Abstract
Glial cells play important roles in the developing brain during axon fasciculation, growth cone guidance, and neuron survival. In the Drosophila brain, three main classes of glia have been identified including surface, cortex, and neuropile glia. While surface glia ensheaths the brain and is involved in the formation of the blood-brain-barrier and the control of neuroblast proliferation, the range of functions for cortex and neuropile glia is less well understood. In this study, we use the nirvana2-GAL4 driver to visualize the association of cortex and neuropile glia with axon tracts formed by different brain lineages and selectively eliminate these glial populations via induced apoptosis. The larval central brain consists of approximately 100 lineages. Each lineage forms a cohesive axon bundle, the secondary axon tract (SAT). While entering and traversing the brain neuropile, SATs interact in a characteristic way with glial cells. Some SATs are completely invested with glial processes; others show no particular association with glia, and most fall somewhere in between these extremes. Our results demonstrate that the elimination of glia results in abnormalities in SAT fasciculation and trajectory. The most prevalent phenotype is truncation or misguidance of axon tracts, or abnormal fasciculation of tracts that normally form separate pathways. Importantly, the degree of glial association with a given lineage is positively correlated with the severity of the phenotype resulting from glial ablation. Previous studies have focused on the embryonic nerve cord or adult specific compartments to establish the role of glia. Our study provides, for the first time, an analysis of glial function in the brain during axon formation and growth in larval development.
Keywords: glia, Drosophila, brain, lineage
Introduction
The function of the brain relies on populations of neurons whose axon tracts form an invariant and highly specific network of connections. To unravel the mechanisms that control the formation of this axonal network is one of the key questions in developmental neurobiology. The Drosophila brain offers a favorable model for this kind of study. It is composed of a relatively small number of neural lineages, each lineage forming a genetic and developmental unit derived from a single neuroblast. Lineages also form structural units, in the sense that axons belonging to one lineage form a cohesive fascicle that interconnects specific domains (“compartments”) of the brain. In this paper we explore the relationship between axon fascicles and glia in the larval Drosophila brain, and ask in how far glia is involved in the growth and patterning of fascicles.
After delaminating from the neuroectoderm of the early embryo (Urbach et al., 2003; Younossi-Hartenstein et al., 1996), neuroblasts divide in a stem-cell like manner to produce approximately 10–20 primary neurons each. Neurons of one lineage emit axons that fasciculate and form the primary axon tracts (PATs) (Nassif et al., 1998; Younossi-Hartenstein et al., 2006). The PATs arborize in the late embryo to generate a stereotypical set of neuropile compartments which are later ensheathed by the cortex and neuropile glia (Hartenstein et al., 1998; Pereanu et al., 2005; Younossi-Hartenstein et al., 2003). During the larval stage neuroblasts re-initiate mitosis to produce the secondary neurons of the brain (Ito and Hotta, 1992). The secondary neurons also form cohesive bundles (secondary axons tracts, SATs) along the pre-established PATs. The SATs remain unbranched until pupal stages (Nassif et al., 2003; Pereanu and Hartenstein, 2006). In the pupal brain, SATs send dendritic and axonal arbors into the existing compartments of the larval brain; in addition, they establish a number of new, adult-specific compartments (Pereanu W., personal communication).
A wealth of experimental work has shown that axonal pathway choice depends on both intrinsic cues and extrinsic signals. Intrinsic cues are proteins (e.g., membrane receptors, cytoskeleton associated proteins) expressed by a given neuron in response to transcription factors inherited from the neuronal progenitor. Extrinsic cues are diffusible or local signals presented to the growing axon by cells that it encounters. During pathway guidance, two major sources of signals have been identified: pioneer neurons and glial cells. Pioneer neurons are early differentiating neurons whose cell bodies and axons are laid out in a scaffold along which the bulk of later born neurons fasciculate (Goodman CS, 1993; Nassif et al., 1998). Glial cells, which are born at around the same stage as pioneer neurons, also form segmentally iterated landmarks, or elongated tracks, that may be involved in axonal directional guidance. This has been studied in the Drosophila and grasshopper embryonic nerve cord (Jacobs and Goodman, 1989); for example, a subset of neuropile glia called “longitudinal glia” are required for the proper formation of connectives (Hidalgo and Booth, 2000; Hidalgo et al., 1995; Jacobs, 1993; Takizawa and Hotta, 2001); midline glia are required for commissure formation controlled, in part, by Neurexin IV and Wrapper-based adhesion (Stork et al., 2009; Wheeler et al., 2009) and robo/slit interactions (Kidd et al., 1999; Klambt and Goodman, 1991). In the embryonic peripheral nervous system, surface glia are required for the guidance of peripheral nerves (Gorczyca et al., 1994; Sepp et al., 2001) and embryonic motor neurons (Takizawa and Hotta, 2001). In the visual system, maturation of the lamina glia is required for correct R axon projection patterns (Yoshida et al., 2005).
Pioneer tracts and PATs of the Drosophila embryonic brain appear prior to the formation of glial sheaths, suggesting that glia may not be significantly involved as a guidance mechanism during this phase of development (Younossi-Hartenstein et al., 2006). This is very different from the situation when secondary axon tracts extend throughout the brain during the larval phase of development. At this stage, three major classes of glial cells have differentiated and are closely associated with the emerging secondary lineages and their axon tracts. Surface glia, consisting of large, squamous cells, surrounds the brain. Specialized intercellular junctions, called septate junctions, form the substrate of the blood-brain-barrier (Stork et al., 2008). Signals derived from surface glia have been implicated in the control of neuroblast proliferation (Ebens et al., 1993). Cortex glia is formed by highly branched cells that are intermingled with neuronal cell bodies in the brain cortex (Pereanu et al., 2005). Lamelliform processes of cortex glia extend in all directions away from the cell body and form sheaths around neuronal somata (trophospongium; (Hoyle et al., 1986; Pereanu et al., 2005). Apical processes of cortex glial cells also interact with surface glia and neuroblasts; basal processes form part of the thick glial lamella (“neuropile glial sheath”) that separates cortex from neuropile. The neuropile glial sheath incorporates another type of glia, called neuropile glia. Somata of neuropile glial cells are flattened and aligned at the cortex-neuropile glial boundary. Apart from contributing to the neuropile glial sheath, fine processes protrude into the neuropile where they form a dense glial reticulum that is in contact with dendrites, axons and synapses of the neuropile (Pereanu et al., 2005).
Several aspects of the relationship between glia and neural lineages of the larval brain have been addressed in previous studies (Dumstrei et al., 2003; Younossi-Hartenstein et al., 2006; Younossi-Hartenstein et al., 2003). It was shown that cortex glia forms chambers that enclose individual neuroblasts and their newly born progeny (ganglion mother cells, undifferentiated neurons). As neurons mature in deeper layers of the cortex, they are individually wrapped by the cortex glia. Cortex glia also envelops the secondary axon tracts in the cortex. Interactions between cortex glia and neurons, in part mediated by the DE-cadherin adhesion protein, are essential for the normal layering of neurons in the cortex (Dumstrei et al., 2003). When entering the neuropile, secondary axon tracts penetrate the neuropile glial sheath; glial envelopes or tracks also accompany SATs within the neuropile to various degree. However, the interaction between lineage associated axon tracts and neuropile glia within the larval brain has not yet been studied. We speculated that this interaction may play an essential role in axon tract formation. Thus, within the neuropile, SATs of neighboring lineages frequently converge and form larger fascicles; in other cases, tracts may come close to each other and cross, without fasciculation. It stands to reason that glial processes that do or do not isolate SATs could influence SAT pathway choices.
To address this question, we first mapped the association between SATs and glia in the third instar brain using confocal and transmission electron microscopy. We next selectively eliminated cortex and neuropile glia in the central brain and analyzed SAT morphology. We find that neuropile glia either wraps SATs, borders tract systems, or surround SATs as a fine reticulum. Simultaneous elimination of both cortex and neuropile glia leads to axon guidance and fasciculation phenotypes in the larval and adult brain, suggesting that glia of the brain, like those of the ventral nerve cord, is required for growth and guidance of the SATs.
Materials and Methods
Markers and Stocks
The larval secondary neurons and axon tracts were labeled with an antibody against the Neurotactin protein (BP106; Developmental Studies Hybridoma Bank) and adult axon tracts were labeled with an antibody against the Neuroglian protein (BP104; Developmental Studies Hybridoma Bank). An antibody against the Drosophila N-Cadherin protein (DN-Ex#8, Developmental Studies Hybridoma Bank) was used to mark the neuropile in both larva and adult. Apoptotic cells were labeled with anti-cleaved Caspase-3 antibody (Cell Signaling Technology, Cat # 9661S). All glia were labeled with an antibody against the Repo protein (8D12; Developmental Studies Hybridoma Bank). Nirvana2-GAL4;UAS-GFP (Sun et al., 1999); Bloomington #6795) was used to drive protein expression in the cortex and neuropile glia. The adult DALcl1 lineage was visualized using the STAT92E10XGFP reporter line, a gift from Dr. GH Baeg (Bach et al., 2007). Apoptosis was induced using UAS-rpr (Bloomington #5823) as well as UAShid,UASrpr;;UASlacZ (Zhou et al., 1997). UASlacZ was used for controls. Conditional inhibition of UAS/GAL4 was accomplished via incorporation of the noc/Cyo;tubGAL80[ts] line (Bloomington #7018).
Immunohistochemistry and histology
Antibodies were used at the following dilution: anti-Neurotactin 1:10, anti-Neuroglian 1:40, anti-DNCadherin 1:20, anti-cleaved Caspase-3 1:1000, and anti-Repo 1:10. Secondary antibodies were Alexa 546-congugated anti-mouse Ig 1:500 (Molecular Probes, A11030); Alexa 546-conjugated anti-rat Ig 1:500 (Molecular Probes A11081); Cy3-conjugated anti-rabbit Ig 1:200 (Jackson Immuno Research, 711-165-152); Cy5-conjugated anti-mouse Ig 1:100 (Jackson Immuno Research 115-175-166); and Cy5-conjucated anti-rat Ig 1:100 (Jackon Immuno Research, 112-175-102). For antibody labeling, standard procedures were followed (Ashburner, 1989). In brief, brains were dissected in 1XPBS pH 7.4 and fixed in 4% formaldehyde 30 min at room temperature or overnight at 4 degrees Celsius. Fixed brains were washed with 1X PBS at least 30 min and either stored in methanol at −20 degrees Celsius or further processed. After at least three 15 min washes in 1XPBT (tritonX .1–.3%), brains were incubated in 10% normal goat serum (Capralogics Inc, GS0500) for at least one hour at room temperature, followed by primary antibody incubation overnight at 4 degrees Celsius. Brains were then washed at least 30 minutes in 1XPBT three times, incubated again in 10% NGS for at least one hour, and left in secondary antibody for two hours at room temperature or overnight at 4 degrees Celsius. Finally, brains were washed at least 15 minutes, four times, in 1X PBT and mounted on glass slides in vectashield (Vector Laboratories, H-1000). Brains were viewed in the confocal microscope (20X or 40X objective; MRC 1024ES microscope with Radiance 2000 using Laser sharp 2000, version 5.2 build 824 software; Bio-Rad, Hercules, CA). Confocal sections were taken at 2 micrometer intervals for all preparations. We used Imgaej1.41d for image analysis and to generate merged confocal sections.
