Abstract
Loss of heterozygosity (LOH), a causal event in cancer and human genetic diseases, frequently encompasses multiple genetic loci and whole chromosome arms. However, the mechanisms by which such extensive LOH arises, and how it is suppressed in normal cells is poorly understood. We have developed a genetic system to investigate the mechanisms of DNA double-strand break (DSB)-induced extensive LOH, and its suppression, using a non-essential minichromosome, Ch16, in fission yeast. We find extensive LOH to arise from a new break-induced mechanism of isochromosome formation. Our data support a model in which Rqh1 and Exo1-dependent end processing from an unrepaired DSB leads to removal of the broken chromosome arm and to break-induced replication of the intact arm from the centromere, a considerable distance from the initial lesion. This process also promotes genome-wide copy number variation. A genetic screen revealed Rhp51, Rhp55, Rhp57 and the MRN complex to suppress both isochromosome formation and chromosome loss, in accordance with these events resulting from extensive end processing associated with failed homologous recombination repair.
Keywords: break-induced replication, copy number variation, DSB, homologous recombination, isochromosome
Introduction
Loss of heterozygosity (LOH), in which the remaining functional allele of a gene is lost, can lead to tumourigenesis through loss of tumour suppressor function (Knudson, 1993). LOH is detected at high frequency in both sporadic and hereditary cancers, and often extends several megabases encompassing whole chromosome arms (Lasko et al, 1991). Such extensive LOH may result from a variety of chromosomal rearrangements, including mitotic non-disjunction, truncations, interstitial deletions and translocations (Jasin, 2000). DNA double-strand breaks (DSBs) are potential initiating events leading to chromosomal aberrations (Pfeiffer et al, 2000). DSBs can result from exposure to DNA damaging agents, such as ionizing radiation, but can also arise spontaneously as a result of normal DNA metabolism (Shrivastav et al, 2008).
Cells have evolved two distinct pathways to repair DSBs: non-homologous end-joining (NHEJ) and homologous recombination (HR). HR is initiated by resection of the broken ends, followed by strand invasion of a homologous template (Krogh and Symington, 2004). Resection during DSB repair requires the MRX/MRN complex (Mre11–Rad50–Xrs2 in Saccharomyces cerevisiae (Sc), Mre11–Rad50–Nbs1 in Schizosaccharomyces pombe (Sp) and Homo sapiens (Hs)) (Paques and Haber, 1999; Llorente et al, 2008), Exo1(Tsubouchi and Ogawa, 2000; Tomita et al, 2003; Llorente and Symington, 2004) and Sae2Sc/Ctp1Sp/CtIPHs, (Clerici et al, 2005; Limbo et al, 2007; Sartori et al, 2007; Huertas et al, 2008). In addition, roles for Sgs1Sc/BLMHs helicase, and Dna2Sc in resection have recently been identified (Gravel et al, 2008; Mimitou and Symington, 2008; Zhu et al, 2008). After such end processing, a Rad51 (Rhp51Sp) nucleoprotein filament is formed, which is facilitated by Rad52 (Rad22Sp) and the heterodimeric Rad55–Rad57 (Rhp55Sp-Rhp57Sp) mediator complex (Sung, 1997a, 1997b). The nucleoprotein filament facilitates homology search and strand invasion leading to the formation of a displacement (D) loop structure (Sugawara et al, 2003; Wolner et al, 2003). Current models suggest that the invaded strand is a substrate for three distinct subpathways of HR: double-strand break repair (DSBR), synthesis-dependent strand annealing (SDSA) and break-induced replication (BIR) (Paques and Haber, 1999; Llorente et al, 2008).
