Abstract
In plant innate immunity, the surface-exposed leucine-rich repeat receptor kinases EFR and FLS2 mediate recognition of the bacterial pathogen-associated molecular patterns EF-Tu and flagellin, respectively. We identified the Arabidopsis stromal-derived factor-2 (SDF2) as being required for EFR function, and to a lesser extent FLS2 function. SDF2 resides in an endoplasmic reticulum (ER) protein complex with the Hsp40 ERdj3B and the Hsp70 BiP, which are components of the ER-quality control (ER-QC). Loss of SDF2 results in ER retention and degradation of EFR. The differential requirement for ER-QC components by EFR and FLS2 could be linked to N-glycosylation mediated by STT3a, a catalytic subunit of the oligosaccharyltransferase complex involved in co-translational N-glycosylation. Our results show that the plasma membrane EFR requires the ER complex SDF2–ERdj3B–BiP for its proper accumulation, and provide a demonstration of a physiological requirement for ER-QC in transmembrane receptor function in plants. They also provide an unexpected differential requirement for ER-QC and N-glycosylation components by two closely related receptors.
Keywords: EFR, ER-quality control, pathogen-associated molecular patterns, pattern-recognition receptor, SDF2
Introduction
Plants initially sense microbes through perception of pathogen (or microbe)-associated molecular patterns (PAMPs/MAMPs) by pattern-recognition receptors (PRRs) located on the cell surface leading to PAMP-triggered immunity (PTI) (Jones and Dangl, 2006; Zipfel, 2008). Arabidopsis thaliana detects a variety of PAMPs including fungal chitin, and bacterial flagellin and EF-Tu, or their peptide surrogates flg22 and elf18, respectively (Gomez-Gomez and Boller, 2000; Zipfel et al, 2006). Although recognition of fungal chitin and of an unknown bacterial PAMP depend on CERK1, a LysM domain receptor kinase (LysM-RK) (Miya et al, 2007; Wan et al, 2008; Gimenez-Ibanez et al, 2009), the related leucine-rich repeat receptor kinases (LRR-RKs) FLS2 and EFR are the PRRs for flagellin and EF-Tu, respectively. The LRR-RK BAK1 is rapidly recruited by FLS2 in a ligand-dependent manner to initiate downstream signalling (Chinchilla et al, 2007; Heese et al, 2007). Flagellin and EF-Tu recognition leads to MAP kinase activation, defence gene induction, production of reactive oxygen species in an oxidative burst, callose deposition, synthesis of the defence hormone salicylic acid (SA) and seedling growth inhibition (SGI) (Schwessinger and Zipfel, 2008). PAMP treatment leads to enhanced resistance to adapted pathogens (Zipfel et al, 2004, 2006; Ferrari et al, 2007), whereas defects in PAMP recognition lead to enhanced susceptibility to adapted and non-adapted pathogens (Zipfel et al, 2004, 2006; de Torres et al, 2006; Hann and Rathjen, 2007), showing a contribution of PTI to both basal and non-host resistance. Pathogenic virulence effectors evolved to directly target PRRs and their associated proteins to cause disease (Göhre et al, 2008; Shan et al, 2008; Xiang et al, 2008; Gimenez-Ibanez et al, 2009), further showing the importance of PTI for plant innate immunity.
FLS2 and EFR are transmembrane glycoproteins that need to transit through the secretory pathway to mature and reach their final destination at the plasma membrane. After translocation into the endoplasmic reticulum (ER), newly synthesised polypeptides interact with different chaperones that will assist them to fold properly and to avoid aggregation in a process called ER-quality control (ER-QC) (Anelli and Sitia, 2008). Unfolded proteins are retained in the ER until they are properly folded, or ultimately destroyed by ER-associated degradation (ERAD) in the cytosol (Vembar and Brodsky, 2008). Most of our knowledge on ER-QC is based on studies in yeast and mammals, while plant ER-QC mechanisms are still not well characterized (Vitale and Boston, 2008). Studies in mammals and yeast revealed that ER-QC relies on three main different pathways. The first, is specific to glycoproteins and depends on the folding sensor UDP-glucose:glycoprotein glucosyltransferase (UGGT) plus the lectins calnexin (CNX) and calreticulin (CRT) (Williams, 2006). The second relies on retention of misfolded proteins by the luminal-binding protein BiP, a member of the Hsp70 family of chaperones. In this system, the ER-localised co-chaperone Hsp40 protein ERdj3 first directly binds to the misfolded substrate. ERdj3 then recruits BiP and activates BiP ATPase activity present in its N-terminus, leading to interaction of the C-terminal region of BiP with the substrate and the release of ERdj3b (Jin et al, 2008, 2009). The BiP retention system acts independently of, or subsequent to, the CNX/CRT cycle (Buck et al, 2007). The third system involves the formation of disulfide bonds between free thiol groups in non-native proteins by protein disulfide isomerases and thiol oxidoreductases (Reddy et al, 1996; Anelli et al, 2003, 2007).
Despite the major contribution of PTI to plant innate immunity, our knowledge of the molecular events underlying PRR biogenesis, PAMP perception by PRRs, and downstream signalling is limited. We report here on an ER protein complex comprising stromal-derived factor-2 (SDF2), ERdj3B and BiP required for the proper biogenesis of the PRR EFR, and reveal an unexpected differential requirement of EFR and FLS2 for ER-QC and glycosylation components.
Results and discussion
Identification of sdf2 mutants in a forward-genetic screen for Arabidopsis elf18-insensitive mutants
To identify new regulators of EFR function in Arabidopsis, we used the property of elf18 to cause SGI (Zipfel et al, 2006) (Figure 1A) and screened ∼35 000 T-DNA activation-tagging transgenic lines and ∼137 500 ethyl methane sulphonate (EMS)-mutagenised M2 seeds (all in Col-0 background). We identified 160 elf18-insensitive (elfin) mutants. Sequencing the EFR locus in these mutants identified 57 efr mutants corresponding to 37 different alleles (data not shown).
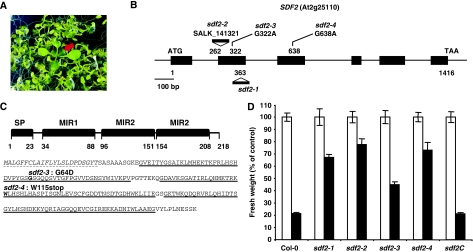
Figure 1.