Electron Microscopy
Larval brains were dissected in cold PBS and fixed in 4% paraformaldehyde, 2.5% glutaraldehyde in 0.1M phosphate buffer, pH 7.3, for 24 hours at 4°C. The brains were post-fixed for 60 minutes with 1% osmium tetroxyde in 0.1M phosphate buffer, pH7.3, at 4°C. Specimens were washed several times with distilled water and dehydrated in graded acetone series at 4°C. Specimens were incubated in 1:3, 2:2, 3:1 Epon:acetone ratio for 2 hours, 3 hours, and overnight, respectively. Brains were transferred to unpolymerized Epon and incubated overnight. They were then transferred to molds, oriented and placed at 60°C for 16 hours to permit polymerization of Epon. Blocks were sectioned at 0.06µm and mounted on slot grids and treated with uranyl acetate and lead citrate.
Inducing glial apoptosis
We generated either UAS-rpr; nrv2-Gal4,UAS-GFP/Cyo or UAShid,UASrpr;nrv2-GAL4,UAS-GFP/CyO;tubGal80ts larva for the experimental group and UASlacZ;nrv2GAL4,UAS-GFP/Cyo;tubGal80ts larva for the control group using the stocks listed above. For the initial experiments, embryos were laid and raised the entire time at 29 degrees Celsius on standard fly food. To initiate late-onset apoptosis in glia, tubGal80ts-containing embryos were laid at 18 degrees Celsius until wandering larval stage, at which point they were transferred to 29 degrees Celsius until dissection.
Calculating the survival curve
Flies were left to lay eggs overnight at 29°C on standard food. In duplicate or triplicate, ten first instar larva were manually picked from control and hid,rpr expressing progeny and placed on 60×15 mm standard food plates and returned to 29 degrees Celsius. Notes were recorded on growth and survival over the span of ten days. The experiment was repeating three times, and averages were taken.
Generation of three-dimensional models
We traced Neurotactin-positive tracts with the Labelfield module of Amira 3.1.1 (Mercury Computer Systems, Inc.). Labeled tracts were then three-dimensionally rendered and smoothed using the Amira SurfaceGen module.
Results
Association of glia and neural lineages at third instar
To observe the association of lineages with the surrounding glia at third instar, we expressed GFP in cortex and neuropile glia using the nirvana2 (nrv2)-GAL4 line and visualized the lineages with an antibody against Neurotactin, a transmembrane glycoprotein that is highly enriched in the nervous system (Hortsch et al., 1990). Using this marker, all secondary lineages are individually recognized (Pereanu and Hartenstein, 2006).
We first investigated the wrapping of SATs by glia in the brain cortex. The cortex of a third instar brain, from deep to superficial layers, is composed of the embryonic-born (primary) cell bodies, early larval born (secondary) cell bodies, neuroblasts, cortex glia, and surface glia. In the deep layers of the cortex, the axons of the secondary neurons within each lineage fasciculate to form the SATs. The SATs are wrapped individually by cortex glia as they project toward the cortex/neuropile boundary (Fig. 1A). Even closely associated SATs are segregated by a thin glial septum, as visualized by confocal (Fig. 1E–G) and electron microscopy (Fig. 2A).
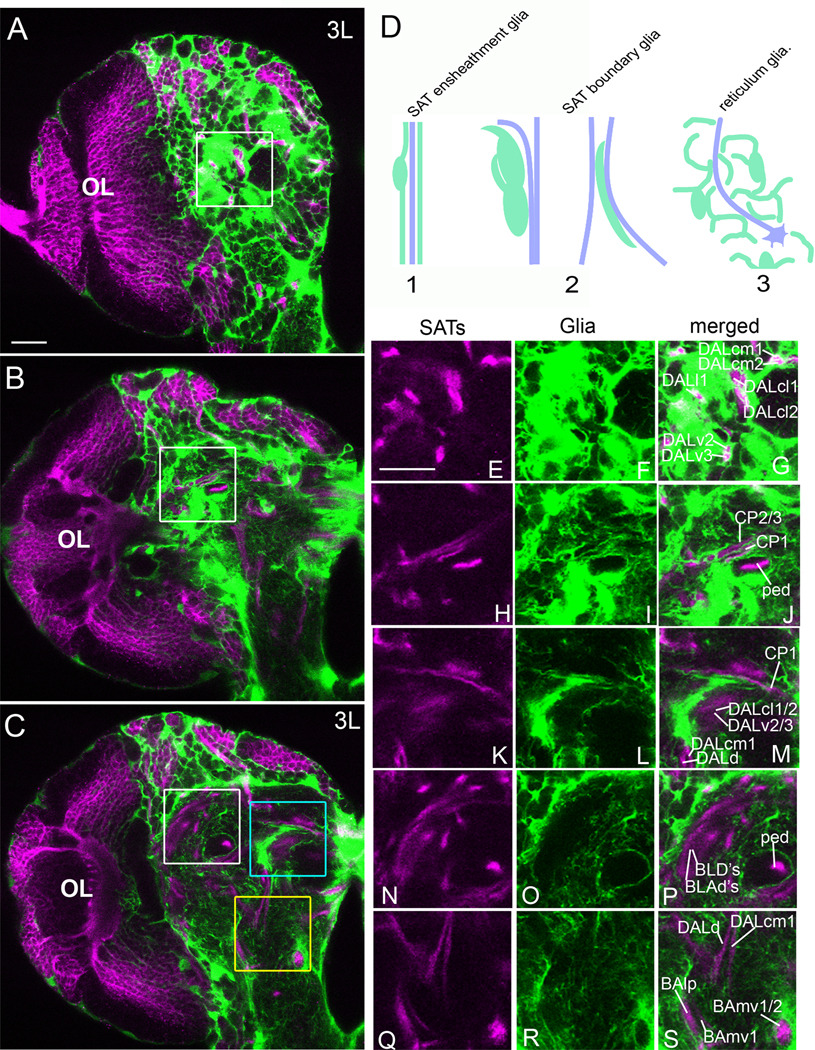
Figure 1. SAT-based pattern of cortex and neuropile glia.
(A–C) Confocal sections (2µm thickness) of a nrv2-GAL4,UAS-GFP third instar brain stained with anti-Neurotactin (magenta). (A) Tangential section of the anterior cortex (medial to the right and dorsal to the top). The area defined by the white square is magnified to show complete wrapping of the DAL SATs in the cortex (E–G). (B) Cross section at the level of the anterior peduncle. The area defined by the white square is magnified to show complete wrapping of the peduncle (ped) and separation of the CP SATs by a glial sheath (H–J). (C) Posterior section at the level of the posterior peduncle. The area defined by the blue square is enlarged to show the CP1 SAT along a thin glial process and a thick glial border on the dorsal side of the DALcl and DALv tracts (K–L). The area defined by the white square is enlarged to show the posterior peduncle (ped), and the dorsal glia border along the BLD and BLAd SATs with little glia between the tracts (N–P). The area defined by the yellow square is enlarged to show complete wrapping of the proximal BAmv1/2 SATs, a glial border along the BAlp SAT, and the terminal DALd and DALcm1 SATs in the neuropile glial reticulum (Q–S). (B) Cartoon depicting types of glia-SAT interaction (glia are drawn in green, axons are denoted by purple lines). Type 1 interactions include complete wrapping of the SAT. The type 2 cartoon depicts SAT contact by condensation (“track”) of glial processes. Type 3 interactions are the most prevalent and include no obvious glial condensation around the SAT. Scale bars: 20 µm.
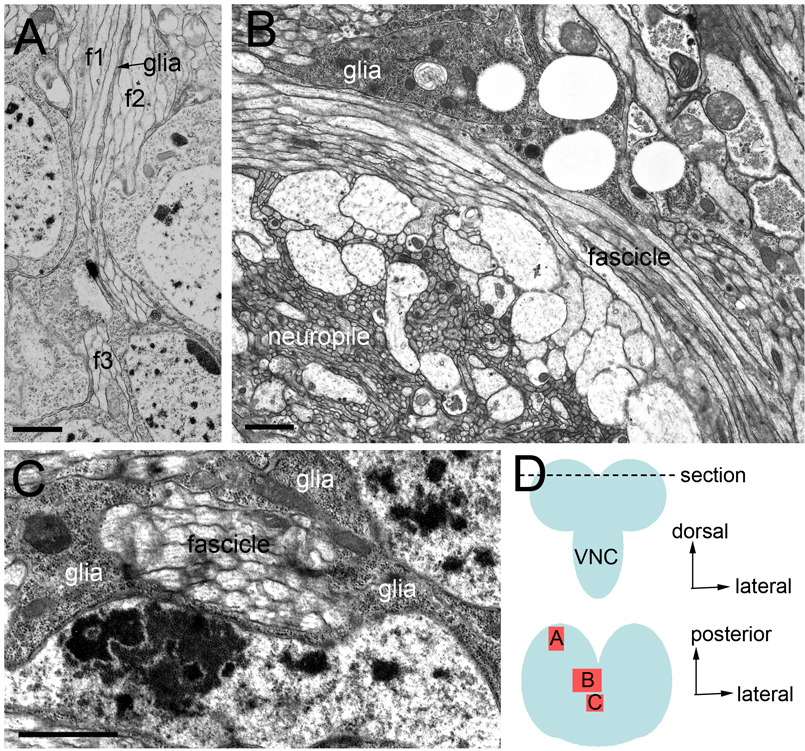
Figure 2. Electron micrographs of third instar brain illustrating the relationship between glia and SATs.
In all sections, glia is distinguished from neuronal processes by a combination of the following: an increased electron density, a lack of organized microtubule bundles, irregular diameters, or the presence of sheath like processes. (A) Section through larval cortex showing a thin glial process separating adjacent axon fascicles (f1–3). (B) Section through a midline portion of the larval brain showing an association of glia with a bend in the axon fascicle as it enters the midline. (C) Transverse section through an axon fascicle at the larval midline highlighting complete wrapping by neuropile glia. (D) cartoon depicting approximate level of EM sections (top, anterior-posterior view) and areas imaged within the section (bottom, dorsal view). VNC ventral nerve cord. Scale bars: 1µm.