In the classic model of HR (DSBR), DNA synthesis is associated with extension of the D-loop, thus facilitating second end capture. After branch migration and ligation, this results in the formation of a double-Holliday junction, which is subsequently resolved either with or without crossovers associated with gene conversion (Szostak et al, 1983). Although crossovers between identical sister chromatids do not compromise genome integrity, HR between homologous chromosomes results in gene conversion and can lead to extensive LOH (Jasin, 2000). During SDSA, the invading strand is expelled from the homologous template after DNA synthesis, resulting in the re-annealing of the two broken chromosome arms. As SDSA minimizes crossovers, it is postulated to occur more frequently during mitotic growth. BIR or recombination-induced replication initiates extensive replication after processing and strand invasion of a single broken end. BIR is thought to be important for repair of stalled or collapsed replication forks, and in telomere maintenance in the absence of telomerase. Both Rad51-dependent and -independent forms have been documented, and recently BIR has been shown to require both leading and lagging strand DNA polymerases (Malkova et al, 1996, 2005; Davis and Symington, 2004; Cullen et al, 2007; Lydeard et al, 2007). BIR can also cause chromosomal rearrangements and extensive LOH by long-distance replication using a homologous chromosome template, ectopic sites, and by template switching (Malkova et al, 1996; Smith et al, 2007; VanHulle et al, 2007; Llorente et al, 2008).
In this study, we have investigated the mechanisms by which a site-specific DSB can give rise to extensive LOH, and how such mechanisms are suppressed, using a non-essential minichromosome in fission yeast. Unexpectedly, we find that a single DSB can lead to extensive LOH resulting from isochromosome formation, in which the broken chromosome arm is resected to the centromere and replaced by an inverted copy of the intact arm. In addition, we have identified roles for homologous recombination genes and the MRN complex in suppressing isochromosome formation. These findings together support a model in which extensive end processing arising from failed or inefficient HR repair leads to chromosomal rearrangements, and extensive LOH.
Results
Extensive LOH is associated with loss of the broken chromosome arm
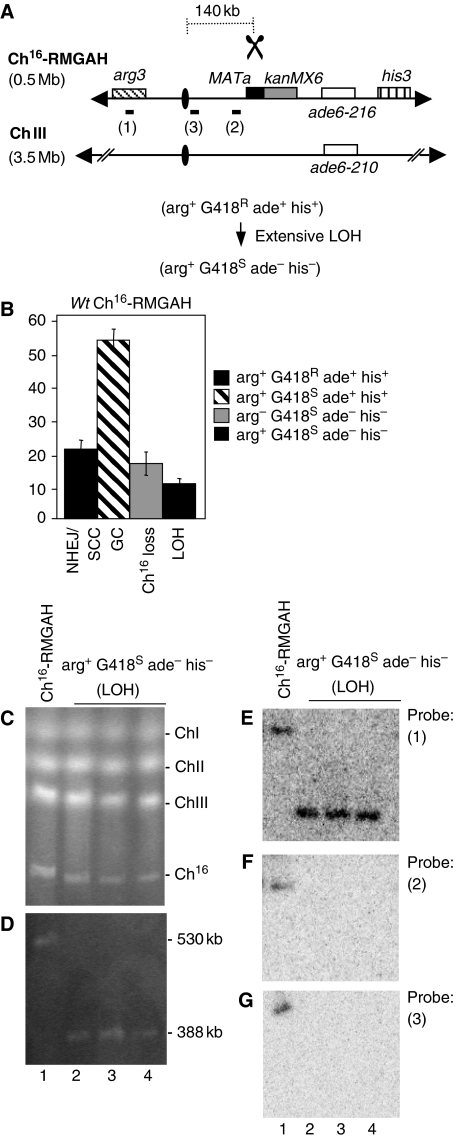
To screen for suppressors of extensive break-induced LOH, a strain carrying a non-essential minichromosome (Ch16), experimentally derived from the centromeric region of chromosome III (ChIII) (Niwa et al, 1986), was adapted such that extensive break-induced LOH, resulting from loss of the distal arm of Ch16 could be detected using a colony-sectoring assay. An HO endonuclease target site, MATa, together with an adjacent kanMX6 gene, encoding G418 resistance, was integrated into the right arm of Ch16, centromere-proximal to the ade6-M216 heteroallele and his3 marker. An arg3 marker was integrated into the left arm of the minichromosome, 300 kb away from the break site, to form Ch16-RMGAH (Figure 1A). In this context, extensive break-induced LOH