Identification of sdf2 mutants. (A) Growth phenotype of elfin (elf18-insensitive) seedlings. Five-day-old Arabidopsis seedlings were covered with liquid MS medium containing 1% sucrose (MS 1%) supplemented with 50 nM elf18 peptide. Seedling growth inhibition was scored qualitatively 1 week after treatment. The red arrowhead indicates an elfin mutant. (B) Schematic representation of the SDF2 gene (At2g25110) with positions of the T-DNA insertions and point mutations. Exons are depicted as black boxes. (C) SDF2 protein organisation. Top, schematic representation of SDF2 with the predicted signal peptide (SP) and MIR domains represented as black boxes. Bottom, SDF2 protein sequence. SP, dashed underlines; MIR domains, underlined. The positions of the sdf2 EMS alleles are indicated in bold. (D) Seedling growth inhibition triggered by elf18 in wild type (Col-0) and sdf2 alleles. Five-day-old Arabidopsis seedlings were transferred to liquid MS 1% without (white bars) or with 100 nM elf18 (black bars). Seedling fresh weight was quantified 1 week after treatment. Sdf2C corresponds to sdf2-2/SDF2p::SDF2-3xHA. Results are average±s.e. (n=6). Similar results were observed in at least three independent experiments.
We isolated three allelic elfin mutants at a new locus by screening the activation-tagging transgenic population. Genetic analyses indicated that the mutation is recessive, suggesting that the phenotype is conferred by a loss-of-function mutation. All three mutants carried the same T-DNA insertion at the At2g25110 locus encoding the Arabidopsis orthologue of the murine stromal cell-derived factor-2 gene, AtSDF2, and were therefore named sdf2-1 (Figure 1B–D; Supplementary Figure 1). In addition, an independent homozygous T-DNA insertion line (SALK_141321), sdf2-2, and two further sdf2 alleles identified in the EMS elfin collection by DNA sequencing (sdf2-3 and sdf2-4) were similarly impaired in elf18-triggered SGI (Figure 1B–D; Supplementary Figure 1). No developmental or growth defects were observed in the sdf2 mutants under our growth conditions (data not shown). Finally, transgenic sdf2-2 seedlings expressing the SDF2–3xHA fusion protein under the control of the SDF2 promoter regained elf18 sensitivity (Figure 1D; Supplementary Figure 1), showing that mutation in SDF2 is responsible for the observed elfin phenotype.
AtSDF2 is a single copy gene in Arabidopsis and clear orthologs exist in all eukaryotes, except fungi (Supplementary Figure 2). Although SDF2 is highly conserved in eukaryotes, no mutant phenotype has been reported in any organism so far. AtSDF2 is a small protein of 218 amino acids (24 kDa) consisting of a 23 amino-acid (aa) predicted N-terminal signal peptide (SP) and three repeats of the MIR domain (Figure 1C) found in mannosyltransferases, the inositol-3-phosphate receptor (IP3R) and the ryanodine receptor (RyR) (Ponting, 2000). Although many eukaryotic proteins contain MIR domains, SDF2 is the only MIR domain-containing protein in plants.
Loss of SDF2 strongly affects EFR function
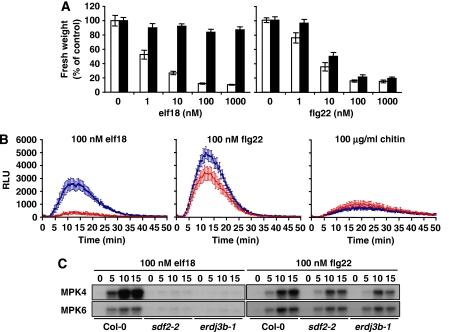
As SDF2 is required for elf18-triggered SGI, we tested whether it is involved in other PTI responses. We first tested whether sdf2-2 was also impaired in the flg22-triggered SGI. Flg22 sensitivity over a range of concentrations was slightly reduced in sdf2-2 seedlings as compared with wild type in this assay, but to a lesser extent than with elf18 (Figure 2A). Elf18 and flg22 typically induce an oxidative burst and MAP kinase activation within minutes of treatment in wild-type plants (Figure 2B and C). Although the oxidative burst induced by elf18 was strongly diminished in sdf2-2 leaves, it was less reduced after flg22 treatment (Figure 2B). In contrast, the oxidative burst triggered by the fungal PAMP chitin, which depends on the LysM-RK CERK1 (Miya et al, 2007; Wan et al, 2008), was not impaired at all in sdf2-2 mutants (Figure 2B), indicating that SDF2 is not required for chitin responses. Activation of MAP kinases 4 and 6 (MPK4 and MPK6) was almost completely abolished after elf18 treatment, whereas weakly decreased in response to flg22 in sdf2-2 seedlings, when compared with wild type (Figure 1C).
Figure 2.
Sdf2 mutant is compromised in PTI responses triggered by elf18 and, to a certain extend, flg22. (A) Seedling growth inhibition triggered by elf18 or flg22 in wild-type Col-0 (white bars) and sdf2-2 (black bars) seedlings. Five-day-old Arabidopsis seedlings were transferred to liquid MS 1% supplemented with the indicated concentrations of peptides. Seedling fresh weight was quantified 1 week after treatment. Results are average±s.e. (n=6). (B) Oxidative burst triggered by 100 nM elf18, 100 nM flg22 or 100 mg/ml chitin in wild-type Col-0 (blue) and sdf2-2 (red) leaf discs measured in relative light units (RLU). Results are average±s.e. (n=12). (C) Activation of the MAP kinases MPK4 and MPK6 in response to 100 nM elf18 or 100 nM flg22 in Col-0, sdf2-2 and erdj3b-1 seedlings (2 weeks old). Two-week-old Arabidopsis seedlings in liquid MS 1% were treated with 100 nM elf18 or flg22. Seedlings were flash frozen in liquid nitrogen at time points indicated. MPK4 and 6 were affinity purified using specific antibodies and used for in vitro kinase reactions performed with the myelin basic protein (MBP) as a substrate in the presence of [γ-32P]ATP.
