP treatment in adults from King’s Creek and Atlantic Wood Superfund Site.
DNA damage in response to i.p. injection of corn oil or 10 mg/kg BaP treatment were measured in liver (A), muscle (B), and brain (C) of adult killifish. BaP-treated group in the King’s Creek population exhibited higher levels of DNA damage compared to the control group (p < 0.001). No differences were seen in the Elizabeth River population. Open bars indicate mtDNA, and closed bars indicate nDNA. * indicates significant difference (p < 0.05) compared to the control group. Error bars represent standard error of means.
3.3. DNA damage in response to BaP in larvae
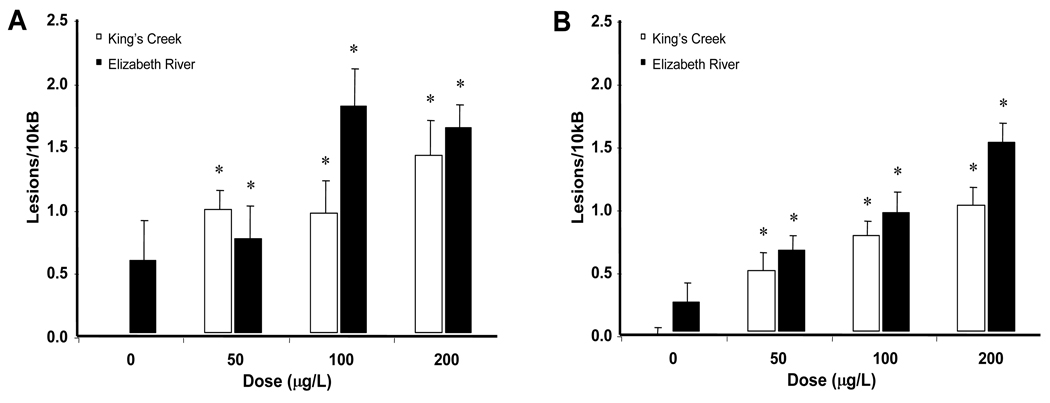
When larvae from the two sites were compared, the lesion frequencies of both mtDNA (Figure 3A) and nDNA (Figure 3B) were significantly increased by BaP treatment (p < 0.001). This increase was dose-dependent (p = 0.001). There was significantly higher damage in the mtDNA compared to the nDNA (p = 0.002), and population also had a significant effect on the differences (p < 0.001). However, there was no significant interaction between any of the independent variables. Post-hoc analysis showed that all treatment groups except the Elizabeth River control group were significantly different from the King’s Creek control group (p ≤ 0.05).
Figure 3. DNA damage levels in larval killifish from King’s Creek and Atlantic Wood Superfund Site.
There is an overall significant difference in response BaP treatment (p < 0.001). BaP treatment induced higher rates of DNA damage in the mitochondria (A) compared to the nucleus (B) (p = 0.002). Fish from the Superfund site exhibited higher basal levels of DNA damage and were less affected by BaP treatment compared to reference site fish (p < 0.001). * indicates significant difference (p < 0.05) compared to the King’s Creek control group according to Fisher’s LSD. Error bars represent standard error of means.