resulting from the HO endonuclease expression would be expected to result in arg+ G418S ade− his− cells, which at high levels could be detected as red sectoring colonies on plates containing low levels of adenine following loss of the ade+ marker (Materials and methods). To examine break-induced LOH, HO endonuclease was expressed from a plasmid (pREP81X-HO) in the absence of thiamine, and colony phenotypes were quantified. Following DSB induction, 20% of the colonies were arg+ G418R ade+ his+, consistent with repair through end joining (NHEJ) or sister chromatid conversion (SCC); 53% of the colonies were arg+ G418S ade+ his+, consistent with repair by interchromosomal gene conversion (GC); and 16% of the colonies were arg− G418S ade− his− as a result of minichromosome loss through failed DSB repair. In addition, 10% of colonies were arg+ G418S ade− his− consistent with having undergone extensive LOH, but were not easily visualized using a colony sectoring assay (Figures 1B and 6A). This level of break-induced LOH was significantly higher than that previously observed (Cullen et al, 2007). Extensive LOH was break dependent, as loss of the markers distal to the break site was not observed after incubation for 48 h with a blank plasmid. To investigate the mechanism of break-induced LOH in Ch16-RMGAH, the chromosomes of 25 arg+ G418S ade− his− colonies were examined by pulsed field gel electrophoresis (PFGE). Although no size changes were detected in endogenous chromosomes of these colonies (Figure 1C), PFGE revealed that these colonies possessed a significantly smaller chromosomal element of 388 kb, instead of the uncut 530-kb Ch16-RMGAH (Figure 1D, lanes 2–4).
Figure 1.
LOH is associated with broken chromosome arm loss in wild-type background. (A) Schematic of Ch16-RMGAH. Ch16-RMGAH, ChIII, centromeric regions (ovals), complementary heteroalleles (ade6-M216 and ade6-M210; white), and the his3 marker (vertical stripes), ∼50 kb centromere-distal to ade6-M216, are as previously shown (Cullen et al, 2007). The MATa site (black) with an adjacent kanMX6 resistance marker gene (grey) was inserted into spcc23B6.06 ∼30 kb centromere-proximal to ade6-M216. The arg3 marker was inserted into spcc1795.09 on the left arm of the minichromosome. Derepression of pREP81X-HO (data not shown) generates a DSB at the MATa target site (scissors). The distance from the MATa site to the centromere is shown. In Ch16-RMHAH, kanMX6 is replaced by hph (B) Percentage DSB-induced marker loss in wild-type backgrounds using strains TH2130-3 and TH2357 (Supplementary Table 5). The levels of non-homologous end joining/sister chromatid conversion (NHEJ/SCC), gene conversion (GC), minichromosome loss (Ch16 loss), and LOH are shown. s.e.m. values are indicated. (C) PFGE analysis of chromosomal DNA from wild-type strain containing Ch16-RMGAH (TH2130; lane1), and individual wild-type arg+ G418S his−ade− strains isolated after DSB induction (lane 2–4). (D) High-resolution PFGE analysis of the strains described above. Southern blot analysis of the PFGE shown in (D) probed with arg3 (E; probe 1), spcc4b3.18 (F; probe 2) and tel1 ∼10 kb centromere-proximal to the MATa site (G; probe 3).