Loss of SDF2 leads to enhanced disease susceptibility to bacteria and fungi
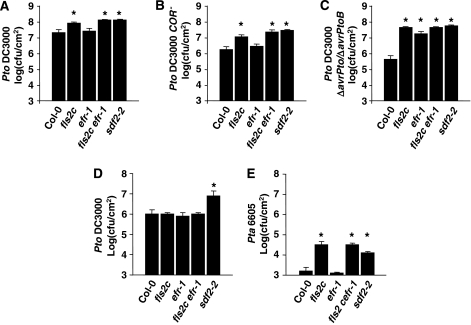
Next, we tested whether SDF2 is required for innate immunity. First, we compared the ability of elf18 and flg22 to induce resistance to the virulent bacterial strain Pseudomonas syringae pv. tomato DC3000 (Pto DC3000). Elf18- and flg22-induced resistances were both reduced in sdf2-2 leaves compared with wild type (Supplementary Figure 3). Spray-inoculated fls2c mutant plants are more susceptible to Pto DC3000 than wild type, whereas efr-1 plants are not (Figure 3A). Sdf2-2 plants were however hyper-susceptible to Pto DC3000, comparably to fls2c plants (Figure 3A). PTI defects can be subtle, but can be detected more sensitively with weakly virulent bacterial strains lacking effector molecules. The bacterial effectors AvrPto and AvrPtoB directly target FLS2, EFR and BAK1 to suppress PTI (Göhre et al, 2008; Shan et al, 2008; Xiang et al, 2008), whereas the phytotoxin coronatine (COR) suppresses PAMP-induced stomatal closure, allowing bacterial entry into the leaf apoplast (Melotto et al, 2006). When spray-inoculated, both Pto DC3000 COR− and Pto DC3000 ΔavrPto/ΔavrPtoB strains caused reduced disease symptoms and multiplied less than Pto DC3000 on wild-type plants (∼1 and 2 log units, respectively) (Figure 3B and C). However, Arabidopsis efr-1, fls2c and fls2c efr-1 mutants displayed more severe disease symptoms and allowed more bacterial growth when spray-infected with these strains (Figure 3B and C), especially in the case of Pto DC3000 ΔavrPto/ΔavrPtoB that grew almost as much as Pto DC3000 in these lines (Figure 3C). We sometimes observed that efr-1 and fls2c efr-1 were more susceptible to Pto DC3000 strains compared with wild type and fls2c, respectively (data not shown), suggesting that FLS2 and EFR can act additively in perception of this bacterium. We found that, although sdf2-2 plants were not substantially affected in their flg22 responses (Figure 2), they were hyper-susceptible to both Pto DC3000 COR− and Pto DC3000 ΔavrPto/ΔavrPtoB to a similar level as fls2c efr-1 (Figure 3B and C). Fls2 and efr mutants do not display enhanced susceptibility to Pto DC3000 when infiltrated into the leaves (Zipfel et al, 2004, 2006). Unexpectedly, we observed that sdf2-2 was however more susceptible to Pto DC3000 after this infection procedure (Figure 3D). FLS2, but not EFR, is involved in the non-host resistance to the non-adapted bacterial strain Pseudomonas syringae pv. tabaci 6605 (Li et al, 2005) (Figure 3E). Surprisingly, we found that the sdf2-2 mutation allowed growth of this strain to similar levels as on fls2c and fls2c efr-1 plants (Figure 3E). Therefore, our results clearly show that sdf2 mutants are more susceptible to bacterial infection than efr mutants.
Figure 3.
Sdf2 mutant is more susceptible to bacterial pathogens. (A–C) Pre-invasive bacterial susceptibility assay. Five-week-old Col-0, fls2c, efr-1, fls2c efr-1 and sdf2-2 plants were sprayed with Pseudomonas syringae pv. tomato (Pto) DC3000 (A), Pto DC3000 COR− (B) or Pto DC3000 ΔavrPto/ΔavrPtoB (C) (OD600 of 0.02, supplemented with 0.04% Silwett L-77) and covered for the remaining of the experiment. Bacterial counts were assessed at 3 dpi. Results are average±s.e. (n=8). (D, E) Post-invasive bacterial susceptibility assay. Leaves of 5-week-old plants were infiltrated with Pto DC3000 (OD600=0.0002) (D) or Pseudomonas syringae pv. tabaci (Pta) 6005 (OD600=0.002) (E). Bacterial populations were determined at 3 dpi. Results are average±s.e. (n=4). For all above experiments, similar results were observed in at least three independent experiments and asterisks indicate P<0.05 by t-test.
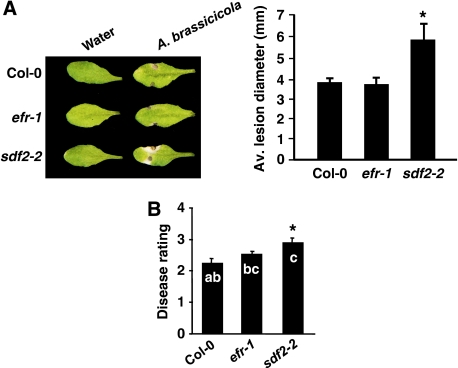
We tested whether the enhanced disease susceptibility of sdf2-2 could result from a defect in SA signalling, as SA positively regulates resistance to Pseudomonas, and as the SDF2 gene was identified earlier as a potential direct target of the SA signalling regulator NPR1 (Wang et al, 2005). SDF2 was however not required for secretion of the PR1 protein (a marker of NPR1-mediated responses) and bacterial resistance induced by the SA analogue benzothiadiazole (BTH) (Supplementary Figure 4). Thus, the hyper-susceptibility of sdf2-2 plants to bacteria is not due to a defect in SA-mediated resistance. SDF2 was also not required for the immunity triggered by recognition of the bacterial effectors AvrRpt2 and AvrRps4 (Supplementary Figure 5). Although sdf2-2 was not affected in chitin sensitivity, we tested its susceptibility to fungal pathogens. Notably, sdf2-2 was more susceptible to the virulent necrotrophic fungi Alternaria brassicicola and Plectosphaerella cucumerina (Figure 4). Thus, SDF2 is required for EFR, and to a lesser extent for FLS2, function. In addition, the enhanced disease susceptibility phenotype of sdf2-2 to bacteria and fungi suggests that SDF2 may have an important function in regulating other, yet unknown, PRRs involved in bacterial and fungal recognition, or conceivably other components of plant defence.
Figure 4.
Sdf2 mutant is more susceptible to fungal pathogens. (A) Susceptibility to A. brassicicola. Left, macroscopic symptoms and disease rating at 5 dpi. Right, lesion diameter at 5 dpi. Similar results were observed in four independent experiments and asterisks indicate P<0.05 by one-way ANOVA with Bonferroni post hoc test. (B) Susceptibility to P. cucumerina. Left, macroscopic symptoms 14 dpi. Right, average disease rating (DR±s.d.) of the indicated genotypes at 14 dpi. DR varies between 0 (no symptoms) and 5 (dead plants). Letters indicate P⩽0.05 by ANOVA with Bonferroni post hoc test. Data are from one of two independent experiments that gave similar results.
SDF2 localises to the ER in a complex with ERdj3B and BiP
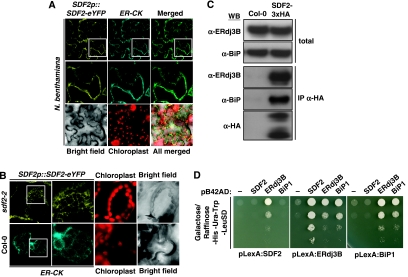
Plant SDF2 proteins contain a predicted SP, but no ER or Golgi retention signals, whereas SDF2 proteins from other eukaryotes carry a C-terminal K/HDEL motif (Supplementary Figure 2) classically associated with the retention of soluble proteins in the ER. To localise AtSDF2 within the plant cell, we transiently co-expressed AtSDF2–eYFP fusion protein under the control of its native promoter with the ER marker ER-CK (35S::CFP-HDEL) (Nelson et al, 2007) in Nicotiana benthamiana. Confocal microscopy revealed overlapping ER localisation for both expressed proteins (Figure 5A). The same ER localisation pattern was observed in sdf2-2 plants transgenic for AtSDF2p::SDF2-eYFP (Figure 5B), suggesting that SDF2 localises to the ER.