We next asked if the SATs remain segregated by glia in the neuropile. We observe three scenarios of SAT/glia interaction: (1) complete wrapping; (2) partial bordering (“glial tracks”); (3) no stereotyped association (see cartoon Fig. 1D).
A minority of SATs, or collectives of SATs, is completely wrapped by glia either along a portion of the tract or in its entirety. Key examples include the peduncle (ped) and lobes of the mushroom body, formed by four adjacent lineages (Ito et al., 1997); MB1-4; Fig. 1B,C, H–J, N–P), the proximal portion of BAmv1/2 (Fig. 1C,Q–S), and all SATs that cross the midline in one of the commissural fascicles (Fig. 1C). Full encapsulation of the axon fascicle is readily found in the third instar midline region by electron microscopy in which glia processes fully wrap a transverse section of an SAT (Fig. 2C).
Another subset of SATs extends along tracks or sheets of glia, without being completely surrounded. For example, we observed adjacent SATs that initially project parallel to each other before ultimately diverging and terminating in different neuropile compartments. On their initial, parallel trajectory, such SATs are typically isolated from each other by glial sheets. More distally, the sheath partially disappears, leaving only tracks of glia along which the SATs grow. Examples illustrated in Figure 1 include the SATs formed by CP1-3 (Fig 1H–J), DALv2-3 (Fig 1K–M), DALcl1-2 (Fig 1K–M), and BLD1-4 (Fig 1N–P). Tracts that turn medially into the commissures are particularly rich in glia ensheathment. In electron microscopy sections, for example, we observe fascicle bending adjacent to glia at the midline (Fig. 2B).
Most SATs do not encounter condensations of glia processes, in the form of complete sheaths or tracks, once they reach into the neuropile. Rather, these SATs follow more or less straight trajectories through compartments formed by the tangled axons and dendrites of primary neurons. Neuropile glia forms a reticulum of fine processes that permeates all compartments and also contacts the SATs of this group, but does not form obvious sheaths or tracks around them. The distal segment of the DALd and DALcm1 SATs are shown as example (Fig. 1C,Q–S). Note that in many cases, the association of glia and axon fascicle changes as one moves along a given SAT. For example, the DALcm1 SAT projects posteriorly in the cortex while ensheathed by cortex glia (Fig 1G), makes a sharp turn ventrally upon passing through the neuropile glial sheath, projects ventrally along a thick track of glia (Fig 1M), and finally, along the last leg of its projection, is not in contact with any glial condensation (Fig 1S). A comprehensive map of posterior, central, and anterior confocal sections of a third instar brain co-labeled with nrv2-GAL4>UAS-GFP and anti-Neurotactin are shown in Supplementary Figure 1 with labeled secondary axon tracts (SATs).
Elimination of cortex and neuropile glia by nrv2-GAL4>UAS-hid,UAS-rpr
To understand the role of glia during SAT guidance and morphology, we selectively eliminated the cortex and neuropile glia at a stage immediately prior to SAT formation. Towards this end, we ectopically expressed Hid and Reaper with nrv2-GAL4, a technique also utilized in previous studies (Pereanu et al., 2007a). This driver line is only expressed in neuropile and cortex glia, and not in surface glia (Fig. 3A, inset). Therefore, the blood-brain-barrier formed by surface glia (Pereanu et al., 2005; Stork et al., 2008) remains intact. This is essential for larval survival; simultaneous elimination of all glial types (Bernardoni et al., 1997; Pereanu et al., 2005) or the abnormal formation of surface glia alone (Bainton et al., 2005; Schwabe et al., 2005) results in embryonic lethality. It is also worth noting that primary lineages, which form before the onset of glial differentiation, are unaffected in nrv2-GAL4>UAS-hid,UAS-rpr (UAShid, rpr) larval brains. For the remainder of this study, the term “glia-less” refers to the elimination of larval cortex and neuropile glia, while surface glia remains unaffected throughout development (Fig. 3I repo-positive, GFP-negative, cleaved Caspase-negative cells).
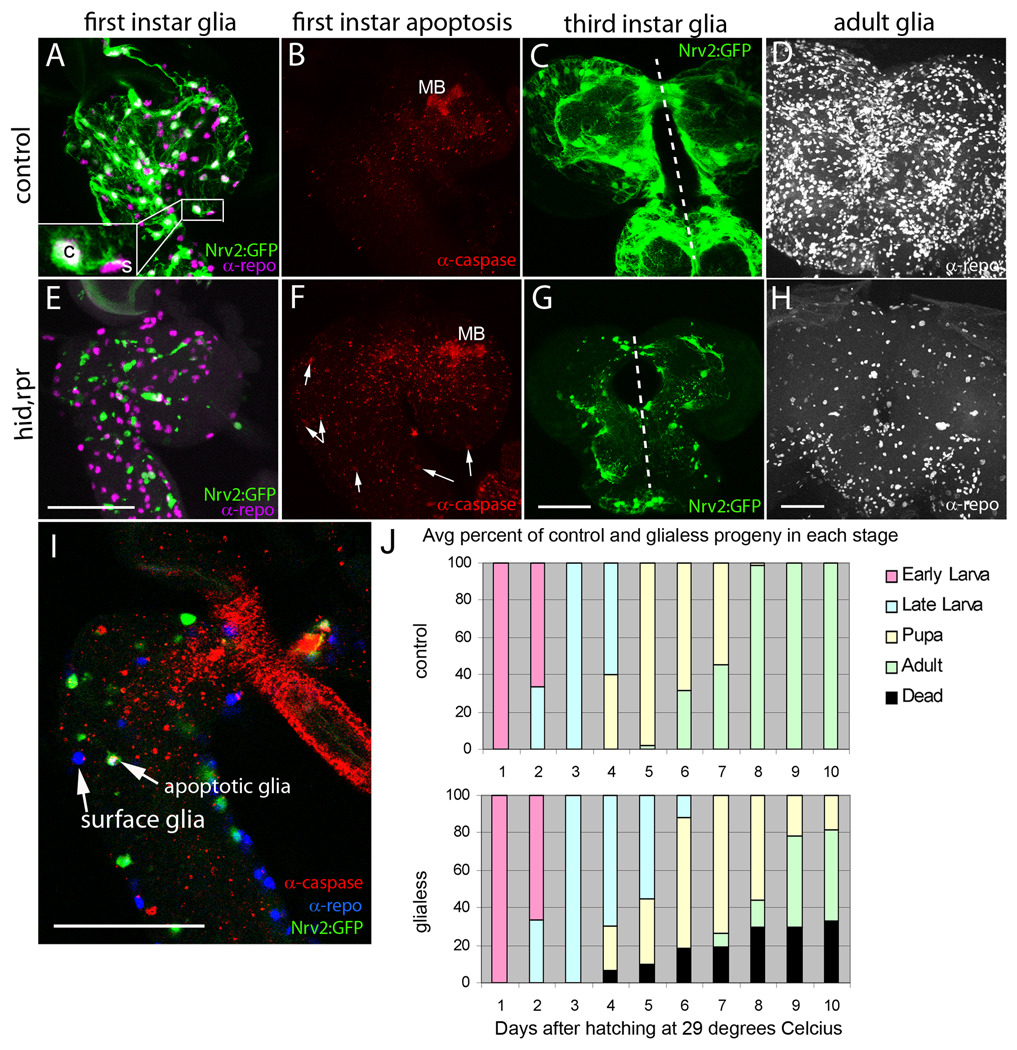
Figure 3. Inducing glia apoptosis.
(A–D) profile of normal glia development from larval to adult stages compared to (E–H) a profile of induced glia apoptosis from larval to adult stages. (A,B) Lateral view of a nrv2-GAL4,UAS-GFP first instar brain and ventral nerve cord stained with anti-Repo (A, magenta) and anticleaved Caspase-3 (B, red) to label all glial nuclei and apoptotic cells, respectively. Only neuropile and cortex glia (c) express GFP while surface glia (s) remain GFP-negative (magnified inset). A sparse amount of apoptotic cells are visualized, with the exception of large amounts of apoptosis in the mushroom body. (E,F) Lateral view of a first instar brain and ventral nerve cord with nrv2-GAL4 driven expression of Hid and Rpr as well as GFP. Glia nuclei are labeled with anti-Repo (E, magenta) and apoptotic cells are labeled with anti-cleaved Caspase-3 (F, red). Note the decrease in GFP-positive cells while the nuclei of surface glia are still present (E) and the increase in apoptosis within the brain and nerve cord (F, arrows). In A,B,E,F dorsal is to the right and posterior is down. (I) lateral view of a first instar brain labeled with nrv2-GAL4>UAS-GFP (green), anti-Repo (blue), and anti-cleaved Caspase-3 (red). Note that the glia positive for both Caspase and Repo expresses GFP while the GFP-negative surface glia is Repo-positive but Caspase-negative. (C,G) anterior-posterior section (dorsal is up, and dotted line indicates the brain midline) of a third instar brain expressing only GFP (C) or Hid, Rpr, and GFP (G) in cortex and neuropile glia. Note the disorganized appearance of any remaining GFP-positive cells. (D,H) Anterior-posterior view of adult brains stained with anti-Repo. (D) twenty-micrometer section of the central portion of a wild-type adult brain compared to (H) the equivalent central portion of a brain expressing Hid and Rpr under nrv2-GAL4. Note the decrease in the number of Repo-positive cells by eclosion in brains expressing Hid and Rpr in cortex and neuropile glia. Remaining repo-positive cells around the periphery are the unaffected surface glia. (J) Growth profile of nrv2-GAL4>UAS-hid,rpr larva showing the percentage of progeny in each stage of development over the course of 10 days at 29 degrees Celsius. Scale bars: 50 µm.
In nrv2-GAL4,UAS-GFP embryos, GFP expression becomes strong in the late embryonic stages (Sun et al., 1999), a time when glial sheaths in the cortex and neuropile have differentiated. Correspondingly, UAS-hid,rpr induced apoptosis begins around hatching (Fig. 3E,I). Glia-less first instar larvae are overtly normal; no difference can be visualized by crawling speed or behavior. The dissected brain, however, shows a decrease in GFP-positive glial processes throughout the brain (Fig. 3E) as compared to their WT counterpart (Fig. 3A). By contrast, GFP-negative, Repo-positive surface glia are unaffected (Fig. 3I). An increased amount of cleaved Caspase, an indicator of apoptosis (Kumar and Doumanis, 2000), is seen in the nrv2-GAL4>UAS-hid,rpr first instar brain and nerve cords (Fig. 3F, arrows). By second instar, the glia-less larvae begin to crawl more slowly, appear uncoordinated, and become generally less active. At this stage, we see massive reduction of cortex and neuropile glia. Glial apoptosis can be quite variable; frequently, one hemisphere of the same brain retains considerably more glia than the other (Fig. 3G). By early third instar, control larvae crawl into the food; however, glia-less larva remain atop the food, are very inactive, and are much smaller in length and girth than their WT counterparts, most likely due to decreased feeding.