To determine the structure of this new chromosomal element Southern blot analysis was carried out. An arg3 probe was able to anneal to both the parental Ch16-RMGAH minichromosome and to the chromosomal elements from three individually isolated arg+ G418S ade− his− colonies, consistent with the arg+ phenotype of these cells (Figure 1E, probe 1). Similarly, a chk1 probe was able to anneal to both parental minichromosomes and to the new chromosomal elements (see Figure 3B). These data indicated that the left arm of the minichromosome was maintained in the shorter chromosomal element. Numerous probes targeted to regions distal to the break site, while annealing to the parental Ch16-RMGAH minichromosome, failed to anneal to this chromosomal element in any arg+ G418S ade− his− colony (our unpublished results). Similarly, probes targeted ∼10 kb centromere-proximal to the break site (probe 2) or close to the centromere (<10 kb away) on the right arm of Ch16 (probe 3) annealed to the parental minichromosome, but failed to anneal to the new chromosomal element (Figure 1F and G, compare lanes 1 with 2–4). These findings indicated that extensive LOH was associated with the complete loss of the right arm of the minichromosome in arg+ G418S ade− his− colonies. The stability of this chromosomal element was found to be equivalent to that of the parental minichromosome (Supplementary Table 1).
LOH associated with chromosome arm loss is break site independent
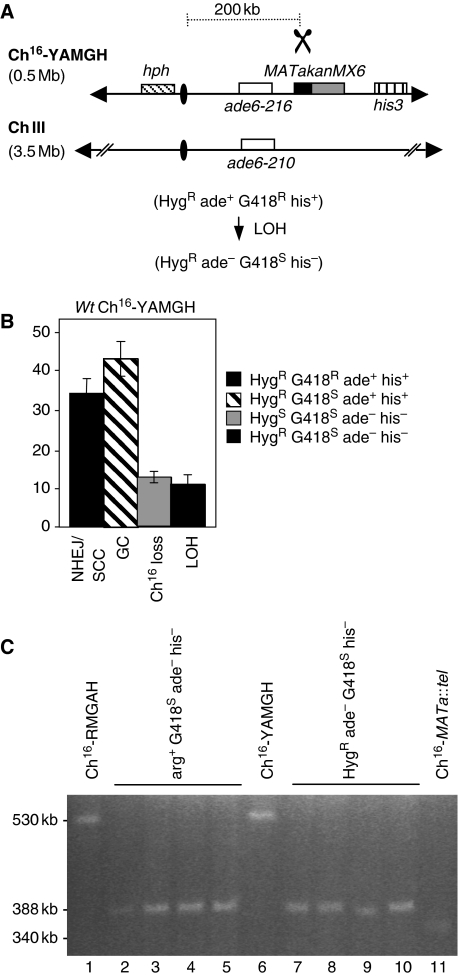
This 388-kb break-induced chromosomal element derived from Ch16-RMGAH was distinct from the previously observed ∼2-Mb chromosomal element, ChX, derived from break-induced LOH of Ch16-MGH, in which the centromere-proximal arm was maintained (Cullen et al, 2007). As the MATa site in Ch16-RMGAH (integrated into spcc23b6.06) was 57-kb centromere-proximal to the site in Ch16-MGH (rad21), it is possible that these apparently different mechanisms of break-induced LOH were locus specific. Alternatively, these differences could reflect the different positions of the arg3 marker on the left arm of Ch16-RMGAH, and the ade6-216 marker on the right arm of Ch16-MGH, which were used to detect extensive break-induced LOH (compare Figures 1A and 2A). To distinguish between these possibilities, a hygromycin (hyg) resistance gene (hph) was introduced into the left arm of the minichromosome Ch16-MGH to form Ch16-YAMGH (Figure 2A). After break induction in cells carrying Ch16-YAMGH, 12% of the colonies were found to be HygR ade− G418S his−, thus showing extensive LOH levels that were comparable with those of extensive LOH observed in Ch16-RMGAH (Figure 2B). Further, PFGE analysis of the minichromosomes derived from HygR ade− G418S his− colonies were of a size identical to those derived from Ch16-RMGAH (Figure 2C). These results indicate that both level and mechanism of LOH associated with loss of the broken minichromosome arm were break site independent. The size of minichromosomal elements observed in both break-induced LOH populations from Ch16-RMGAH and Ch16-YAMGH were larger than what would have been expected if they had just lost their right arm (∼180 kb), and were also larger than a minichromosome derived from Ch16-RMGAH which had undergone de novo telomere addition at the break site (∼340 kb; Figure 2C compare lanes 2–5 and 7–10 with lane 11). These findings together suggest that break-induced extensive LOH had resulted from a new mechanism.