Figure 5.
SDF2 exists in an ER complex with ERdj3b and BiP. (A) Subcellular localisation of SDF2 in N. benthamiana. The constructs SDF2p::SDF2-eYFP and ER-CK (35S::HDEL-CFP) were transiently co-expressed by Agrobacterium-mediated transformation in N. benthamiana leaves. Confocal analysis of transformed cells was performed at 2 dpi. The middle row of panels are zoom-ins. (B) Subcellular localisation of SDF2 in transgenic sdf2-2/SDF2p::SDF2-eYFP Arabidopsis leaves. The second column of panels corresponds to zoom-ins. (C) Co-immunoprecipitation of SDF2 with ERdj3B and BiP in vivo. Protein extract from sdf2-2/SDF2p::SDF2-3xHA or wild-type Col-0 seedlings were subjected to immunoprecipitation with anti-HA affinity matrix. Western blot analysis was performed with anti-ERdj3B, anti-BiP and anti-HA antibodies. (D) ERdj3B serves as an adaptor between SDF2 and BiP. Yeast cultures were grown overnight in the -His-Ura-Trp liquid SD medium supplemented with glucose. Cultures were spun down and resuspended in water to OD600=1. A series of 10-fold dilutions was plated on -His-Ura-Trp-Leu SD medium supplemented with galactose and raffinose to assess the expression of the LEU reporter gene. Images were taken after 3 days incubation at 30°C.
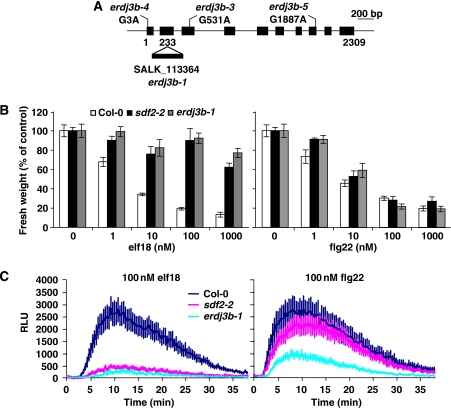
As SDF2 does not possess an ER retention signal, its ER localisation could be due to its interaction with ER resident proteins. To test this hypothesis, we searched for SDF2 interactors in planta. Immunoprecipitation of proteins associated with SDF2–3xHA and subsequent mass spectrometry analysis revealed the soluble luminal protein ERdj3B (At3g62600) as the major SDF2 interactor (Supplementary Figure 6). ERdj3B, an ER-localised member of the HSP40 co-chaperone family, is one of the two Arabidopsis orthologs of the mammalian ERdj3 and yeast Scj1p proteins (Yamamoto et al, 2008). The SDF2–ERdj3B interaction was confirmed in planta with a specific anti-ERdj3B antibody (Figure 5C). Although ERdj3B directly interacted with SDF2 in the yeast two-hybrid (Y2H) system, its Arabidopsis paralog ERdj3A did not (Figure 5D; Supplementary Figures 7 and 8). In mammals, SDF2L exists in complex with ERdj3 and the luminal-binding protein BiP, an ER-localised member of the HSP70 family of chaperones (Meunier et al, 2002; Bies et al, 2004; Jin et al, 2008). Similarly, we found that the SDF2 immunocomplex also contains BiP in Arabidopsis (Figure 5C). There are three BiP isoforms in Arabidopsis (BiP1–3) (Noh et al, 2003). In Y2H assays, BiP1–3 interacted with ERdj3B, as well as with ERdj3A, but not with SDF2 (Figure 5D; Supplementary Figure 7). These results indicate that, in Arabidopsis, SDF2 exists in a complex with ERdj3B and BiP1–3, in which ERdj3B may act as a bridge between SDF2 and BiP. Notably, SDF2 was not found in complex with other ER resident proteins under the conditions tested (Supplementary Figures 8 and 9), showing that the association of SDF2 with ERdj3B and BiP was specific. As both SDF2 and ERdj3B lack an ER retention signal, their ER localisation might be due to interaction with BiPs. Three erdj3b alleles were independently identified by map-based cloning and sequencing of elfin mutants (Figure 6A; Supplementary Figure 10). Analysis of a T-DNA null allele later confirmed that erdj3b-1 plants were strongly affected in the elf18-triggered SGI, oxidative burst and MAP kinase activation, resembling sdf2-2 plants (Figures 6B and C and 2C). Erdj3b-1 was also weakly impaired in flg22 responses (Figures 6B and C and 2C) but not in chitin responses (Supplementary Figure 11). These results reveal that ERdj3B is also required for EFR and to a lesser extent FLS2 function.
Figure 6.
Erdj3b mutants show a phenotype similar to sdf2. (A) Schematic representation of the ERdj3B gene (At3g62600) with position of the T-DNA insertions and point mutations. Exons are depicted as black boxes. (B) Seedling growth inhibition triggered by elf18 or flg22 in wild-type Col-0 (white bars), sdf2-2 (black bars) and erdj3b-1 (grey bars). Results are average±s.e. (n=6). (C) Oxidative burst triggered by 100 nM elf18 or 100 nM flg22 in wild-type Col-0 (blue), sdf2-2 (purple) and erdj3b-1 (cyan) leaf discs measured in relative light units (RLU). Results are average±s.e. (n=12).
The unfolded protein response (UPR) is activated by conditions that overload the ER-QC (Vembar and Brodsky, 2008; Vitale and Boston, 2008). Interestingly, we found that both sdf2-2 and erdj3b-1 were more sensitive to the drug tunicamycin, an inhibitor of N-glycosylation that triggers the UPR (Supplementary Figure 13), providing additional evidence for SDF2 and ERdj3B being involved in ER-QC. We also analysed functions for BiP1–3 in elf18 responses. Single bip1, bip2 or bip3 mutants all showed wild-type levels of elf18 responses (Supplementary Figure 12). Although the single bip2 mutant was shown earlier to be fully impaired in SA-induced resistance (Wang et al, 2005), BiP1 and BiP2 are highly similar and might be functionally redundant, whereas BiP3 is more distantly related (Noh et al, 2003). We therefore generated a bip1-1 bip2-1 double mutant, which showed a wild-type response to elf18 as well (Supplementary Figure 12). Silencing of all three Arabidopsis BiP genes recently proved to be lethal (Hong et al, 2008) thus preventing us from testing the possible requirement of all three BiPs in elf18 responses.