3.4. 8-oxo-dG analysis
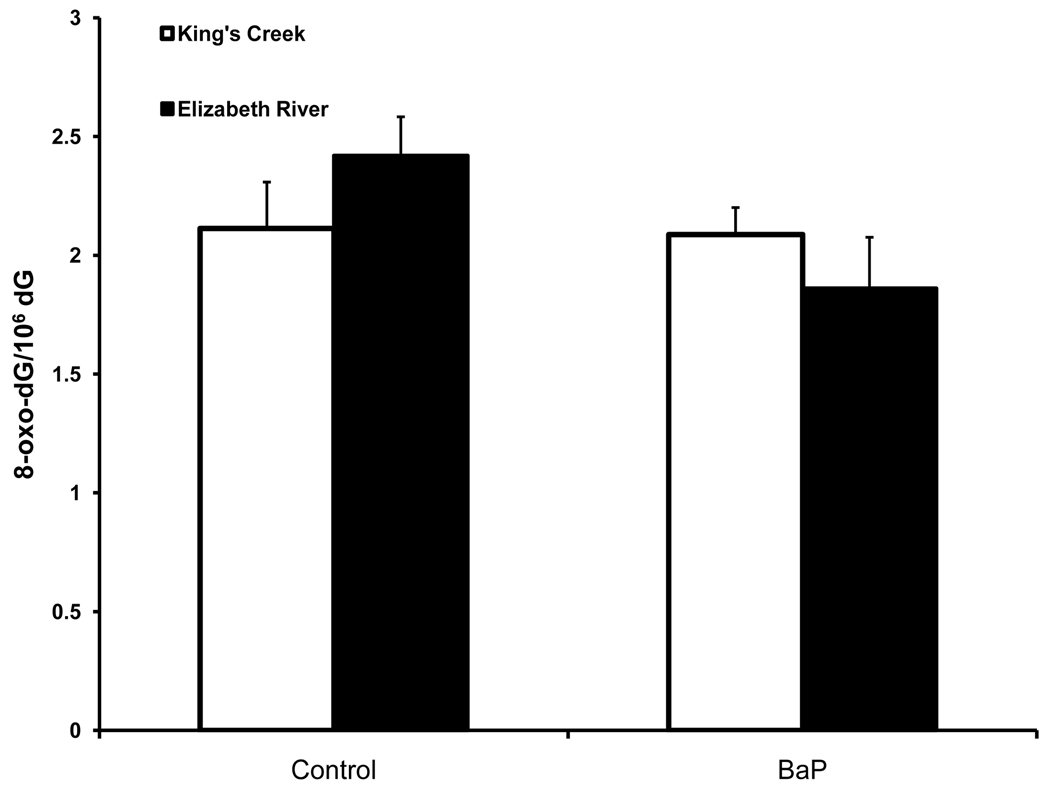
To further investigate the nature of the DNA damage detected with the LA-QPCR method, we performed LC-MS/MS analysis on adult male killifish dosed with BaP to investigate the presence of 8-oxo-dG, a biomarker of oxidative DNA damage. There were no significant differences in 8-oxo-dG frequency among the groups (p = 0.304) (Figure 4). Neither treatment nor population had significant effects (p = 0.834 and p = 0.173 respectively). This result indicates that oxidative DNA damage may not be the major form of DNA damage.
Figure 4. 8-oxo-dG adduct levels in adult killifish liver treated with BaP.
8-oxo-dG adduct levels did not differ between treated groups (p = 0.304). Error bars represent standard error of means.
4. Discussion
We examined basal levels of DNA damage, as well as the effect of BaP treatment on nDNA and mtDNA damage in killifish from the Elizabeth River Superfund site and a reference site. The Elizabeth River population had higher levels of DNA damage compared to the reference population both in the adults brought to the laboratory and in the F1 generation reared in the laboratory. These differences were seen in both mtDNA and nDNA. Our previous results on field-caught samples using the same DNA damage detection method showed similar results as the current study in that significantly higher levels of mtDNA and nDNA damage were seen in the liver of the Elizabeth River fish compared to the reference fish (Jung et al., 2009). Using 32P-post labeling analysis, Rose et al (2000) found DNA adducts in spleen, liver, and anterior kidney in adult killifish collected from the Elizabeth River.
An interesting distinction between the previous studies and this study is that, previous studies assessed the damage levels right after field capture, whereas in this study, fish from both populations were reared in the laboratory for at least four weeks for depuration purposes. Therefore, the fish from the Elizabeth River appear to retain substantial levels of DNA damage well after removal from contaminant exposure. Generally, bulky adducts formed by covalent binding of PAH-metabolites are repaired by NER. Studies using mammalian cell cultures show that even though BaP-adducts can be readily repaired, different types of PAH-adducts as well as different co-exposures with other chemicals can affect repair efficiencies (Maier et al., 2002; Lagerqvist et al., 2008). Elizabeth River fish are exposed to a complex mixture of PAHs that can form various bulky adducts. Future studies will explore how such exposure history can affect the efficiency of DNA repair in these populations.
F1 generation Elizabeth River killifish reared in the laboratory still had higher levels of DNA damage in the control group when compared to the King’s Creek fish. Wills et al (2009) reported that F1 generation fish from the Elizabeth River population exhibited slower metabolism of PAHs and that these fish seem to retain the parent compound much longer than F1 fish from the reference site. Given these data, it is plausible to infer that what we see in this study in terms of DNA damage is due to the fact that the Elizabeth River killifish are still retaining PAHs their body, and there is a possibility that there was maternal transfer of the chemicals into the eggs in the Elizabeth River fish.