In general, the glia-less third instar larvae either die or remain in the larval stage longer than the control larva, which pupate by day 4 of development at 29 degrees Celsius. The length of time a glia-less larva will take to pupate can take up to 7 days at 29 degrees Celsius. The variable degree of glial ablation likely accounts for the variability in growth and survival; ultimately an average of fifty percent of glia-less progeny eclose (Fig. 3J). The glia-less adults most likely account for the least affected larvae, retaining a sufficient level of glia to progress through development to eclosion; however the adults are uncoordinated and very short-lived, surviving no longer than a few days.
Glia association predicts a glial requirement for SAT growth and guidance
The most prominent result of glial ablation is abnormalities in the pattern of secondary axon tracts. Phenotypes can be classified as misguidance, truncation, or absence of the SAT. Under “absent” SATs we include cases where axons are truncated already in the cortex, or where misguidance of the SAT and its neighbors is so severe that we cannot determine its identity. The percentage of effected tracts among SATs analyzed range from 0% to 94% (Fig 4, lower graph).
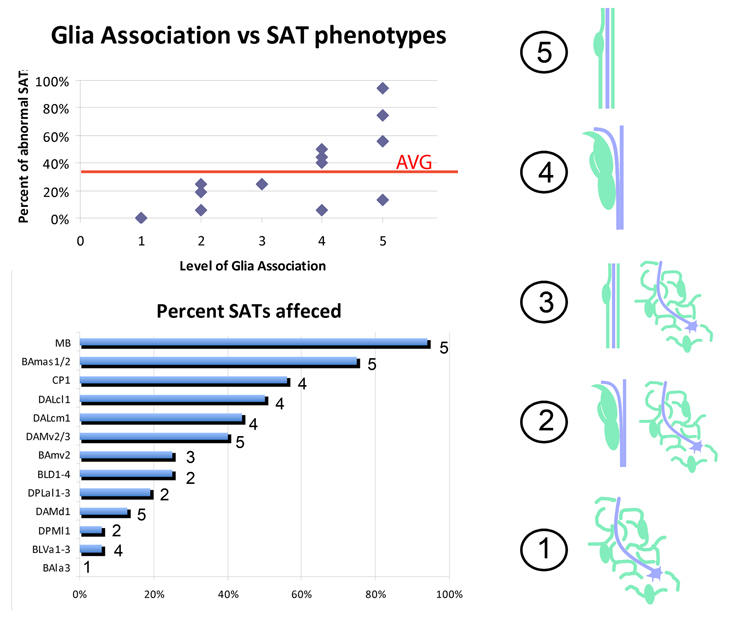
Figure 4. Glia ranking correlates with SAT phenotypes upon glia elimination.
(right) a numerical value is given to each type of SAT-glia interaction (glia are drawn in green, axons are denoted by a purple line). A ranking of 5 indicates the SAT is completely ensheathed by glia. 4 denotes glia bordering SAT tracts. 3 includes SATs that are first ensheathed but later have no apparent glial wrapping. 2 includes SATs that are first bordered but then show no stereotypical glial association. A ranking of 1 indicates that the SAT never has a stereotypical association with glia and instead entirely projects through the neuropile reticulum. (top) for all SATs analyzed, their glial ranking is plotted against the percentage of those SATs that exhibited an abnormal phenotype at third instar when cortex and neuropile glia were eliminated. The average number of SATs affected among all SATs is indicated by a red line. (bottom) graph indicating the percentage of SATs affected for each specific lineage with the given glial ranking shown to the right of each bar.
If SATs are ranked according to the amount of glia associated with them in the neuropile (see previous section), this ranking correlates positively with the percentage of phenotypes seen per SAT (Fig. 4, upper graph). In other words: SATs that normally are associated with a lot of glia are generally affected more frequently, and show more severe phenotypes, than SATs that normally do not have much glial contact. For the ranking we score glia-SAT association in the following manner: A score of 5 indicates that an SAT is completely wrapped by glia; 4: SATs that extend along glial tracks or sheets; 3: SATs that are wrapped by glia over a short segment of their trajectory; 2: SATs are contacted by glial track over a short segment of SAT trajectory; 1: no overt association with glia (Fig. 4, cartoon). Table 1 summarizes SAT morphologies, trajectories, and scores their association with neuropile glia.
Table 1. Secondary Axon Tracts Analyzed.
The following table lists the lineages selected for this study, their fascicle trajectories into the larval neuropile compartments, any stereotypical association they have with the neuropile glia, and their ranking of glial association (glia rank). Glia rankings indicate the following: 5) full glia wrapping, 4) full glia bordering, 3) partial wrapping, 2) partial bordering, 1) no obvious association with reticulum. BAla, basoanterior lineages, lateral subgroup; BAmas, basoanterior lineages, medial ascending subgroup; BAmv, basoanterior lineages, ventromedial subgroup; BLVa, ventral basolateral lineages, anterior subgroup; DPMl, medial dorsoposterior lineages, lateral subgroup; DAMd, medial dorsoanterior lineages, dorsal subgroup; DAMv, medial dorsoanterior lineages, ventral subgroup; DPLal, lateral dorsopoterior lineages, antero-lateral subgroup; BLD, dorsal basolateral lineages; DALcm, lateral dorsoanterior lineages, centro-medial subgroup; DALcl, lateral dorsoanterior lineages, centro-lateral subgroup; CP, centro-posterior lineages; MB, mushroom body lineages; BPM, basal-posterior-medial compartment; DACC1, dorsal anterior commisure 1; DAC, dorsal anterior commisure; trDL, tritocerebral dorsal lateral tract.
| Secondary Axon Tract(s) |
Trajectories | Association with Glia | Glial Score |
|---|---|---|---|
| BAla3 | straight posteriorly into BPM compartment | encounters only reticulum | 1 |
| BAmas1/2 | dorsal along median bundle into anterior and dorsal DACC1 commissure | mostly ensheathed by midline glia | 5 |
| BAmv2 | curves posteromedially, splitting into a dorsal and medial tract in the BPM compartment | breifly ensheathed, enters reticulum | 3 |
| BLVa1-3 | anteriorly and dorsally along medial optic anlage | travels along neuropile glia boundary | 4 |
| DPMl1 | anteroventrally, flanking medial caylx | briefly bordered ending in reticulum | 2 |
| DAMd1 | straight medially crossing in the DAC | always ensheathed by midline glia | 5 |
| DAMv2/3 | posteromedially corssing in the DAC | always ensheathed by midline glia | 5 |
| DPLal1-3 | curves posteroventrally toward lateral peduncle and dorsal again | breifly bordered ending in reticulum | 2 |
| BLD1-4 | dorsomedically into the trDL tract system | bordered as larger tract, no glia inside | 2 |
| DALcm1 | one medial projecting tract and one lateral projecting tract that curves ventral over the base of the peduncle | proximally ensheathed, bordered in center, terminates in reticulum | 4 |
| DALcl1 | ventrally onto the spur, splitting into a dorsal and ventral branch | proximally ensheathed, bordered in center, terminates in reticulum | 4 |
| CP1 | anteromedially, crossing the peduncle and entering the diagonal commisure | proximally ensheathed and tightly bordering distally | 5 |
| MB | anteriorlly, splitting into a dorsal and medial lobe | always ensheathed | 5 |
The SATs most strongly affected were those of the four mushroom body lineages that form the peduncle and lobes of the mushroom body. The mushroom body SATs join together at the cortex neuropile boundary (see Fig 1A). Once in the neuropile, the four SATs merge to form the peduncle; neuropile glia completely surround the peduncle, but do not project processes between the mushroom body SATs. The peduncle travels in a straight posterior to anterior direction. Near the anterior end of the neuropile, the peduncle branches into a dorsal and medial lobe. In glia-less brains, we find three distinct abnormal mushroom body morphologies: 1) mushroom body SAT splaying/misguidance, 2) mushroom body and non-mushroom body-SAT merging, and 3) premature (intracortical) mushroom body SAT fasciculation (Fig 5).
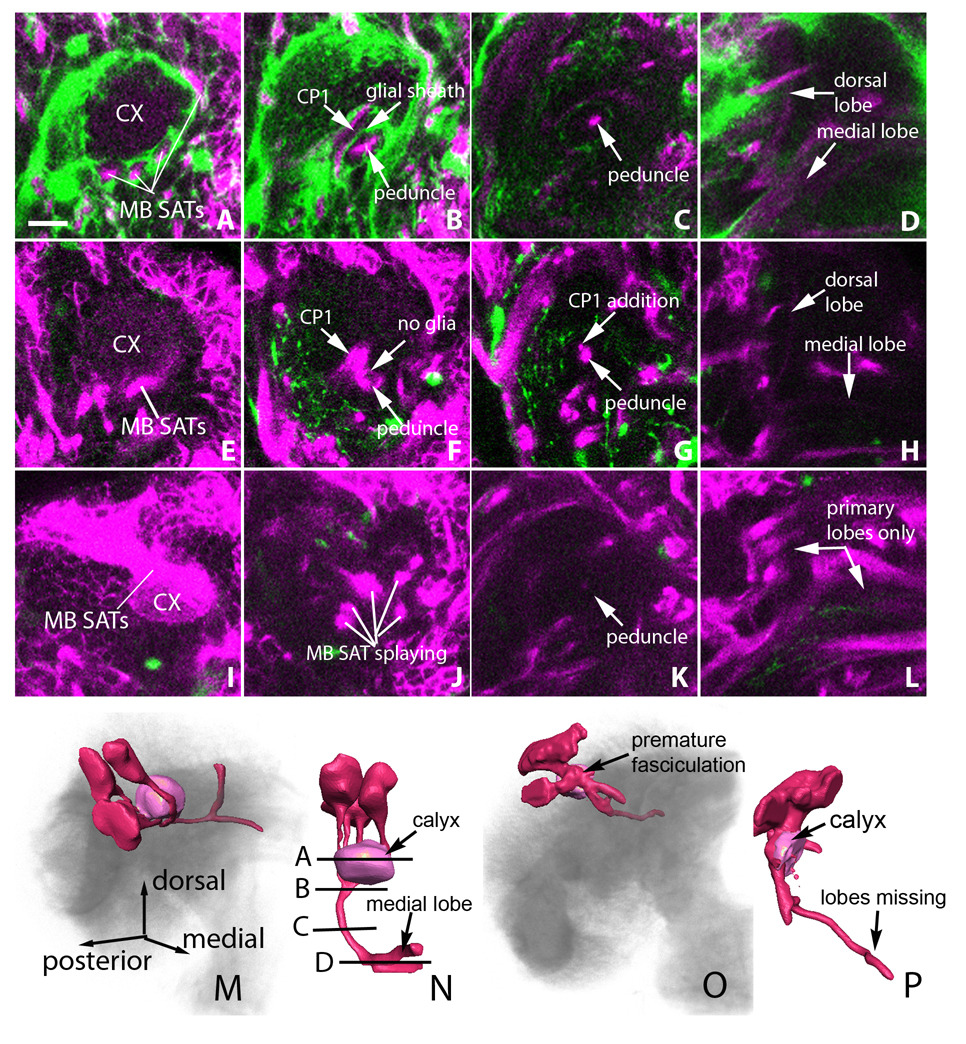
Figure 5. Glia elimination causes abnormal mushroom body fasciculation.