SDF2 is required for EFR biogenesis
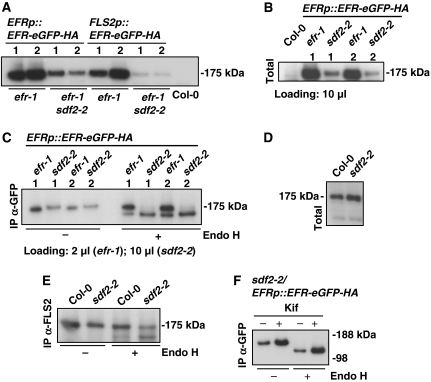
On the basis of its association with ERdj3B and BiP, we hypothesised that SDF2 is also part of the ER-QC. We therefore tested whether SDF2 could regulate EFR directly. As EFR gene expression was not affected in sdf2-2 (Supplementary Figure 14A), we next determined EFR protein levels in vivo. We generated efr-1, sdf2-2 and efr-1 sdf2-2 lines transgenic for the EFRp::EFR-eGFP-HA construct to enable detection of physiological levels of EFR protein. Expression of the tagged EFR complemented the efr-1 mutation (data not shown) and we detected a single band running at ∼175 kDa, the expected size for the glycosylated form of EFR-eGFP-HA in several independent primary transformants (Supplementary Figure 14B). However, the EFR protein level in total extracts was strongly reduced in the efr-1 sdf2-2 and sdf2-2 backgrounds (Supplementary Figure 14B; Figure 7A and B), showing a requirement of SDF2 for EFR protein accumulation. The enzyme endoglycosidase H (Endo H) cleaves off ER-specific glycans, whereas complex glycans that are matured during their transit through the Golgi apparatus are resistant to Endo H cleavage. Therefore, Endo H treatment allows the differentiation of plasma membrane localised glycoproteins from their ER localised counterparts. Using Endo H assays, we found that, although transgenic sdf2-2 plants express some levels of EFR protein, it is completely cleaved by Endo H (Figure 7C), revealing that the expressed EFR protein is retained in the ER. In contrast, most of the EFR protein expressed in the efr-1 background is resistant to Endo H, revealing its localisation at the plasma membrane. Consistently, cross-linking and binding studies with radiolabelled elf18 peptide indicated that sdf2-2 has fewer binding sites than wild type (Supplementary Figure 15A), corroborating the reduced elf18 sensitivity of the sdf2-2 mutant (Figures 1 and 2). Although FLS2 accumulated to similar levels in sdf2-2 and WT total extracts (Figure 7D), half of FLS2 seems to be retained in the ER in sdf2-2 (Figure 7E). This is consistent with the decreased flg22 sensitivity and binding of sdf2-2 (Figure 2A and B; Supplementary Figures 3 and 15).
Figure 7.
SDF2 controls EFR protein accumulation. (A) Effect of the sdf2-2 mutation on EFR protein levels. Total protein extracts were prepared from T2 transgenic lines (the numbers above the lanes indicate independent lines). Col-0 was used as a negative control. Equal amounts of plant tissue were used in all cases. (B) Relative abundance of EFR expressed under its native promoter in efr-1 and sdf2-2 genetic backgrounds. (C) Effect of the sdf2-2 mutation on EFR glycosylation state. Endo H assays were performed with the EFR protein from efr-1/- and efr-1 sdf2-2/EFRp::EFR-eGFP-HA. EFR was immunoprecipitated using anti-GFP agarose beads (Caltag Medsystems) and incubated with (+) or without (−) Endo H for 1 h at 37°C. (D) Immunoblot of FLS2. Total protein extracts were prepared from Col-0 and sdf2-2 plants as described in (A). FLS2 was detected with a polyclonal anti-FLS2 antibody. (E) Effect of sdf2-2 on the FLS2 glycosylation state. FLS2 was immunoprecipitated using the polyclonal anti-FLS2 antibody and protein G sepharose beads (Sigma). The Endo H assay was performed as in (C). (F) EFR expressed in the sdf2-2 background is stabilised by kifunensine (Kif). Despite the increase in the protein level on treatment with Kif, EFR expressed in sdf2-2 remains sensitive to Endo H. Two-week-old seedlings were incubated for 20 h in the 1/2 MS medium with (+) or without 50 μM Kif. The protein extracts were prepared as in (A) and the Endo H assay was performed as in (C). In (A–C, F) EFR was detected with the anti-GFP antibody (TP401, AMS Biotechnology).
Interestingly, treatment with kifunensine, an inhibitor of ER mannosidase I that prevents ER exit and consequently ERAD (Tokunaga et al, 2000), increased EFR protein levels in sdf2-2 (Figure 7F), whereas the 26S proteasome inhibitor MG132 had no effect (Supplementary Figure 16). Despite increased levels after kifunensine treatment, Endo H assays revealed that EFR is still retained in the ER in the sdf2-2 background (Figure 7F). These results revealed that EFR is actively degraded in sdf2-2 and that this degradation occurs outside of the ER. Future work will be required to determine whether EFR is degraded in the cytosol as most ERAD substrates or in the vacuole as recently shown for a subset of plant ERAD substrates (Foresti et al, 2008).
EFR function specifically depends on SST3A-mediated N-glycosylation
An intriguing question is why FLS2 function is affected less than EFR by mutations in ER-QC components, whereas CERK1 function is not affected at all. This is particularly surprising as EFR and FLS2 are structurally similar and belong to the subfamily XII of LRR-RKs. One possible explanation for the apparent specificity could be due to a receptor level threshold required for responsiveness. EFR protein levels might be much lower than FLS2 protein levels. Therefore, mutations in ER-QC components would reduce EFR protein amounts to a level that severely impacts elf18 responses, whereas FLS2 protein amounts would still be sufficient to ensure flg22 responses. However, expression of EFR under the control of the FLS2 promoter in efr-1 and efr-1 sdf2-2 did not revert the reduced EFR accumulation because of the sdf2 mutation (Figure 7A). Therefore, the requirement of SDF2 for EFR is not due to a decreased protein expression compared with FLS2, and might rather be due to intrinsic properties of EFR.
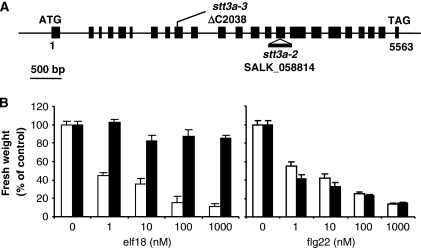
Notably, we have identified in our elfin population alleles of STT3A (Staurosporin and temperature sensitive-3A), coding for a component of the ER oligosaccharyltransferase (OST) complex involved in N-glycosylation of nascent proteins (Koiwa et al, 2003). Unexpectedly, the characterisation of stt3a mutants revealed that STT3a-mediated N-glycosylation is indispensable for EFR, but dispensable for FLS2 function (Figure 8; Supplementary Figure 17). Although plant extracts of stt3a mutants still contain glycosylated proteins, the pattern of N-glycosylation is different (Koiwa et al, 2003), suggesting that only a subset of proteins are STT3a substrates. In mammals, STT3a is important for co-translational N-glycosylation of nascent proteins, whereas STT3b rather has a function in post-translational N-glycosylation (Ruiz-Canada et al, 2009). Arabidopsis stt3b mutants were not affected in elf18- or flg22-triggered SGI (data not shown), showing that STT3A is the key glycosylation determinant for EFR. Although both EFR and FLS2 carry numerous putative glycosylation sites, they may have different glycosylation states (e.g. position, number of glycosylated sites, glycan structures) that could impact their interaction with and dependence on the ER-QC machinery.