In response to BaP treatment, King’s Creek fish showed expected increases in DNA damage in all tissues regardless of the genome type. There was a 2-fold to 7-fold increase in the level of DNA damage depending on the tissue type and the source of DNA. It is notable that the muscle and brain showed higher relative BaP-induced DNA damage than the liver. In contrast to the reference site population, there were no significant differences in DNA damage estimates for BaP-treated and control Elizabeth River fish with BaP-treated group showing less than 1.5-increase compared to the control group. It should be noted that, although the relative damage due to BaP-treatment is different, the actual levels of DNA damage in the Elizabeth River population, whether BaP-treated or not, are comparable to the BaP-treated King’s Creek fish. Interestingly, a similar trend was seen in the experiment with the F1 generation. F1 fish from both King’s Creek and Elizabeth River had significantly higher levels of DNA damage in response to BaP (p < 0.001). However, the magnitude of increase was different in the two populations. Elizabeth River F1 fish had between 1.1 and 5.7 fold increase in lesion frequencies in response to BaP treatment, whereas the fold increase in the King’s Creek population was from 18.2 to 76.9. This indicates that, although the F1 fish from both populations showed higher level of lesion formation with BaP treatment in larvae compared to the adults, the Superfund site larvae fish appear to exhibit some protection from further DNA damage by BaP treatment. This protection may be caused by slower metabolism of BaP into reactive metabolites due to the refractory CYP1A induction of the F1 generation seen in previous studies (Meyer and Di Giulio, 2002; Meyer et al., 2002).
A previous study with killifish adapted to a site with high concentrations of dioxin-like compounds also observed that adapted fish had reduced DNA adduct formation compared to reference site fish when treated with BaP (Nacci et al., 2002). The authors suggested that constant exposure to aryl hydrocarbon receptor (AhR) agonists would result in a refractory AhR pathway, and therefore reduced metabolism of such compounds. As a result, there would be reduced DNA adduct formation in these fish when exposed to chemicals that are metabolized via the AhR pathway. This would result in reduced DNA damage from treatment with genotoxins that are activated through the AhR pathway. However, due to constant exposure to these chemicals, there would still be higher levels of DNA damage in situ. Analysis of PAH metabolism in the F1 generation of Elizabeth River fish supports this hypothesis (Wills et al., 2009).
McFarland et al. (1999) found that brown bullhead (Ameiurus nebulosus) from sites heavily contaminated with PAHs had refractory CYP1A activity, as well as decreased DNA single strand breakage in comparison to fish at sites where chronic exposure to PAH began more recently. The authors argued that there might be increased DNA repair and cell turnover rates in the population with a longer exposure history. This may explain our data in which the Elizabeth River population exhibits no difference in DNA damage following BaP treatment, although the time frame of 72 hrs may be inadequate for this to occur. However, this does not explain the high damage level in the Elizabeth River population “depurated” in lab. Nevertheless, it would be interesting to explore whether there are inherent differences in response to DNA damaging agents and whether these differences are dependent on the AhR-pathway.
There was no difference between the two populations in the abundance of the 8-oxo-dG adduct following BaP exposure. This indicates that the majority of the relative increase in DNA damage we see in BaP-dosed fish is due to covalent binding of BaP metabolites with DNA, rather than from oxidative damage due to reactive oxidative metabolite formation. This was supported by our preliminary analysis of the BPDE-dG adduct measurements in King’s Creek fish treated with BaP (data not shown). Previous studies have shown that Elizabeth River fish have a higher antioxidant capacity than reference fish (Meyer et al., 2003a; Bacanskas et al., 2004). However, this does not seem to play a role in BaP-DNA adduct formation in these fish. Nevertheless, whether oxidative DNA damage is caused by exposure to complex mixture has not been explored. Further analysis using more sensitive techniques will allow us to determine the extent of oxidative DNA damage that occur in the Elizabeth River killifish.