Figure 2.
Extensive LOH is break-site independent. (A) Schematic of Ch16-YAMGH. Centromeric regions (ovals), complementary heteroalleles (ade6-M216 and ade6-M210; white), the MATa site (black) with an adjacent kanMX6 resistance marker (grey) and the his3 marker (vertical stripes) are shown, as previously described (Cullen et al, 2007). The distance from the MATa site to the centromere is shown. The hph resistance marker gene was inserted into chk1+ on the left arm of Ch16-MGH (Cullen et al, 2007) to form Ch16-YAMGH. Derepression of pREP81X-HO (data not shown) generates a DSB at the MATa target site (scissors). (B) Percentage DSB-induced marker loss in wild-type background using strains TH3315-8 and TH3319-20 (Supplementary Table 5). The levels of non-homologous end joining/sister chromatid conversion (NHEJ/SCC), gene conversion (GC), minichromosome loss (Ch16 loss), and LOH are shown. s.e.m. values are indicated. (C) High-resolution PFGE analysis from wild-type Ch16-RMGAH (TH2130; lane1), individual wild-type arg+ G418S his− ade− (LOH) strains isolated after DSB induction (lanes 2–5), wild-type Ch16-YAMGH (TH3317; lane 6), individual wild-type HygR ade− G418S his− (LOH) strains isolated after DSB induction (lanes 7–10), and Ch16-MATa∷tel (lane 11).
Break-induced LOH results from isochromosome formation
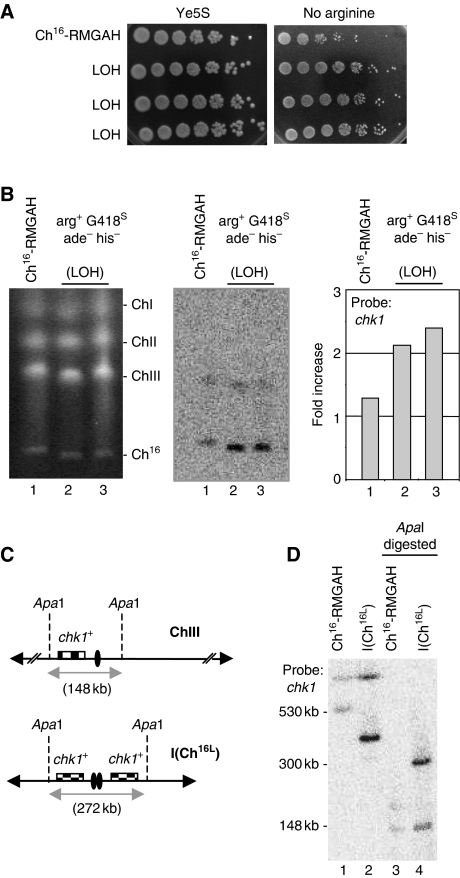
Duplication of the left arm of the minichromosome and centromere to form an isochromosome could account for the unexpected large size of this chromosomal element. Consistent with this, an increased growth rate on plates lacking arginine (arg−), but not Ye5S plates, was observed in arg+ G418S ade− his− colonies compared with the parental strain carrying Ch16-RMGAH, suggesting that the arg3 gene on the left arm of the minichromosome had been duplicated (Figure 3A). In addition, Southern blot analysis using a chk1 probe, which is specific to the left arm of both ChIII and Ch16-RMGAH, indicated that the LOH minichromosomal element showed an increased signal intensity compared with the parental Ch16, after normalization against the endogenous ChIII chk1 signal. This result is again consistent with duplication of the left arm of the minichromosome in the LOH colonies (Figure 3B).