Figure 8.
Stt3a affects EF-Tu but not flagellin responsiveness. (A) Schematic representation of the STT3A gene with positions of the single nucleotide deletion and the T-DNA insertion. Exons are depicted as black boxes. stt3a-3 and stt3a-2 (Koiwa et al, 2003) are indicated. (B) Seedling growth inhibition triggered by elf18 or flg22 in wild-type Col-0 (white bars) and stt3a-2 (black bars) seedlings. Five-day-old Arabidopsis seedlings were transferred into liquid MS 1% supplemented with the indicated concentrations of peptides. Seedling fresh weight was quantified 1 week after treatment. Results are average±s.e. (n=6).
A subset of ER-QC components is specifically required for the biogenesis of transmembrane PRRs
In addition to reporting a function for SDF2 and ERdj3B in plant innate immunity, we and others have found that the Arabidopsis CRT3 and UGGT are also required for EFR function, as loss of either CRT3 or UGGT leads to complete loss of EFR accumulation (Li et al, 2009; Saijo et al, 2009). Altogether, this shows that EFR relies on both the CNX/CRT3 and SDF2/ERdj3B/BiP systems for its proper function. BiP and CRT exist in an abundant large complex in tobacco (Denecke et al, 1995; Crofts et al, 1998). CRT3, SDF2, ERdj3B, BIP and potentially UGGT may therefore exist in the same complex to regulate proper EFR folding. Such large chaperone complexes have been reported in mammals (Meunier et al, 2002), but we have failed so far to detect CRT3 or UGGT in the SDF2 immunocomplex. Further work is therefore required to investigate the existence of such large complex.
Our infection data strongly suggest that SDF2 may also regulate other unknown PRRs involved in bacterial and fungal recognition, or conceivably other defence components, whose identity will be interesting to investigate in future experiments. The specificity of the reported mutations, however, show that SDF2 and ERdj3B are specifically involved in plant innate immunity and clearly argues against a general defect in protein secretion. It would be interesting to test whether they are also having a function in other stress responses, for example, abiotic stresses, and to identify which other members of the Hsp70 and Hsp40 families for example ensure folding of proteins involved in other plant physiological functions.
This work provides the first demonstration of a requirement for the ER-QC in generating functional transmembrane receptors in plants. Recent studies reported a function for the ER-QC in retaining mutant variants of the brassinosteroid receptor BRI1, a LRR-RK, within the ER (Jin et al, 2007; Hong et al, 2008). However, in these studies, the function of wild-type BRI1 was not affected by mutations in ER-QC components. This is in clear contrast with our findings that mutations in a subset of ER-QC components affect wild-type EFR.
Although most cytoplasmic nucleotide-binding domain and LRR-containing (NLR) immune receptors involved in effector recognition rely on the HSP90–SGT1 chaperone complex (Shirasu, 2009), we propose that a subset of surface-exposed PRRs, including EFR require one or several ER complexes comprising SDF2, ERdj3B, BiP and potentially CRT3 and UGGT for their accumulation and subcellular localisation. We speculate that as EFR is only found in the Brassicaceae, whereas FLS2 has been identified in several dicots and monocots (Takai et al, 2008; Zipfel, 2008), EFR may have evolved more recently than FLS2, and thus its amino-acid sequence is less capable of folding properly in the absence of ER-QC components. It is conceivable that evolution of new recognition specificities may result in proteins that detect novel ligands but that have not been selected for high protein stability, and thus require extra “buffering” (Queitsch et al, 2002).
Materials and methods
Plant materials and growth
A. thaliana ecotype Columbia (Col-0) was the background for all mutants and transgenic lines used in this study. The Arabidopsis plants used in this study were grown as one plant per pot at 20–21°C with an 10 h photoperiod, or on plates containing MS salts medium (Duchefa), 1% sucrose, and 1% agar with a 16 h photoperiod.
The T-DNA insertion lines SALK_141321 (sdf2-2), SALK_113364 (erdj3b-1), SALK_063999 (bip1-1), SALK_093564 (bip1-2), SALK_47956 (bip2-1 gift from X Dong), bip3-2, GABI_075D06, and SALK_024133 (bip3-1) were obtained from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). The fls2c efr-1 double mutant was generated by crossing fls2c (Zipfel et al, 2004) with efr-1 (Zipfel et al, 2006). Primers used to genotype the T-DNA lines are listed in Supplementary Table I.
Elfin forward-genetic screen
To isolate elf-18 insensitive (elfin) plants, we screened ∼35 000 T-DNA activation-tagging transgenic lines (Molinier et al, 2004) (gift from J Molinier) and ∼137 500 EMS-mutagenised M2 seeds (Tedman-Jones et al, 2008) (all in Col-0 background). Five-day-old seedlings germinated on MS 1% agar plates were submerged with liquid MS 1% supplemented with 50 nM elf18. At 8 to 10 days after treatment, SGI was visually scored and putative elfin mutant plant was transferred to the soil. Progenies of ∼300 putative elfin mutants were retested in the next generation by a quantitative SGI assay, as described in Zipfel et al (2006). At the end, 160 elfin mutants were confirmed, including 57 that carry mutations in EFR.
The genomic DNA flanking the T-DNA insertion in the sdf2-1 mutant was cloned using thermal asymmetric interlaced-PCR (TAIL-PCR), based on Liu et al (1995).
To identify the erdj3b-3 mutation, elfin plants in the Columbia-0 (Col-0) background were crossed to wild-type plants of the Landsberg erecta (Ler-0) ecotype. Genomic DNA from elfin seedlings in the segregating F2 population was extracted and used for subsequent PCR analysis. The mutation was mapped to chromosome III between the markers NGA6 and NGA112 using a set of known simple sequence length polymorphism markers (data not shown). Fine mapping delimited the mutation to a 51-kb interval between the cleaved amplified polymorphic sequence markers CHIII-23122 and CHI-23173. Sequencing the whole region in the mutant identified a single-nucleotide mutation in ERdj3B (At3g62600).
Generation of transgenic plants
SDF2p::SDF2-eYFP construct was generated by PCR amplification of the SDF2 genomic fragment, including the promoter and the coding region, with 5′-CACCAAAATTAATTTCCATGATGTAAAATTA-3′ and 5′-CTTGCTGCTCTCATTAAGGG-3′ primers and resulting PCR product was cloned into the pENTRY-D/TOPO vector using the pENTR Directional TOPO Cloning Kit (Invitrogen). The resulting clone was sequenced and the insert transferred into the GATEWAY-compatible vector pGWB40 (Nakagawa et al, 2007) using GATEWAY LR CLONASE II enzyme (Invitrogen). The final construct was electroporated into Agrobacterium tumefaciens strain GV3101.