One question that we wanted to explore was whether there were differences in the damage level of mtDNA and nDNA in response to BaP treatment. On the organism level, loss of mitochondrial integrity would translate to decreased energy production, and therefore decreased energy allocation on growth and reproduction. (Kooijman and Bedaux, 1996; Woodford et al., 1998). Unlike nDNA, which less than 5% is actively transcribed, the entire mtDNA is continuously involved in transcription. Therefore, damage in the mtDNA can result in deleterious functional consequences (Copeland et al., 2002; Stuart and Brown, 2006; Kroemer, 2006), as well as organismal level consequences (Brandon et al., 2006). In this study, we observed at least equal or higher levels of DNA damage in the mitochondria as in the nucleus in response to BaP treatment both in adults and in larvae. Therefore, we can speculate that the integrity of mtDNA is affected by BaP treatment and that this may have implications for normal mitochondrial function and other cellular functions in which mitochondria are involved. In addition, Elizabeth River control groups in both adults and larvae showed higher levels of mtDNA damage. This implies that the Superfund site fish population may be chronically dealing with a lower energy budget compared to reference site fish, which may affect their general well being in addition to more severe toxic effects.
It should be noted that our results are in agreement with our previous study with killifish (Jung et al., 2009), but contradict previous results from mammalian studies. Studies in mammalian models have shown that there is as much as 50 to 500 times higher BaP adduct formation in mtDNA than in nDNA (Allen and Coombs, 1980; Backer and Weinstein, 1980; Sawyer and Van Houten, 1999a). Although there were no statistically significant differences between the genome types in BaP-induced damage in adults, we did observe higher basal levels of damage in the mtDNA compared to nDNA in both adult and in F1 larvae. Mitochondria have no capacity for NER that can remove bulky adducts caused by PAHs (Shadel and Clayton, 1997; Sawyer and Van Houten, 1999b). Although it is debatable whether fish have lower NER capability compared to their mammalian counterparts (Regan et al., 1982; Weimer et al., 2000; Willett et al., 2001; David et al., 2004; Notch and Mayer, 2009), it may be that there is universally reduced NER in the fish nucleus, and that the damages that we detected in both genomes were due largely to bulky adducts. However, it remains to be seen whether mitochondria are more efficient in DNA repair against oxidative damage and have a different adduct profile from nDNA.
Of the three tissues examined, the difference in response to BaP in the two populations was most pronounced in the brain. The BaP-treat King’s Creek fish had the highest fold increase in comparison to the control group of the same population (4.9 fold increase in mtDNA and 5.7 fold increase in nDNA). Although little is known concerning BaP metabolism in brain tissue of fish, Ericson et al. (1999) showed that BaP adducts in the brain of juvenile northern pike (Esox lucius) are not much higher, but are more persistent relative to other tissues. In addition, CYP1A proteins have been identified in localized areas such as the globus pallidus in the rat brain (Kapitulnik et al., 1987; Strobel et al., 2001). This indicates that there may be higher rate of BaP metabolism in specific areas of the brain. Neurological effects of PAHs have been gaining in research attention recently (Saunders et al., 2006; McCallister et al., 2008; Gesto et al., 2009; Perera et al., 2009). High levels of DNA damage in the brain could have long-term behavioral effects, such as foraging behavior as well as flight response from predators, likely decreasing survival. Further investigation may increase our understanding of whether exposure to BaP and other chemicals may affect important behavioral aspects in aquatic animals.
In conclusion, killifish from the Elizabeth River Superfund site show high levels of basal nuclear and mitochondrial DNA damage compared to fish from the reference site, even after several weeks of depuration. However, the level of damage induced due to BaP treatment was comparable to the corn oil-treated group of the same population as well as the BaP-treated group of the killifish from a relatively unpolluted site. This same trend is seen in the F1 larvae of the Superfund site population that were hatched in the laboratory. Our results indicate that mitochondria are important targets of BaP toxicity in teleosts. Additional studies are needed to determine the consequences of this mtDNA damage in terms of mitochondrial protein production, aerobic metabolism, and organismal fitness.