Posterior to anterior sections of wild-type third instar mushroom bodies (A–D) are compared to mushroom bodies that develop in nrv2-GAL4>UAS-hid,rpr conditions (E–L). (A) posterior larval section showing glia labeled with GFP (green) and anti-Neurotactin (magenta) labeled SATs. Note glial separation of the four mushroom body SAT tracts as they wrap around the calyx (CX). (E,I) mushroom bodies that develop in the absence of glia show premature SAT bundling at the level of the calyx. (B) larval section at the point where the four mushroom body tracts merge. Note the thin glial process that separates the CP SATs from the peduncle. (F,J) larval sections in glia-less brains showing peduncle-CP tract merging (F) or mushroom body tract splaying (J) phenotypes. (C) In a more anterior section, the peduncle is seen as a single tract containing all four mushroom body lineage SATs. (G) An additional tract can be seen adjacent to the peduncle stemming form the CP-peduncle merge that occurred anteriorly. (K) After mushroom body splaying, the peduncle is seen as a very thin, faint tract in more anterior sections. (D, H) In the posterior brain, the mushroom body SATs split into a dorsal and medial lobe. (L) In the brain presenting mushroom body splaying, the dorsal and medial lobes are missing. Note that the negative space indicates the presence of a dorsal and medial lobe generated from the primary axon tracts which formed before glia elimination but are not labeled by anti-Neurotactin. (M,N) Three dimensional rendering of the mushroom body from panels A–D. (O,P) Three dimensional rendering of the mushroom body from panels I–L. In M and O, dorsal is up and medial is to the right. In N and P, posterior is up and dorsal is to the right. In all panels, sections are two micrometers thick. Scale bar: 10 µm.
The variability in mushroom body phenotypes is consistent with the variability in glia elimination in general. (1) The mushroom body SAT splaying is characterized by random de-fasciculation and arbitrary trajectories away from the peduncle (Fig. 5I–L). (2) In addition to keeping the mushroom body SATs bundled and directing their straight forward trajectory, glia insulate these SATs from adjacent axon tracts. In glia-less brains we observe axons from the CP SATs, that normally cross over the peduncle in front of the calyx, de-fasciculate from their own SATs and join the peduncle (Fig. 5E–H). Mushroom body and non-mushroom body SAT fusion are never observed in control samples (Fig 5A–D), where a glial sheath segregates the mushroom body SATs from the CP SATs (Fig. 5B). (3) We frequently observed premature bundling of the mushroom body SATs in the brain cortex (Fig. 5I,O). Normally, the four lineages separately project into the neuropile, forming four tracts surrounding a central sphere of dendritic arbors, the calyx. In the glia-less brain, the lineages merge too soon in the cortex, frequently forming a misshapen, superficial, calyx (Fig. 5I,P).
To investigate the effect of glia ablation on other lineages, we selected representative SATs of the nine families of lineages and studied their pattern in 16 glia-less hemispheres of third instar brains. The lineages selected had SATs that were easily identified in wild-type brains by trajectories that stood out (i.e. they are non-linear, members of larger tract systems, or cross the midline). The lineages selected are listed in Table 1. These lineages can be grouped into four categories: 1) medial with high glial association (BAmas1/2, DAMd1, DAMv2/3, CP1); 2) medial with lower glial association (BAmv2); 3) lateral with high glial association (DALcm1, DALcl1, BLVa1-3); and finally 4) lateral with a low glial association (BAla3, DPMl1, DPLal1-3, BLD1-4). In the following we will consider the effect of glia loss on these categories of lineages.
Midline SAT growth and guidance in glia-less brains
Most SATs that cross the midline are completely wrapped by glia (score of 5) and, similar to the mushroom body SATs, show frequent abnormalities in glia-less brains. Of the medial glia-rich lineages, the CP1 SAT in particular shows a dramatic phenotype in which its medial projecting tract makes a sharp turn ventral at the midline rather than crossing to the contralateral lobe (37.5%, Fig. 6A,A” green box, Fig. S2A–C). CP1 also has a branch that extends anteriorly and ventral to the peduncle, and 18% of the ventral tracts showed misguidance/fasciculation abnormalities (data not shown). In other medial glia-rich lineages, we observe missing tracts (Fig. S2A, yellow arrows indicate DAMd tracts missing in right hemisphere), and abnormal morphology of SATs in the glia-less midline (Fig. 6A,A” green box) as compared to the control brain (Fig. 6B,B” green box). Lineages with a lower glial association along the majority of the distal SAT, such as BAmv2, show a smaller percentage of abnormal projections. In 25% of the BAmv2 tracts, its two projections fail to separate upon turning dorsal in glia-less brains (Fig. 6C,C” yellow box) compared to control brains (Fig. 6D,D” yellow box).
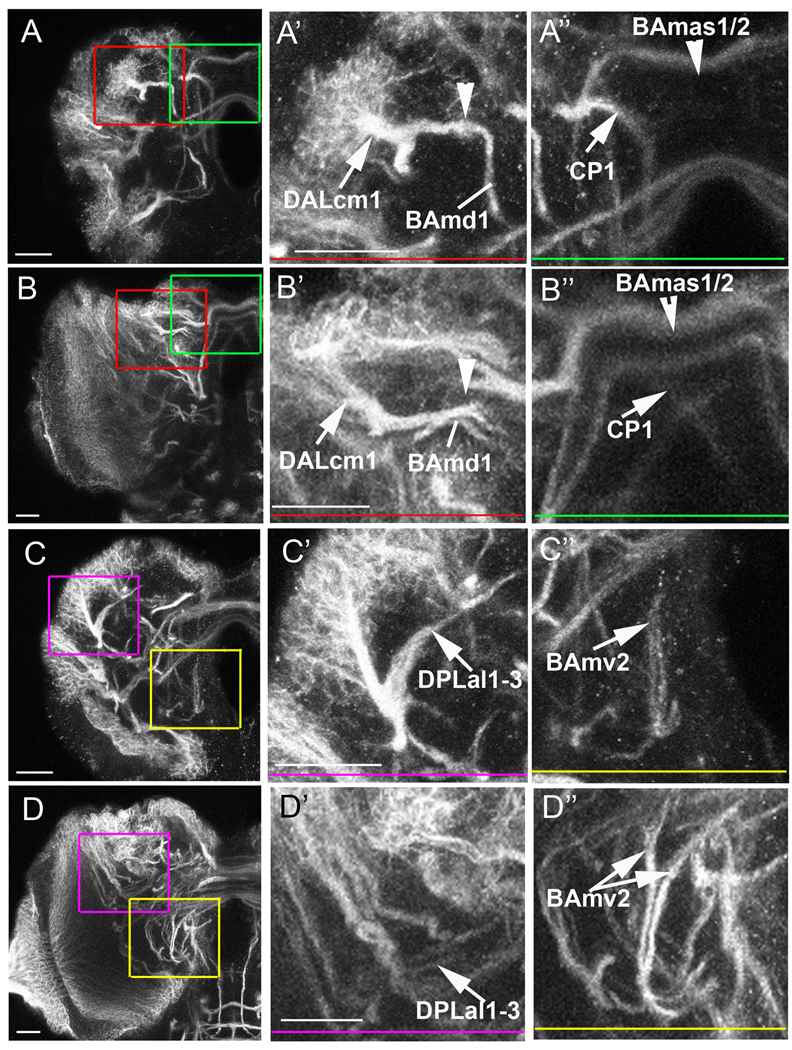
Figure 6. Truncation and misguidance of SATs in the absence of glia.
Ten-micrometer thick sections of glia-less (A,C) and wild-type (B,D) third instar brains stained with anti-Neurotactin. Enlargements of boxed areas are shown to the right with colored bars indicating area of enlargement. In each case, the wild-type projection is shown below the aberrant projections. (A,B) anterior section of a larval brain at the level of the category 5 DALcm1 (A’,B’), BAmas1/2 and CP1 (A”, B”) SATs. Note that the DALcm1 tract truncates early when it meets the dorsal projection of the BAmd1 lineage (A’, arrowhead). Normally, the DALcm1 SAT meets the BAmd1 SAT and continues to project medially (B’, arrowhead). The BAmas1/2 and CP1 SATs are compared in A” and B”. (A”) In the glia-less setting, the BAmas1/2 SAT only projects the more dorsal tract across the midline (A”, arrowhead), while the normal projection contains both a dorsal and ventral crossing (B”, arrowhead). The CP1 SAT projects in a curved trajectory down the medial portion of the ipsilateral hemisphere in the glia-less brain (A”, arrow). However, in normal conditions the CP1 tract diagonally crosses the midline into the contralateral hemisphere (B”, arrow). (C,D) Posterior section of the larval brain at the level of the DPLal1-3 lineages and the BAmv2 SAT terminal endings. (C’) The DPLal1-3 tracts project along the edge of the neuropile in glia-less brains (arrow). (D’) In control brains, the DPLal1-3 SATs enter the neuropile and make a U-shape dorso-medial turn (arrow). (C”) The terminal endings of the BAmv2 tract fail to separate upon glia elimination (arrow); whereas there is a clear BAmv2 SAT bifurcation in the control preparation (D”, double arrow). Scale bars: 20 µm.
Based on the observation that midline-projecting larval SATs exhibit a high level of abnormal projections, we next ask if midline-projecting fascicles of the adult brain also show aberrant trajectories. Axon fascicles in the adult brain are visualized with anti-Neuroglian. In most cases, SATs of several lineages merge during pupal stages. It is therefore difficult without specific markers to verify the structural abnormalities of SATs diagnosed for the glia-less larval brains. In regard to the medial lineages, adult defects can be inferred. Several of the fascicles to which SATs of the medially projecting lineages contribute are abnormal, typically missing or truncated on one side (Fig. 7B, arrows) as compared to a control adult brain (Fig. 7A).