SDF2p::SDF2-3xHA construct was generated by PCR amplification of the SDF2 genomic fragment, including the promoter and the coding region, with 5′-TACAATTGAAAATTAATTTCCATGATGTAAAATTA-3′ and 5′-GGATCCGAGACGCTTGCTGCTCTCATTAAGGG-3′ primers. The resulting PCR product was cloned into the pGEMTeasy vector by the T/A ligation. The clone was sequenced. The insert was then released by digesting with MfeI and BsmBI and ligated into the epiGreenB5 binary vector digested with EcoRI and BamHI. As a result, the Pro35S::GUS cassette in epiGreenB5 was replaced with SDF2p::SDF2 producing the SDF2p::SDF2-3xHA fusion.
EFRp::EFR-eGFP-HA construct has been generated as following: the genomic fragment comprising the EFR promoter plus the coding region was PCR-amplified from Col-0 genomic DNA using 5′-ATCCCGGGATCTAGACGATTAAGTAATTGAG-3′ and 5′-ATGGATCCCATAGTATGCATGTCCGTATTTA-3′ primers, and cloned into the pGEMTeasy vector by the T/A ligation. The resulting clone was sequenced. The EFRp::EFR insert was further subcloned into the epiGreenB(eGFP-HA) vector: pGEMTeasy:EFRp::EFR clone was digested with EcoRI+SacI and SacI+BamHI in two separate reactions and the two released fragments of the insert were ligated into the epiGreenB(eGFP-HA) vector, digested with EcoRI and BamHI, using the three-way ligation method.
FLS2p::EFR-eGFP-HA construct has been generated as following: the FLS2 promoter was PCR-amplified from Col-0 genomic DNA using 5′-ATCAATTGcccttttcggacattctaaatat-3′ and 5′-CATGTCGATTATAAAAAGATAAATCTATAGACGAAGTCATATGT-3′. The EFR 5′-UTR plus the coding region were PCR-amplified from Col-0 genomic DNA using 5′-Atgacttcgtctatagatttatctttttataatcgacatgaagc-3′ and 5′-ATGGATCCCATAGTATGCATGTCCGTATTTA-3′ primers. A fusion between FLS2p and EFR was created using the chimeric PCR method and the resulting PCR fragment was cloned into the pGEMTeasy vector by the T/A ligation. The resulting clone was sequenced. The FLS2p::EFR insert was further subcloned into the epiGreenB(eGFP-HA) vector: pGEMTeasy:FLS2p::EFR clone was digested with MfeI+BamHI and the released insert was ligated into the epiGreenB(eGFP-HA) vector, digested with EcoRI and BamHI.
epiGreenB(eGFP-HA) was constructed as following: a PCR fragment comprising eGFP followed by a single HA tag (eGFP-HA) was cloned into the pBin19g vector (Rivas et al, 2004) using BamHI and XbaI restriction enzymes to replace the single HA tag with eGFP-HA in this vector. The resulting pBin19(eGFP-HA) vector was digested with BamHI and HindIII enzymes, and the released fragment containing eGFP-HA-NOS was cloned into the epiGreenB5 vector using the same restriction enzymes. As a result, the 3xHA tag in epiGreenB5 was replaced with eGFP-HA producing the epiGreenB(eGFP-HA) vector.
Both epiGreenB(eGFP-HA):EFRp::EFR-eGFP-HA and epiGreenB5::SDF2p::SDF2-3xHA constructs were electroporated into A. tumefaciens strain GV3101 carrying the pSOUP helper plasmid. All constructs were transformed into relevant Arabidopsis mutant lines using the floral dipping method (Clough and Bent, 1998). The transformants were selected on the MS agar medium supplemented with 10 μg/ml kanamycin in the case of SDF2p::SDF2-eYFP and spraying soil grown seedlings with BASTA in the case of SDF2p::SDF2-3xHA and EFRp::EFR-eGFP-HA.
PTI assays
SGI and oxidative burst assays were performed as described earlier (Zipfel et al, 2006) except that luminescence was measured using a Varioscan Flash plate reader. MAPK assays were performed as described (Meskiene et al, 2003) using antibodies against MPK4 and MPK6 (Sigma).
Infection assays
Bacterial assays were performed as described earlier (Zipfel et al, 2004). Inoculations with A. brassicicola and P. cucumerina were performed as described earlier (Hernández-Blanco et al, 2007; van Esse et al, 2008).
For the BTH-induced secretion of PR-1, intercellular wash fluid was collected as described earlier (Wang et al, 2005) from equal amounts of leaves treated for 24 h with water or BTH (300 μM). For BTH-induced resistance, water or BTH (300 μM; black bars) were sprayed onto leaves from 5-week-old plants 24 h before infiltrating Pto DC3000 (OD600=0.0002). Bacterial populations were determined at 2 dpi.
Transient expression in N. benthamiana
A. tumefaciens strains were grown in L medium supplemented with appropriate antibiotics overnight. Next morning cultures were spun down and resuspended in 10 mM MgCl2 to OD600=0.6. A. tumefaciens strains carrying ER-CK (pBin20:35S::CFP-HDEL) and pGWB40:SDF2p::SDF2-eYFP were mixed 1:1 and syringe-infiltrated into 3-week-old N. benthamiana leaves. The confocal analysis was performed at 2 dpi.
Confocal microscopy
Analysis of intracellular fluorescence was performed by confocal laser-scanning microscopy on a Leica DM6000B/TCS SP5 confocal microscope (Leica Microsystems CMS GmbH, Germany) using a 20 × objective. Fluorophores were excited using an argon laser at 458 nm (CFP) or 514 nm (YFP). Images were collected in the multi-channel mode and the overlay images were generated using the Leica analysis software LAS AF1.8.2.
Y2H assays
Y2H assays were performed using MATCHMAKER LexA two-hybrid system (Clontech). cDNAs encoding proteins tested were PCR amplified using specific primers. In all cases, the SP was omitted. The amplified PCR fragments were first cloned into the pGEMTeasy vector using the T/A ligation, sequenced and then subcloned, using restriction enzymes specified in the table, into pLexA and pB42AD vectors pre-digested with EcoRI and XhoI. Protein–protein interactions were tested in the yeast strain EGY48 according to the manufacturer's instructions. Oligos used for cloning of SDF2, Erdj3B and BiP1 cDNAs are described in Supplementary Table III.
Protein co-immunoprecipitations
Protein co-immunoprecipitations were performed as described earlier (Moffett et al, 2002) using leaves of 5-week-old transgenic Arabidopsis plants using the anti-HA affinity matrix (Roche). Immunoprecipitates were analysed by western blot with anti-HA, anti-ERdj3B (Yamamoto et al, 2008), anti-SHD (Klein et al, 2006), anti-CRT (Pagny et al, 2000) and anti-BiP (Pedrazzini et al, 1997) antibodies (gifts from R Boston, S Nishikawa, and A Vitale).