Acknowledgements
We thank Yoonkyung Cha for assistance with sample processing, and Katja Strauch and Valeryi Afonin for assistance and advice in DNA-adduct analysis. This research was supported by the Superfund Basic Science Research Program (P42 ES10356 and P42 ES05948) and NIEHS Integrated Toxicology and Environmental Health Program (T32ES07031) at Duke, and the UNC Center for Environmental Health and Susceptibility (P30 ES010126).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abele D, Puntarulo S. Formation of reactive species and induction of antioxidant defence systems in polar and temperate marine invertebrates and fish. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 2004;138:405–415. doi: 10.1016/j.cbpb.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Allen JA, Coombs MM. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980;287:244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- Bacanskas LR, Whitaker J, Di Giulio RT. Oxidative stress in two populations of killifish (Fundulus heteroclitus) with differing contaminant exposure histories. Mar. Environ. Res. 2004;58:597–601. doi: 10.1016/j.marenvres.2004.03.048. [DOI] [PubMed] [Google Scholar]
- Backer JM, Weinstein IB. Mitochondrial DNA is a Major Cellular Target for a Dihydrodiol-Epoxide Derivative of Benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- Berneburg M, Kamenisch Y, Krutmann J, Röcken M. 'To repair or not to repair-no longer a question': repair of mitochondrial DNA shielding against age and cancer. Exp. Dermatol. 2006;15:1005–1015. doi: 10.1111/j.1600-0625.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: Dynamic Organelles in Disease, Aging, and Development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–569. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- David WM, Mitchell DL, Walter RB. DNA repair in hybrid fish of the genus Xiphophorus. Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 2004;138:301–309. doi: 10.1016/j.cca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol. Aspects Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- EPA. Record of Decision (ROD) Abstract - Atlantic Wood Industries, Inc. 1995 Available: http://www.epa.gov/reg3hwmd/super/sites/VAD990710410/planproposed/AWI_July_2007_Proposed_Plan_Text.pdf.
- EPA. Record of Decision (ROD) - Atlantic Wood Industries, Inc. 2007 Available: http://www.epa.gov/reg3hscd/super/sites/VAD990710410/rod/rod2007.htm.
- Ericson G, Noaksson E, Balk L. DNA adduct formation and persistence in liver and extrahepatic tissues of northern pike (Esox lucius) following oral exposure to benzo[a]pyrene, benzo[k]fluoranthene and 7H-dibenzo[c,g]carbazole. Mutat. Res.-Fund. Mol. M. 1999;427:135–145. doi: 10.1016/s0027-5107(99)00088-3. [DOI] [PubMed] [Google Scholar]
- Gesto M, Tintos A, Soengas JL, Míguez JM. [β]-Naphthoflavone and benzo(a)pyrene alter dopaminergic, noradrenergic, and serotonergic systems in brain and pituitary of rainbow trout (Oncorhynchus mykiss) Ecotoxicol. Environ. Saf. 2009;72:191–198. doi: 10.1016/j.ecoenv.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Sayer JM, Jerina DM, Copeland WC. Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 2004;32:397–405. doi: 10.1093/nar/gkh213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell SI, Hameedi J. Magnitude and extent of contaminated sediment and toxicity in Chesapeake Bay. NOAA Technical Memorandum NOS NCCOS. 2007;47(ixix):1–215. [Google Scholar]
- Huc L, Rissel M, Solhaug A, Tekpli X, Gorria M, Torriglia A, Holme JA, Dimanche-Boitrel MT, Lagadic-Gossmann D. Multiple apoptotic pathways induced by p53-dependent acidification in benzo[a]pyrene-exposed hepatic F258 cells. J. Cell Physiol. 2006;208:527–537. doi: 10.1002/jcp.20686. [DOI] [PubMed] [Google Scholar]