Figure 3.
Extensive LOH arises from isochromosome formation. (A) Spot dilutions of wild-type Ch16-RMGAH (TH2130) and three individual wild-type arg+ G418S his− ade− strains (LOH) on Ye5S and EMM plus uracil, histidine, adenine and thiamine (no arginine) plates. (B) Left panel: PFGE analysis of chromosomal DNA from wild-type Ch16-RMGAH (TH2130; lane1) and individual wild-type arg+ G418S his− ade− (LOH) strains isolated after DSB induction (lanes 2 and 3). Middle panel: Southern blot of the PFGE probed with chk1. Right panel: quantification of the Southern blot indicating the fold increase over the chk1+ background present on ChIII. (C) Positions of relevant ApaI sites are indicated for the native chromosome III (ChIII; 148 kb apart), and for an isochromosome (ICh16L; ∼272 kb apart). Centromeric regions indicated by ovals. Checked boxes indicate the position of chk1. (D) Southern blot analysis of chromosomal DNA digested using ApaI and probed with chk1.
To further confirm the duplication of the left arm, restriction digestions using ApaI, present only once on the left arm of the minichromosome, were carried out. Digestion of genomic DNA from the parental strain carrying Ch16-RMGAH gave the expected band of 148 kb, when chk1 was used as a probe. If the left arm of the minichromosome was duplicated together with the centromere to form an isochromosome, a band of 272 kb would be expected (Figure 3C). ApaI digestion of genomic DNA derived from an arg+ G418S ade− his− (LOH) colony generated a band consistent with this (Figure 3D, lane 4). A shorter band is also observed in this lane, consistent with the 148-kb band expected from ApaI digestion of the endogenous ChIII.
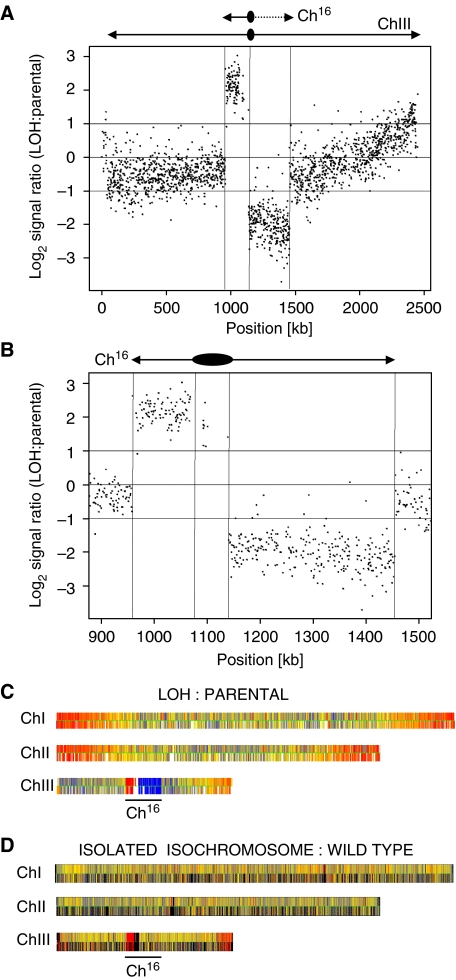
To investigate the degree of duplication in more detail, comparative genome hybridization (CGH) was used to examine the genomes of the arg+ G418S ade− his− (LOH) colonies and the parental strain carrying Ch16-RMGAH. Genomic DNA samples from the two strains were differentially hybridized to ORF and intergenic microarrays covering the coding and non-coding regions of the fission yeast genome (Materials and methods). The hybridization signals for each region obtained from the LOH strain were divided by those from the parental strain to give the hybridization (signal) ratio, a measure of copy number variation across ChIII. CGH analysis of LOH and parental strains carrying Ch16-RMGAH showed reduced log2 hybridization ratios across the right arm of the minichromosome, thus confirming the absence of the right arm of the minichromosome in the LOH colonies (Figure 4A). CGH analysis also indicated increased ratios across the intact left arm of the minichromosome, strongly suggesting that the entire left arm of the minichromosome had been duplicated, consistent with isochromosome formation (Figure 4A and B). The breakpoint at which the right arm of the minichromosome was lost could be estimated from the CGH data. Given the resolution of the platform, the breakpoint was found to lie in a 1.3-kb region containing part of the tRNA Lys.11 and some of its downstream sequences (from position 1 139 584 to 1 140 929 on the endogenous ChIII; Figure 5A). This region lies just outside of the otr3R region (Allshire, 2004), and immediately centromere-proximal to an inverted repeat element, irc3-R. A strikingly similar breakpoint was also obtained after the analysis of HygR ade− G418S his− colonies derived from Ch16-YAMGH (our unpublished results). Remarkably, CGH analysis of some isochromosome-containing strains additionally revealed amplification of endogenous chromosomal regions, most frequently near chromosomal ends (Figure 4C). This was not associated with increased isochromosome numbers per cell (our unpublished results). Further CGH analysis of DNA using an isolated isochromosome against wild-type genomic DNA identified the far right arm of endogenous ChIII to exhibit an increased copy number. As this amplification is absent in the wild-type strain the duplicated endogenous chromosomal region must have been present on the isolated isochromosome (Figure 4D). Thus isochromosome formation can also include duplication of endogenous chromosomal regions leading to genome-wide copy number variation.
Figure 4.
Comparative genome hybridization (CGH) analysis of isochromosomes. (A) CGH, showing the log2 of the signal ratio between wild-type Ch16-RMGAH (TH2125) and a wild-type arg+ G418S his− ade− (LOH) strain (TH4313) across ChrIII isolated after DSB induction. Locations of Ch16 and ChIII centromeres (oval) and telomeres (arrows) are indicated. Data acquisition and normalization were carried out as described in Materials and methods. (B) CGH analysis of LOH and parental strains. Vertical lines indicate the location of the centromere and telomeres. (C) CGH of the above strains showing three endogenous chromosomes. Yellow indicates a 1:1 ratio. Red indicates signal intensity >1. Blue indicates signal intensity <1. (D) CGH of an isolated isochromosome against a wild-type strain without a minichromosome (TH400) showing three endogenous chromosomes. Colour coding as above.
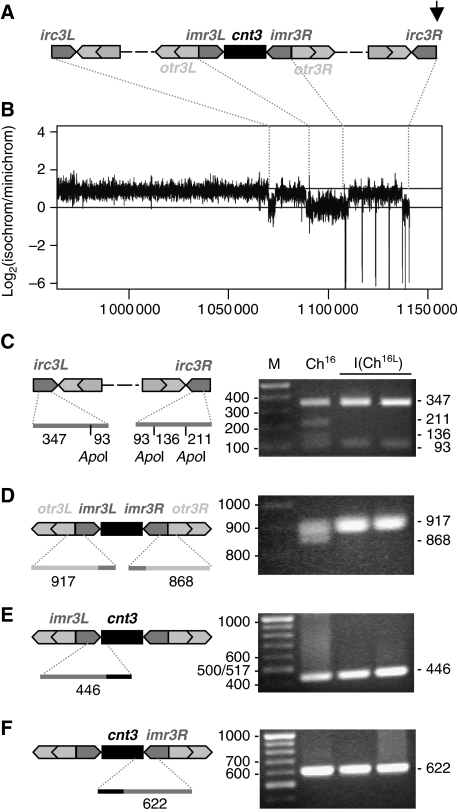
Figure 5.