Endo H assay
EFR-eGFP-HA and FLS2 were immunoprecipitated using the anti-GFP agarose (Caltag Medsystems), and protein G sepharose (Sigma) plus anti-FLS2 polyclonal antibodies (Chinchilla et al, 2007), respectively. The treatment with Endo H (New England Biolabs) was performed for 1 h at 37°C according to the manufacturer's instructions.
Supplementary Material
Supplementary Figures 1–17
Supplementary data, Tables I–III
Review Process
Acknowledgments
This research was supported by Biotechnology and Biological Sciences Research Council grant BB/F021046/1 (CZ), ERA-NET Plant Genomics (JDGJ, AM, BPHJT), the Gatsby Charitable Foundation (CZ, JDGJ), and Swiss National Foundation grant 31003A-120655 (DC). We thank A Jones and L Serazetdinova for their precious help with the mass spectrometry analysis, R Fuchs for his help with the confocal microscopy, D Alger and his team for excellent plant care, L Price and P Dominy for helpful advices on TAIL-PCR, and J Rathjen and R Strasser for their useful discussions and comments on the manuscript. R Boston, X Dong, B Kunkel, J Li, D Mackey, G Martin, B Mauch-Mani, J Molinier, S Nishikawa, and A Vitale are acknowledged for providing materials.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anelli T, Alessio M, Bachi A, Bergamelli L, Bertoli G, Camerini S, Mezghrani A, Ruffato E, Simmen T, Sitia R (2003) Thiol-mediated protein retention in the endoplasmic reticulum: the role of ERp44. EMBO J 22: 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Ceppi S, Bergamelli L, Cortini M, Masciarelli S, Valetti C, Sitia R (2007) Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J 26: 4177–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Sitia R (2008) Protein quality control in the early secretory pathway. EMBO J 27: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies C, Blum R, Dudek J, Nastainczyk W, Oberhauser S, Jung M, Zimmermann R (2004) Characterization of pancreatic ERj3p, a homolog of yeast DnaJ-like protein Scj1p. Biol Chem 385: 389–395 [DOI] [PubMed] [Google Scholar]
- Buck TM, Wright CM, Brodsky JL (2007) The activities and function of molecular chaperones in the endoplasmic reticulum. Sem Cell Dev Biol 18: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Leborgne-Castel N, Pesca M, Vitale A, Denecke J (1998) BiP and calreticulin form an abundant complex that is independent of endoplasmic reticulum stress. Plant Cell 10: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Carlsson LE, Vidal S, Höglund AS, Ek B, van Zeijl MJ, Sinjorgo KM, Palva ET (1995) The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell 7: 391–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, Forsyth A, Robatzek S, Grant M, Boch J (2006) Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J 47: 368–382 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, De Marchis F, de Virgilio M, Klein EM, Arcioni S, Belluci M, Vitale A (2008) Protein domains involved in assembly in the endoplasmic reticulum promote vacuolar delivery when fused to secretory GFP, indicating a protein quality control pathway for degradation in the plant vacuole. Mol Plant 1: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Hann DR, Rathjen JP (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J 49: 607–618 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Blanco C, Feng DX, Hu J, Sánchez-Vallet A, Deslandes L, Llorente F, Berrocal-Lobo M, Keller H, Barlet X, Sánchez-Rodríguez C, Anderson LK, Somerville S, Marco Y, Molina A (2007) Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam KH, Li J (2007) Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell 26: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Awad W, Petrova K, Hendershot LM (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J 27: 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zhuang M, Hendershot LM (2009) ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry 48: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Klein EM, Mascheroni L, Pompa A, Ragni L, Weimar T, Lilley KS, Dupree P, Vitale A (2006) Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J 48: 657–673 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, Bressan RA, Hasegawa PM (2003) The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic Stress. Plant Cell 15: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chu Z-H, Batoux M, Nekrasov V, Roux M, Chinchilla D, Zipfel C, Jones JDG (2009) Specific ER quality control components required for biogenesis of the plant immune receptor EFR. Proc Natl Acad Sci USA (e-pub ahead of print 26 August 2009; doi:10.1073/pnas.0905532106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278: 18945–18952 [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Ramos C, Fritsch O, Hohn B (2004) CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis. Plant Cell 16: 1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Noh SJ, Kwon CS, Oh DH, Moon JS, Chung WI (2003) Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311: 81–91 [DOI] [PubMed] [Google Scholar]
- Pagny S, Cabanes-Macheteau M, Gillikin JW, Leborgne-Castel N, Lerouge P, Boston RS, Faye L, Gomord V (2000) Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell 12: 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzini E, Giovinazzo G, Bielli A, de Virgilio M, Frigerio L, Pesca M, Faoro F, Bollini R, Ceriotti A, Vitale A (1997) Protein quality control along the route to the plant vacuole. Plant Cell 9: 1869–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP (2000) Novel repeats in ryanodine and IP3 receptors and protein O-mannosyltransferases. Trends Biochem Sci 25: 48–50 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Reddy P, Sparvoli A, Fagioli C, Fassina G, Sitia R (1996) Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled Ig light chains. EMBO J 15: 2077–2085 [PMC free article] [PubMed] [Google Scholar]
- Rivas S, Rougon-Cardoso A, Smoker M, Schauser L, Yoshioka H, Jones JD (2004) CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J 23: 2156–2165 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ruiz-Canada C, Kelleher DJ, Gilmore R (2009) Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J (e-pub ahead of print 17 September 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Zipfel C (2008) News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr Opin Plant Biol 11: 389–395 [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Takai R, Isogai A, Takayama S, Che FS (2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol Plant Micro Interact 21: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Tedman-Jones JD, Lei R, Jay F, Fabro G, Li X, Reiter WD, Brearley C, Jones JD (2008) Characterization of Arabidopsis mur3 mutations that result in constitutive activation of defence in petioles, but not leaves. Plant J 56: 691–703 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Brostrom C, Koide T, Arvan P (2000) Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase I. J Biol Chem 275: 40757–40764 [DOI] [PubMed] [Google Scholar]
- van Esse HP, Van't Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, Vervoort J, de Wit PJ, Thomma BP (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S, Brodsky JL (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Boston RS (2008) Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Williams DB (2006) Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci 119: 615–623 [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou JM (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Maruyama D, Endo T, Nishikawa S (2008) Arabidopsis thaliana has a set of J proteins in the endoplasmic reticulum that are conserved from yeast to animals and plants. Plant Cell Physiol 49: 1547–1562 [DOI] [PubMed] [Google Scholar]
- Zipfel C (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20: 10–16 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–17
Supplementary data, Tables I–III
Review Process