- Izzotti A. Gene-environment interactions in non-cancer degenerative diseases. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2009;667:1–3. doi: 10.1016/j.mrfmmm.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Jung D, Cho Y, Meyer JN, Di Giulio RT. The long amplicon quantitative PCR for DNA damage assay as a sensitive method of assessing DNA damage in the environmental model, Atlantic killifish (Fundulus heteroclitus) Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 2009;149:182–186. doi: 10.1016/j.cbpc.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitulnik J, Gelboin HV, Guengerich FP, Jacobowitz DM. Immunohistochemical localization of cytochrome P-450 in rat brain. Neuroscience. 1987;20:829–833. doi: 10.1016/0306-4522(87)90243-0. [DOI] [PubMed] [Google Scholar]
- Ko C-B, Kim S-J, Park C, Kim B-R, Shin C-H, Choi S, Chung S-Y, Noh J-H, Jeun J-H, Kim N-S, Park R. Benzo(a)pyrene-induced apoptotic death of mouse hepatoma Hepa1c1c7 cells via activation of intrinsic caspase cascade and mitochondrial dysfunction. Toxicology. 2004;199:35–46. doi: 10.1016/j.tox.2004.01.039. [DOI] [PubMed] [Google Scholar]
- Kooijman SALM, Bedaux JJM. Analysis of toxicity tests on fish growth. Water Res. 1996;30:1633–1644. [Google Scholar]
- Kroemer G. Mitochondria in cancer. Oncogene. 2006;25:4630–4632. doi: 10.1038/sj.onc.1209589. [DOI] [PubMed] [Google Scholar]
- Lagerqvist A, Håkansson D, Prochazka G, Lundin C, Dreij K, Segerbäck D, Jernström B, Törnqvist M, Seidel A, Erixon K, Jenssen D. Both replication bypass fidelity and repair efficiency influence the yield of mutations per target dose in intact mammalian cells induced by benzo[a]pyrene-diol-epoxide and dibenzo[a,l]pyrene-diol-epoxide. DNA Repair. 2008;7:1202–1212. doi: 10.1016/j.dnarep.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Latimer JS, Zheng J. The Sources, Transport, and Fate of PAHs in the Marine Environment. In: Douben PET, editor. PAHs: An Ecotoxicological Perspective. Chichester: John Wiley & Sons Ltd; 2003. pp. 9–33. [Google Scholar]
- Li N, Sioutas C, Cho A, Schmitz D, Misra C, Sempf J, Wang M, Oberley T, Froines J, Nel A. Ultrafine particulate pollutants induce oxidative stress and mitochondrial damage. Environ. Health Perspect. 2003;111:455–460. doi: 10.1289/ehp.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Schumann BL, Chang X, Talaska G, Puga A. Arsenic co-exposure potentiates benzo[a]pyrene genotoxicity. Mutat. Res.-Gen. Tox. En. 2002;517:101–111. doi: 10.1016/s1383-5718(02)00057-8. [DOI] [PubMed] [Google Scholar]
- Matson CW, Clark BW, Jenny MJ, Fleming CR, Hahn ME, Di Giulio RT. Development of the morpholino gene knockdown technique in Fundulus heteroclitus: A tool for studying molecular mechanisms in an established environmental model. Aquat. Toxicol. 2008;87:289–295. doi: 10.1016/j.aquatox.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallister MM, Maguire M, Ramesh A, Aimin Q, Liu S, Khoshbouei H, Aschner M, Ebner FF, Hood DB. Prenatal exposure to benzo(a)pyrene impairs later-life cortical neuronal function. Neurotoxicology. 2008;29:846–854. doi: 10.1016/j.neuro.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland VA, Inouye LS, Lutz CH, Jarvis AS, Clarke JU, McCant DD. Biomarkers of oxidative stress and genotoxicity in livers of field-collected brown bullhead, Ameiurus nebulosus. Arch. Environ. Contam. Toxicol. 1999;37:236–241. doi: 10.1007/s002449900510. [DOI] [PubMed] [Google Scholar]
- Meyer J, Di Giulio R. Patterns of heritability of decreased EROD activity and resistance to PCB 126-induced teratogenesis in laboratory-reared offspring of killifish (Fundulus heteroclitus) from a creosote-contaminated site in the Elizabeth River, VA, USA. Environ. Res. 2002;54:621–626. doi: 10.1016/s0141-1136(02)00170-8. [DOI] [PubMed] [Google Scholar]
- Meyer J, Di Giulio R. Heritable adaptation and fitness costs in killifish (Fundulus heteroclitus) inhabiting a polluted estuary. Ecol. Appl. 2003;13:490–503. [Google Scholar]