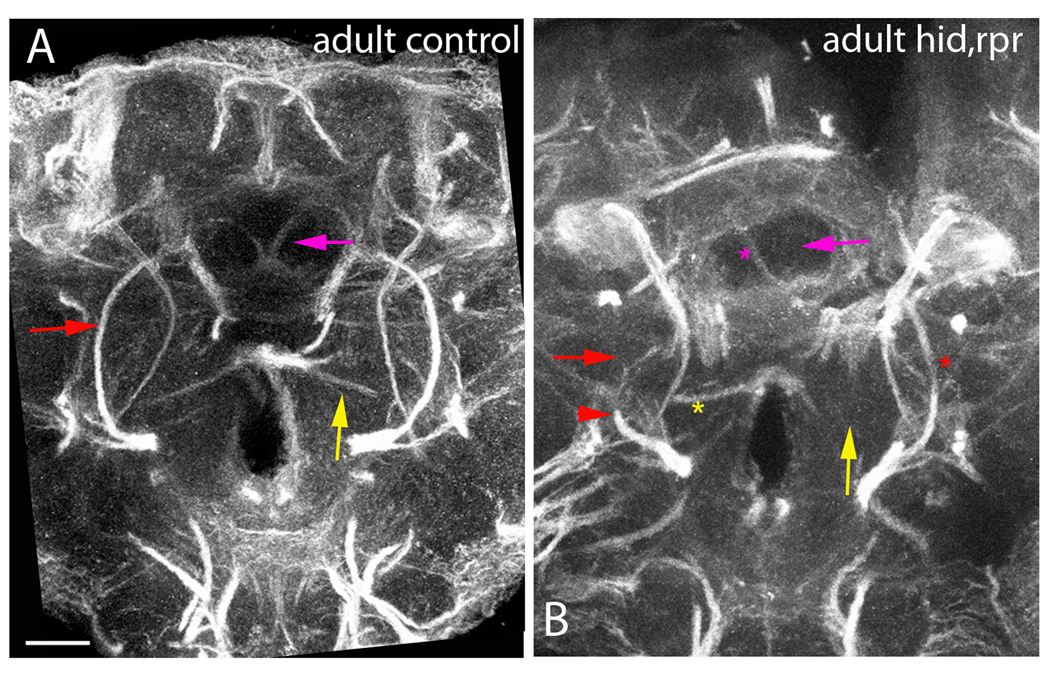
Figure 7. Aberrant Projections in Adult SATs without glia.
Central section of the adult brain stained with anti-Neuroglian at the level of the central-complex. Control adult brain (A) and nrv2-GAL4>UAS-hid,rpr adult brain (B) comparing differences in fascicle projections. In panel A, all arrows point to the stereotypical projections, while in panel B all arrows point to the missing tract. Pink arrows project to SATs that travel medially into the central portion of the ellipsoid body. Red arrows point to the axon fascicle generated by the BAmv1 lineage, and the red arrowhead indicates the truncated BAmv1 tract. Yellow arrows point to a ventrally located axon fascicle that originates in the lateral brain and projects towards the midline. In panel B, an asterisk indicates that the missing projection is still seen in the opposite hemisphere, indicating that we are in a comparable section to the control brain. Scale bar: 25 µm.
Lateral SAT growth and guidance in glia-less brains
Third instar lateral lineages with a high glial association studied include BLVa1-3, DALcm1, and DALcl1. With the exception of BLVa1-3, which is rarely affected (SAT truncation in 6%), all other lateral lineages with glia-rich SATs show a high degree of abnormalities in glia-less brains. In the DALcm1, 31% of tracts are truncated at the point where DALcm1 meets the BAmd1 SAT (Fig. 6A,A’ red box). The remaining percentage of DALcm1 phenotypes (each only observed in one hemisphere) include loss of axon fasciculation and disorganized projection of the medial and ventral DALcm1 tracts (data not shown).
To study DALcl1 SAT morphology, we utilize the STAT92EGFP reporter line (Bach et al., 2007). This reporter is expressed in a large subset of DALcl1 neurons in the adult brain. In control adult brains, the SAT of STATGFP-positive neurons projects dorsal to the mushroom body peduncle (Fig. 8A,A”). In glia-less brains, 60% of the hemispheres show the STAT92EGFP positive tract as a projection ventral to the mushroom body (Fig. 8B,B”). We next looked at the terminal arbors of the tract. The DALcl1 axonal terminals form large, conspicuous terminal boutons in the lateral bulb, a small compartment flanking the central complex (Fig. 8A’). In DALcl1 axon tracts of glia-less brains, we observe ectopic terminal boutons directly ventral to the mushroom body spur, much more laterally than in wild type (Fig. 8B’, arrowhead). However, the majority of the tract terminates in a similar area to the control brain even after extending along a different path.
Figure 8. DALcl1 SAT projects toward the Fan-Shaped Body (FSB) along an aberrant path without glia.
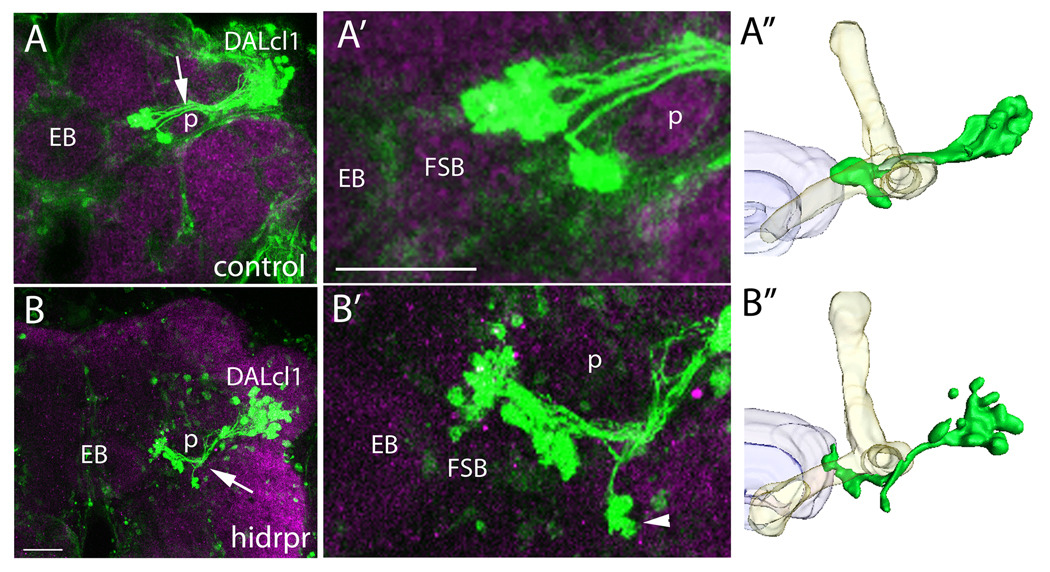
Confocal images of STAT92E10XGFP reporter expression (green) in the dorsal-lateral quadrant of adult brains stained with anti-DN-Cadherin to label the neuropile (magenta). (A,A’) STAT92E induced GFP expression is visualized in the DALcl1 lineage. The DALcl1 SAT projects dorsal to the peduncle (p, arrow) and terminates at the lateral edge of the fan-shaped body (FSB) in a cluster of small glomerular structures. (B,B’) Confocal image of a brain with nrv2-GAL4>UAS-hid,rpr showing STAT92E induced GFP expression in the DALcl1 lineage. In the glia-less setting, the tract projects ventral to the peduncle (arrow) towards the FSB, presenting ectopic terminal arbors in an abnormally lateral and ventral position (arrowhead). (A”,B”) Three-dimensional models showing the DALcl1/2 axon (green) in comparison to the mushroom body (yellow) and the central-complex (purple). Dorsal is up and medial is to the left in all images. EB ellipsoid body, FSB fan-shaped body, p peduncle. Scale bars: 25 µm.
While most of the lateral SATs highly associated with glia have frequent misguidance phenotypes, lateral SATs with a low glial score exhibit defects at a lower rate, and these defects typically consisted in truncation/absence of the SAT. For example, select lateral glial-poor SATs normally enter the neuropile at the lateral edge and turn dorsally into the CPLd compartment (Fig. S2D left hemisphere, white arrow). However, these particular SATs are severely reduced in 25% of the glia-less brains, and in one brain a lump of truncated SATs at the cortex/neuropile boundary is identified (Fig. S2D, right hemisphere, white arrow). Other lateral glial-poor SATs enter at the lateral neuropile and project postero-ventrally toward the lateral face of the peduncle. Figure 6C,C’ (pink box) shows an ectopic trajectory along the lateral boundary of the neuropile from a set of lateral lineages. This trajectory is not evident in any control preparations (Fig. 6D,D’ pink box). Of the remaining lateral glial-poor lineages studied, only one brain contained a missing SAT (Fig. S2D, yellow arrows), and some of the lateral glial-poor lineages failed to show any aberrant phenotypes.
Loss of glia affects compartment morphogenesis
Axonal and dendritic arborizations of lineage fascicles form the compartments of the larval and adult brain. First, in the late embryo and early larva, terminal arbors (both axonal and dendritic) sprouting from primary axon tracts (PATs) form the larval compartments. Most of these persist through metamorphosis into the adult brain; typically, they merely grow and, in some cases, are further subdivided into distinct sub-compartments. Thus, during metamorphosis, secondary axon tracts produce their own terminal arbors. These arbors grow into the pre-existing compartments of the larva. In addition, new, adult-specific compartments are generated. These include the central complex (fan-shaped body FSB, ellipsoid body EB, noduli), the protocerebral bridge, the lateral bulbs (input region of central complex), the antennal lobe, the antenno-mechanosensory and motor center (AMMC), and the optic tubercle.
If glia is removed during larval stages, as in the experiments reported here, one would expect abnormalities to be more prominent in compartments formed de novo from secondary lineages. This turns out to be true for some compartments but not others. For example, in glia-less adults, 73% (n=15) of cases have pronounced defects in the morphology of the EB. Typically, the EB is flattened in the dorso-ventral axis and is not closed ventrally in the absence of cortex and neuropile glia (Fig. 9D–F) as compared to a control preparation (Fig. 9A–C). The next most-affected adult-specific compartment includes the AMMC, in which 25% (n=16) are severely reduced in size if not completely missing. In contrast, the FSB, protocerebral bridge, and optic tubercle show no severe morphological defects (data not shown), although the detailed neural layering and connectivity was not tested; therefore we can not rule-out a functional change in these compartments.