- Meyer JN, Nacci DE, Di Giulio RT. Cytochrome P4501A (CYP1A) in killifish (Fundulus heteroclitus): heritability of altered expression and relationship to survival in contaminated sediments. Toxicol. Sci. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Smith JD, Winston GW, Di Giulio RT. Antioxidant defenses in killifish (Fundulus heteroclitus) exposed to contaminated sediments and model prooxidants: short-term and heritable responses. Aquat. Toxicol. 2003a;65:377–395. doi: 10.1016/j.aquatox.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Volz DC, Freedman JH, Giulio RTD. Differential display of hepatic mRNA from killifish (Fundulus heteroclitus) inhabiting a Superfund estuary. Aquat. Toxicol. 2005;73:327–341. doi: 10.1016/j.aquatox.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Meyer JN, Wassenberg DM, Karchner SI, Hahn ME, Di Giulio RT. Expression and inducibility of aryl hydrocarbon receptor pathway genes in wild-caught killifish (Fundulus heteroclitus) with different contaminant-exposure histories. Environ. Toxicol. Chem. 2003b;22:2337–2343. doi: 10.1897/02-495. [DOI] [PubMed] [Google Scholar]
- Nacci DE, Kohan M, Pelletier M, George E. Effects of benzo[a]pyrene exposure on a fish population resistant to the toxic effects of dioxin-like compounds. Aquat. Toxicol. 2002;57:203–215. doi: 10.1016/s0166-445x(01)00196-5. [DOI] [PubMed] [Google Scholar]
- Niranjan BG, Avadhani NG, DiGiovanni J. Formation of benzo([a])pyrene metabolites and DNA adducts catalyzed by a rat liver mitochondrial monooxygenase system. Biochem. Bioph. Res. Co. 1985;131:935–942. doi: 10.1016/0006-291x(85)91329-4. [DOI] [PubMed] [Google Scholar]
- Notch EG, Mayer GD. Wastewater treatment effluent alters nucleotide excision repair in zebrafish (Danio rerio) Comparative Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 2009;150:307–313. doi: 10.1016/j.cbpc.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Ownby D, Newman M, Mulvey M, Vogelbein W, Unger M, Arzayus L. Fish (Fundulus heteroclitus) populations with different exposure histories differ in tolerance of creosote-contaminated sediments. Environ. Toxicol. Chem. 2002;21:1897–1902. [PubMed] [Google Scholar]
- Perera FP, Li Z, Whyatt R, Hoepner L, Wang S, Camann D, Rauh V. Prenatal Airborne Polycyclic Aromatic Hydrocarbon Exposure and Child IQ at Age 5 Years. Pediatrics. 2009;124:e195–e202. doi: 10.1542/peds.2008-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JD, Carrier WL, Samet C, Olla BL. Photoreactivation in two closely related marine fishes having different longevities. Mech. Ageing Dev. 1982;18:59–66. doi: 10.1016/0047-6374(82)90030-6. [DOI] [PubMed] [Google Scholar]
- Rose WL, French BL, Reichert WL, Faisal M. DNA adducts in hematopoietic tissues and blood of the mummichog (Fundulus heteroclitus) from a creosote-contaminated site in the Elizabeth River, Virginia. Mar. Env. Res. 2000;50:581–589. doi: 10.1016/s0141-1136(00)00252-x. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Saunders CR, Das SK, Ramesh A, Shockley DC, Mukherjee S. Benzo(a)pyrene-induced acute neurotoxicity in the F-344 rat: role of oxidative stress. J. Appl. Toxicol. 2006;26:427–438. doi: 10.1002/jat.1157. [DOI] [PubMed] [Google Scholar]
- Sawyer DE, Van Houten B. Repair of DNA damage in mitochondria. Mutat. Res.-DNA Repair. 1999;434:161–176. doi: 10.1016/s0921-8777(99)00027-0. [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Strobel HW, Thompson CM, Antonovic L. Cytochromes P450 in Brain: Function and Significance. Curr. Drug Metab. 2001;2:199–214. doi: 10.2174/1389200013338577. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Brown MF. Mitochondrial DNA maintenance and bioenergetics. BBA - Bioenergetics. 2006;1757:79–89. doi: 10.1016/j.bbabio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovasc. Res. 2004;64:279–288. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- van der Oost R, Beyer J, Vermeulen NPE. Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ. Toxicol. Phar. 2003;13:57–149. doi: 10.1016/s1382-6689(02)00126-6. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair. 2006;5:145–152. doi: 10.1016/j.dnarep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Van Metre PC, Mahler BJ. Trends in hydrophobic organic contaminants in urban and reference lake sediments across the United States, 1970–2001. Environ. Sci. Technol. 2005;39:5567–5574. doi: 10.1021/es0503175. [DOI] [PubMed] [Google Scholar]
- Van Metre PC, Mahler BJ, Furlong ET. Urban Sprawl Leaves Its PAH Signature. Environ. Sci. Technol. 2000;34:4064–4070. [Google Scholar]
- Van Veld PA, Westbrook DJ. Evidence for depression of cytochrome P4501A in a population of chemically resistant mummichog (Fundulus heteroclitus) Environ. Sci. 1995;3(4):221–234. [Google Scholar]
- Vogelbein WK, Fournie JW, Cooper PS, Van Veld PA. Hepatoblastomas in the mummichog, Fundulus heteroclitus (L.), from a creosote-contaminated environment: a histologic, ultrastructural and immunohistochemical study. J. Fish Dis. 1999;22:419–431. [Google Scholar]
- Vogelbein WK, Fournie JW, Van Veld PA, Huggett RJ. Hepatic neoplasms in the mummichog Fundulus heteroclitus from a creosote-contaminated site. Cancer. Res. 1990;50:5978–5986. [PubMed] [Google Scholar]
- Vogelbein WK, Unger M, Gauthier D. The Elizabeth River Monitoring Program 2006–2007: Association between mummichog liver histopathology and sediment chemical contamination. The Virginia Department of Environmental Quality. 2008 [Google Scholar]
- Walker SE, Dickhut RM, Chisholm-Brause C. Polycyclic Aromatic Hydrocarbons in a Highly Industrialized Urban Estuary: Inventories and Trends. Environ. Toxicol. Chem. 2004;23:2655–2664. doi: 10.1897/03-628. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wassenberg DM, Di Giulio RT. Teratogenesis in Fundulus heteroclitus embryos exposed to a creosote-contaminated sediment extract and CYP1A inhibitors. Mar. Environ. Res. 2004;58:163–168. doi: 10.1016/j.marenvres.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Weimer TL, Reddy AP, Harttig U, Alexander D, Stamm SC, Miller MR, Baird W, Hendricks J, Bailey G. Influence of beta-naphthoflavone on 7,12-dimethylbenz(a)anthracene metabolism, DNA adduction, and tumorigenicity in rainbow trout. Toxicol. Sci. 2000;57:217–228. doi: 10.1093/toxsci/57.2.217. [DOI] [PubMed] [Google Scholar]
- Wilhelm Filho D. Reactive oxygen species, antioxidants and fish mitochondria. Front. Biosci. 2007;12:1229–1237. doi: 10.2741/2141. [DOI] [PubMed] [Google Scholar]
- Willett KL, Lienesch LA, Di Giulio RT. No detectable DNA excision repair in UV-exposed hepatocytes from two catfish species. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001;128:349–358. doi: 10.1016/s1532-0456(00)00206-4. [DOI] [PubMed] [Google Scholar]
- Wills LP, Zhu S, Willett KL, Di Giulio RT. Effect of CYP1A inhibition on the biotransformation of benzo[a]pyrene in two populations of Fundulus heteroclitus with different exposure histories. Aquat. Toxicol. 2009;92:195–201. doi: 10.1016/j.aquatox.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford JE, Karasov WH, Meyer MW, Chambers L. Impact of 2,3,7,8-TCDD exposure on survival, growth, and behavior of ospreys breeding in Wisconsin, USA. Environ. Toxicol. Chem. 1998;17:1323–1331. [Google Scholar]
- Xia T, Korge P, Weiss JN, Li N, Venkatesen MI, Sioutas C, Nel A. Quinones and aromatic chemical compounds in particulate matter induce mitochondrial dysfunction: implications for ultrafine particle toxicity. Environ. Health Perspect. 2004;112:1347–1358. doi: 10.1289/ehp.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Li YB, Trush MA. Characterization of Benzo[a]pyrene Quinone-Induced Toxicity to Primary Cultured Bone Marrow Stromal Cells from DBA/2 Mice: Potential Role of Mitochondrial Dysfunction. Toxicol. Appl. Pharm. 1995;130:108–120. doi: 10.1006/taap.1995.1015. [DOI] [PubMed] [Google Scholar]