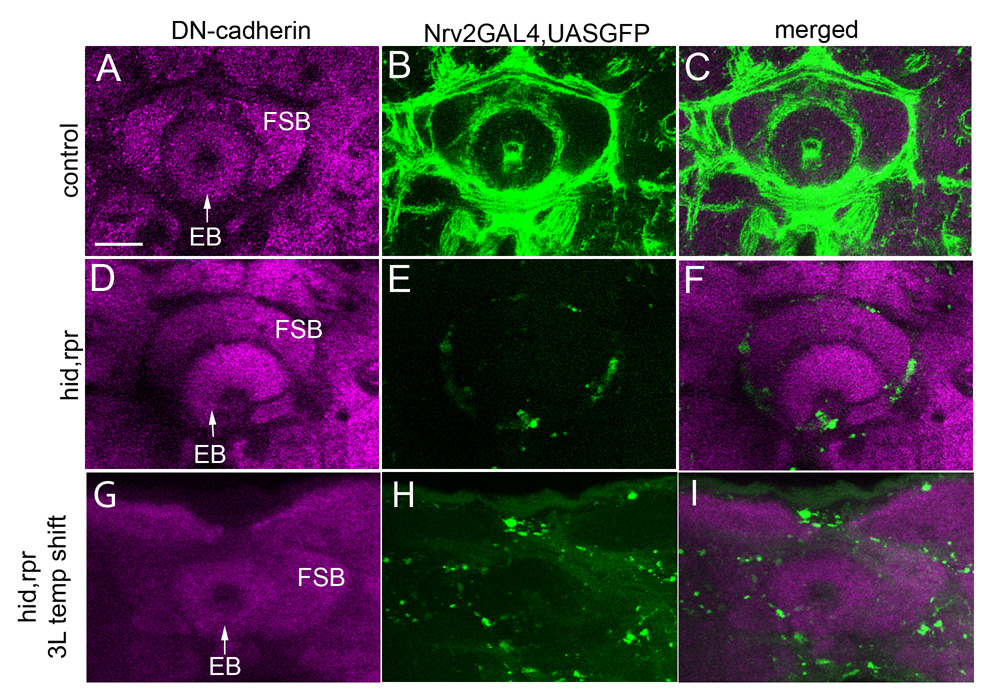
Figure 9. Adult neuropile compartment disruption with glial loss.
Confocal sections of the central-complex in adult brains stained with anti-DN-Cadherin (magenta) to label the neuropile compartments. The left column shows DN-Cadherin staining; the central column shows GFP expression (green) in cortex and neuropile glia, and the right column is the merged image. (A–C) Control nrv2-GAL4>UAS-GFP brain showing a fully closed, circular ellipsoid body (EB) and fan-shaped body (FSB) surrounded by thick glial sheathes. (D–F) Experimental nrv2-GAL4>UAShis,rpr brain exhibiting an open EB devoid of any circular structure (arrow) and a smaller, misshapen FSB. (G–I) Confocal sections from adult brains after incorporation of the tubGAL80 temperature sensitive (ts) transgene into the nrv2-GAL4>UAS-hid,rpr line followed by temperature shifting from 18 degrees Celsius to 29 degrees Celsius at wandering third instar. Note that the EB retains an ellipsoid shape and the FSB is no longer misshapen when glia are not eliminated until late third instar. Scale bar: 25 µm.
The final adult-specific compartment we analyze is the antennal lobe (AL), which is formed during pupal development in place of the degenerated larval BA compartment (Jefferis et al., 2004). In the AL, olfactory neuron terminal arbors organize into glomerular structures and synapse with the dendrites of uni- and multi-glomerular projection neurons, which then project to the mushroom body calyx and the lateral horn (Lai et al., 2008). AL glia elimination in the moth Manduca sexta results in disorganized glomerular architecture (Rossler et al., 1999); however it has been hypothesized that glia do not play a major role in Drosophila AL organization due to a late onset of glia wrapping, after glomeruli have already formed (Jefferis et al., 2004; Oland et al., 2008). Normally, the AL has a well-described architecture with readily identifiable glomeruli that have been carefully mapped and named (Laissue et al., 1999). When we eliminate glia at third instar, 43% (n=14) of the ALs are smaller in size and lack clear glomerular boundaries (Fig. S3D–F) as compared to the control brains (Fig. S3A–C). In the most severe case, the glomeruli aren’t identifiable at all (Fig. S3D–F, right AL). Therefore, cortex and neuropile glia elimination in the larva at least partially disrupts the organization of the adult AL.
We next investigated compartment morphology when cortex and neuropile glia are eliminated after SAT projection. To this end, we utilized a temperature sensitive GAL80 transgene to conditionally express Hid and Rpr only after secondary lineages are born and project axons into the neuropile. Larvae were incubated at 18 degrees Celsius, the temperature at which GAL80ts is active, until late third instar, allowing secondary lineages to form in the presence of glia. At late third instar, the larvae were shifted to 29 degrees Celsius, rendering GAL80ts inactive, to induce glial apoptosis. While late onset of glial death still results in glia elimination in the adult (Fig 9H), neuropile compartments show a normal morphology (Fig. 9G–I). This suggests that the compartment abnormalities caused by glial loss stem from events that occur during larval and pupal development, such as secondary axon tract formation and branching.
Discussion
Drosophila has been extensively used to analyze the developmental roles of glia in nervous system growth and maintenance. Most studies have focused on the embryonic nerve cord, due to its highly organized and relatively simple structure, in an attempt to understand the complex interactions between neurons and glia. Less attention has been given to the more complex larval brain, stemming in part from a lack of morphological characterization of the brain’s structure. Only recently did we develop a map that defines the position and axonal trajectory of each neural lineage within the larval brain (Pereanu and Hartenstein, 2006), providing an anatomical body of reference to which abnormal phenotypes resulting from genetic manipulations can be compared. In the present study, we have addressed the role of glia in larval brain development. Specifically, we establish the degree to which each lineage-related axon tract is associated with glia, to then ask how the pattern of SATs changes in the absence of the cortex and neuropile glia.
Glial ablation affects SAT growth and trajectory in a lineage specific manner
Secondary neurons, which are born during the larval period, form SATs that have to extend over relatively long distances, finding their way amidst a complex array of (primary) axons, dendrites, and glia. The association of glia and SATs varies for different lineages. SATs either 1) remained wrapped within the neuropile or joined other tracts that were then ensheathed as a larger tract system, 2) encountered strands of glial condensations, or 3) had no association with glial sheaths. The association between individual SATs and glia was highly invariant. Thus, if SAT A joined SAT B to form a larger tract system that became wrapped by glia, the same densities of glia could be observed in other brains for the corresponding SATs.
To address the role of glia during SAT fasciculation, growth, and guidance, we selectively eliminate cortex and neuropile glia, the two glial types in contact with growing SATs. Expression of the pro-apoptotic proteins Hid and Rpr were effective in inducing apoptosis of most cortex and neuropile glia by the early or mid larval stage. We can assume that the primary axon tract formation is not disrupted, given that expression of the nrv2-GAL4 driver line does not set in prior to stage 12 (Sun et al., 1999), PAT patterning is complete before glia invade the neuropile (Younossi-Hartenstein et al., 2006), and the first signs of apoptosis appear after hatching. In addition, the surface glia remain intact, providing general ensheathment around the brain, a functional blood-brain barrier, and potential signaling molecules used to control neuroblast proliferation.
Upon the elimination of glia, we see frequent abnormalities in the pattern of SATs. Importantly, the strength of an SAT phenotype appears to correlate with the degree of glial association of that SAT in a wild-type brain. Of all SATs analyzed, the mushroom body (associated with a complete glial sheath) exhibits the most severe defects, including complete SAT misguidance and aberrant fasciculation of neurites from adjacent SATs. In contrast, SATs that were less endowed with glia in the wild type typically had a normal projection pattern.
Our study does not differentiate between a role for glia in producing chemo-attractant signals or acting as a physical scaffold for guidance. In previous studies, ectopic expression of dominant-negative Drosophila E-cadherin in either cortex/neuropile glia or SATs themselves results in non-radial trajectories of SATs into the neuropile (Dumstrei et al., 2003). This suggests a requirement for SAT-glia adhesion as secondary axons project toward the cortex-neuropile boundary. However, the direction of neuroblast division is also disrupted with DE-cadherin knock-down, thus aberrant SAT trajectories may be a secondary effect of abnormal cell body layering within the cortex. In support of a signaling role for glia, the midline guidance defect of the CP1 SAT is reminiscent of the robo-slit phenotypes in the Drosophila ventral nerve cord (Kidd et al., 1999). Whether robo/slit signaling is also used between SATs and glia in the brain is a question that warrants further investigation.
The situation that SATs in part require glia for pathway guidance in the neuropile is different than the embryonic brain in which pioneer axons and PATs are formed in the absence of extensive glial processes. Why are SATs different? One can imagine the scenario in which the PATs generate the neuropile de novo, forcing cell body movement outwards as the central neuropile grows. By first instar, a full neuropile is established, and SATs must guide through a dense maze of neurites, glia, and trachea. It therefore appears that: 1) PATs establish the initial connectivity of the brain, 2) glia grow in around this initial scaffolding, and finally 3) SATs use both the PAT scaffolding and the glial boundaries for guidance into and around the neuropile.
Insects as model systems to study the developmental role of glia
Insects have long been used to evaluate glial-neuronal interactions from embryonic to adult stages. An important focus of these studies was whether or not glia or axon tracts appear first, and in how far axonal pathfinding is disrupted if glia is ablated. In this regard, clear developmental differences have been found between distinct regions of the nervous system, as well as between insect species for homologous nervous system domains.
In the Drosophila ventral nerve cord, two subpopulations of neuropile glia were studied in the context of axonal pathfinding: midline glia and longitudinal glia (reviewed in (Klambt et al., 1996). Both types of glial cells appear around the same stage when pioneer neurons extend their axons. The proper number and positioning of midline glia is clearly required for the formation of commissural axon tracts (Sonnenfeld and Jacobs, 1995). The loss of longitudinal glia (by ablation and in embryos mutant for the gcm gene) primarly affects the defasiculation and fasciculation events of longitudinal pioneer tracts, subsequently affecting the follower neuron trajectories (Hidalgo and Booth, 2000). Note that gcm is required for all classes of glia, including surface glia; thus, in very late gcm mutant embryos, severe disruptions of the entire neuropile result from the fact that, with the onset of embryonic movement, the CNS lacking surface glia is literally “shredded to pieces” (Pereanu et al., 2007b).
In the embryonic Drosophila peripheral nervous system, ablation of the peripheral glia (i.e. exit glia) via targeted overexpression of the cell death genes grim and ced-3 lead to aberrant pathfinding of both motor and sensory axons as they exited the CNS; although the motor neurons eventually overcame the absence of peripheral glia finding correct muscle targets, suggesting a limited role for the peripheral glia in the initial trajectory of motor neurons (Sepp et al., 2001). In grasshopper, ablation of the cell-segment boundary glial guidepost cell lead to more severely aberrant axon trajectories (Bastiani and Goodman, 1986). Ablation of glia surrounding the antennal lobe of adult Manduca generates olfactory axon defasciculation and misguidance (Rossler et al., 1999). Finally, guidance phenotypes have also been observed with disruption of the Drosophila lamina glia (Tayler and Garrity, 2003; Yoshida et al., 2005); R1–R6 photoreceptor axons show aberrant guidance past the lamina into the medulla of the optic lobe (Tayler and Garrity, 2003).
An important aspect of the developmental role of glia added by our study is the focus on the correlation between closeness of axon tract/glia association, and axon tract abnormality in the absence of glia. In other words: the nervous system is formed by a large number of fascicles, and these fascicles vary in their degree of glial wrapping. It would be misleading when carrying out a genetic study to only focus on a single fascicle (or small subset of fascicles), and extrapolate from the phenotype observed for this fascicle onto the brain as a whole. In our analysis, SATs of several lineages, notably those that in normal brains have little glia covering them in the neuropile, show few abnormalities in glia-less brains. By contrast, other lineages were affected in the majority of cases, and typically, these lineages also were the ones whose SATs were associated more closely with glia.
Role of Glia in the formation of neuropile compartments
Neuropile compartments are formed by terminal branches of axons and dendrites and their synapses. For example, the antennal lobe of insects consists of the axonal terminals of sensory neurons located in the antenna, and dendritic terminals of antennal projection neurons and local interneurons located in the deuterocerebrum, aside from a relatively small number of other modulatory neurons. Sensory neurons expressing the same olfactory receptor all converge onto the dendrites of a small number of projection neurons to form an olfactory glomerulus (Laissue and Vosshall, 2008). In many insects, notably Manduca, olfactory glomeruli are individually compartmentalized by neuropile glia, and it has been shown that glia plays a prominent role in establishing the glomeruli organization. According to the prevailing view, glomeruli are initially ordered by the specialized endings of sensory terminals into protoglomeruli; however, glial processes soon invade the space in between protoglomeruli and restrict the arborization of receptor axons (Baumann et al., 1996). Therefore, in Manduca antennal lobes, glia is required for early maintenance of the glomerular map.
A recent analysis of the time course of glial development in the Drosophila antennal lobe (Oland et al., 2008) suggested that in this species, glia plays an even lesser role, since glomeruli are only incompletely, and at a late time point, wrapped by glia (Jefferis et al., 2004; Oland et al., 2008). While glia were never ablated in those studies, later work found that elimination of Neuroglian from midline glia resulted in an inability of ORN axons to cross through the antennal commissural tract to the contralateral lobe (Chen and Hing, 2008). In our study, most adult brains lacking cortex and neuropile glia still form antennal lobes with glomeruli, however in some samples the glomeruli are poorly defined, and cannot be identified by their position and shape in the antennal lobe. The variability in phenotype penetrance likely stems from larvae containing the most glia surviving to eclosion; therefore, a relatively normal looking adult brain could stem from a weak glial apoptosis early in development. Whether the antennal lobe disorganization is a secondary defect due to a lack of definition normally provided by the presence of glia, a result of SATs that normally contribute to the AL misguiding or truncating early, or a maintenance defect in which axons begin inappropriately intermingling among glomeruli is not clear. Perhaps the Drosophila antennal lobe glia is required for glomeruli maintenance even after glomeruli organization is established, and future studies will hopefully address this possibility.
In the post-embryonic midline of Drosophila, ablation of the transient interhemispheric fibrous ring (TIFR), a transient population of midline glia, by ectopic expression of the pro-death gene hid, generates defects in the adult central complex (Hitier et al., 2000). It is unclear, however, the cellular event that is effected to cause the defects. Our study suggests that morphological abnormalities in adult compartments from glial manipulation are due to the misguidance of larval SATs to the correct neuropile compartment in the brain, affecting the formation of adult neuropile structures. Here, we present evidence that glia is an important mediator of axon guidance in the Drosophila larval brain; the mechanism for glia-neuron communication during this process is an exciting area for future investigation.
Supplementary Material
Acknowledgements
We are grateful to the Bloomington Stock Center and Dr. GH Baeg for providing us with fly stocks. This work was supported by NIH Grant RO1 NS29357-15 to V.H., the Ruth L. Kirschstein National Research Service Award GM007185 to S.S., and IMSD grant GM55052 to I.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. A Laboratory Handbook. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. Moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell. 2005;123:145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Bastiani MJ, Goodman CS. Guidance of neuronal growth cones in the grasshopper embryo. III. Recognition of specific glial pathways. J Neurosci. 1986;6:3542–3551. doi: 10.1523/JNEUROSCI.06-12-03542.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann PM, Oland LA, Tolbert LP. Glial cells stabilize axonal protoglomeruli in the developing olfactory lobe of the moth Manduca sexta. J Comp Neurol. 1996;373:118–128. doi: 10.1002/(SICI)1096-9861(19960909)373:1<118::AID-CNE10>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Bernardoni R, Vivancos V, Giangrande A. glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol. 1997;191:118–130. doi: 10.1006/dbio.1997.8702. [DOI] [PubMed] [Google Scholar]
- Chen W, Hing H. The L1-CAM, Neuroglian, functions in glial cells for Drosophila antennal lobe development. Dev Neurobiol. 2008;68:1029–1045. doi: 10.1002/dneu.20644. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation,neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci. 2003;23:3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebens AJ, Garren H, Cheyette BN, Zipursky SL. The Drosophila anachronism locus: a glycoprotein secreted by glia inhibits neuroblast proliferation. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- Goodman CS, D.C . Embryonic development of the Drosophila central nervous system. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Gorczyca MG, Phillis RW, Budnik V. The role of tinman, a mesodermal cell fate gene, in axon pathfinding during the development of the transverse nerve in Drosophila. Development. 1994;120:2143–2152. doi: 10.1242/dev.120.8.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Nassif C, Lekven A. Embryonic development of the Drosophila brain. II. Pattern of glial cells. J Comp Neurol. 1998;402:32–47. [PubMed] [Google Scholar]
- Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Urban J, Brand AH. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. Development. 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- Hitier R, Simon AF, Savarit F, Preat T. no-bridge and linotte act jointly at the interhemispheric junction to build up the adult central brain of Drosophila melanogaster. Mech Dev. 2000;99:93–100. doi: 10.1016/s0925-4773(00)00483-4. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Hoyle G, Williams M, Phillips C. Functional morphology of insect neuronal cell-surface/glial contacts: the trophospongium. J Comp Neurol. 1986;246:113–128. doi: 10.1002/cne.902460108. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Jacobs JR. Perturbed glial scaffold formation precedes axon tract malformation in Drosophila mutants. J Neurobiol. 1993;24:611–626. doi: 10.1002/neu.480240507. [DOI] [PubMed] [Google Scholar]
- Jacobs JR, Goodman CS. Embryonic development of axon pathways in the Drosophila CNS. I. A glial scaffold appears before the first growth cones. J Neurosci. 1989;9:2402–2411. doi: 10.1523/JNEUROSCI.09-07-02402.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Vyas RM, Berdnik D, Ramaekers A, Stocker RF, Tanaka NK, Ito K, Luo L. Developmental origin of wiring specificity in the olfactory system of Drosophila. Development. 2004;131:117–130. doi: 10.1242/dev.00896. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Klambt C, Goodman CS. Role of the midline glia and neurons in the formation of the axon commissures in the central nervous system of the Drosophila embryo. Ann N Y Acad Sci. 1991;633:142–159. doi: 10.1111/j.1749-6632.1991.tb15604.x. [DOI] [PubMed] [Google Scholar]
- Klambt C, Hummel T, Menne T, Sadlowski E, Scholz H, Stollewerk A. Development and function of embryonic central nervous system glial cells in Drosophila. Dev Genet. 1996;18:40–49. doi: 10.1002/(SICI)1520-6408(1996)18:1<40::AID-DVG5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Kumar S, Doumanis J. The fly caspases. Cell Death Differ. 2000;7:1039–1044. doi: 10.1038/sj.cdd.4400756. [DOI] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Laissue PP, Vosshall LB. The olfactory sensory map in Drosophila. Adv Exp Med Biol. 2008;628:102–114. doi: 10.1007/978-0-387-78261-4_7. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Embryonic development of the Drosophila brain. I. Pattern of pioneer tracts. J Comp Neurol. 1998;402:10–31. [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J Comp Neurol. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Oland LA, Biebelhausen JP, Tolbert LP. Glial investment of the adult and developing antennal lobe of Drosophila. J Comp Neurol. 2008;509:526–550. doi: 10.1002/cne.21762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Spindler S, Cruz L, Hartenstein V. Tracheal development in the Drosophila brain is constrained by glial cells. Dev Biol. 2007a;302:169–180. doi: 10.1016/j.ydbio.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Spindler S, Im E, Buu N, Hartenstein V. The emergence of patterned movement during late embryogenesis of Drosophila. Dev Neurobiol. 2007b;67:1669–1685. doi: 10.1002/dneu.20538. [DOI] [PubMed] [Google Scholar]
- Rossler W, Oland LA, Higgins MR, Hildebrand JG, Tolbert LP. Development of a glia-rich axon-sorting zone in the olfactory pathway of the moth Manduca sexta. J Neurosci. 1999;19:9865–9877. doi: 10.1523/JNEUROSCI.19-22-09865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in drosophila. Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld MJ, Jacobs JR. Apoptosis of the midline glia during Drosophila embryogenesis: a correlation with axon contact. Development. 1995;121:569–578. doi: 10.1242/dev.121.2.569. [DOI] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klambt C. Organization and function of the blood-brain barrier in Drosophila. J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Thomas S, Rodrigues F, Silies M, Naffin E, Wenderdel S, Klambt C. Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development. 2009;136:1251–1261. doi: 10.1242/dev.032847. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci U S A. 1999;96:10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa K, Hotta Y. Pathfinding analysis in a glia-less gcm mutant in Drosophila. Dev Genes Evol. 2001;211:30–36. doi: 10.1007/s004270000117. [DOI] [PubMed] [Google Scholar]
- Tayler TD, Garrity PA. Axon targeting in the Drosophila visual system. Curr Opin Neurobiol. 2003;13:90–95. doi: 10.1016/s0959-4388(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Urbach R, Schnabel R, Technau GM. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–3606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- Wheeler SR, Banerjee S, Blauth K, Rogers SL, Bhat MA, Crews ST. Neurexin IV and wrapper interactions mediate Drosophila midline glial migration and axonal ensheathment. Development. 2009;136:1147–1157. doi: 10.1242/dev.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Soustelle L, Giangrande A, Umetsu D, Murakami S, Yasugi T, Awasaki T, Ito K, Sato M, Tabata T. DPP signaling controls development of the lamina glia required for retinal axon targeting in the visual system of Drosophila. Development. 2005;132:4587–4598. doi: 10.1242/dev.02040. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nguyen B, Shy D, Hartenstein V. Embryonic origin of the Drosophila brain neuropile. J Comp Neurol. 2006;497:981–998. doi: 10.1002/cne.20884. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra PM, Hartenstein V. Early development of the Drosophila brain: IV. Larval neuropile compartments defined by glial septa. J Comp Neurol. 2003;455:435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller Hj, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc Natl Acad Sci U S A. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.