Analysis of the isochromosome centromere. (A) Schematic of predicted centromere structure of Cen-Ch16 based on homologous cen3, with the break point of Ch16-RMGAH and Ch16-YAMGH-derived LOH colonies, indicated by an arrow. cnt3, innermost (imr3), outer (otr3) and irc3 repeats are shown. (B) Log2 ratio of the fold coverage from Illumina GAII sequencing across isolated isochromosome and minichromosome DNA (log2 of isochromosome/minichromosome) from strains TH4313 and TH2125, respectively. Sequence data aligned to S. pombe ChIII reference sequence at positions indicated (C) PCR amplification of irc3L and irc3R using irc3-R and irc3-F primers, digested using ApoI, (D) PCR amplification of imr3–otr3 junctions using imr-out and dh primers (E) PCR amplification of imr3L–cnt3 junction using primers cnt3-L and imr3-in (F) PCR amplification of cnt3–imr3R junction using primers cnt3-R and imr3-in, separated on a 2% agarose gel and stained with EtBr. Diagnostic PCR product sizes are shown. PCR amplification was carried out as described previously (Nakamura et al, 2008).
Analysis of the isochromosome centromere
To investigate the centromere structure of the isochromosome further, an isochromosome and parental minichromosome were isolated and sequenced using an Illumina GAII with 36-bp short reads. Sequence analysis of the isochromosome confirmed the absence of the broken right arm and the presence of an intact left arm, although no additional ectopic sequences were identified. Moreover, no sequence junctions were identified at or near the break site, suggesting that duplication of the left arm was not associated with an ectopic invasion of non-homologous sequence. Sequence comparison between the isochromosome and the parental minichromosome revealed over 100 short deletions within the isochromosome centromere ranging from 1 to 264 bp in length. These regions do not to correspond to any obvious sequence structure (our unpublished results). The variation in depth of coverage across the isochromosome and minichromosome was compared, as this is indicative of the copy number at any particular locus. Using the Schizosaccharomyces pombe genome sequence as reference, Figure 5B shows the variation in log2 ratio of coverage. Such analysis indicated that although there was a 100–500-fold overall coverage of both the minichromosome and isochromosome, the left arm of the isochromosome had approximately twice the sequence coverage compared with that of the minichromosome, consistent with the duplication of the left arm. Analysis of the centromeric region indicated that although the otr3L repeats had been duplicated, the imr3L, cnt3 and imr3R regions corresponding to central domain had not been duplicated, suggesting that the isochromosome was functionally monocentric (Pidoux and Allshire, 2004). Furthermore, the irc3L and irc3R regions seem not to have been duplicated. This analysis also identified short repeated sequence gaps in otr3R and irc3R regions, which corresponded, in part, to the deleted regions identified from the sequence analysis, suggesting that these sequences were absent from the isochromosome, and/or that otherwise identical repeats derived from elsewhere were being aligned to this region (Figure 5B). To determine the position at which the right centromeric arm was lost and the left arm was duplicated, PCR analysis was carried out. irc3L differs from irc3R by 120 bp, which contains an ApoI site (Nakamura et al, 2008). PCR amplification and ApoI digestion revealed the presence of the irc3R sequence within the minichromosome, however, this was absent in two isochromosome isolates (Figure 5C). Further PCR analysis revealed that the isolated isochromosomes had lost the otr3R–imr3R junction while retaining the otr3L–imr3L, imr3L–cnt3 and cnt3–imr3R junctions (Figure 5D–F) (Nakamura et al, 2008). These findings together with the CGH analysis and isochromosome size are consistent with isochromosome formation having resulted from loss of the right arm and duplication of the left arm of the centromere and minichromosome either at or near the imr3R junction.
Rhp51, Rhp55 and Rhp57 suppress break-induced isochromosome formation
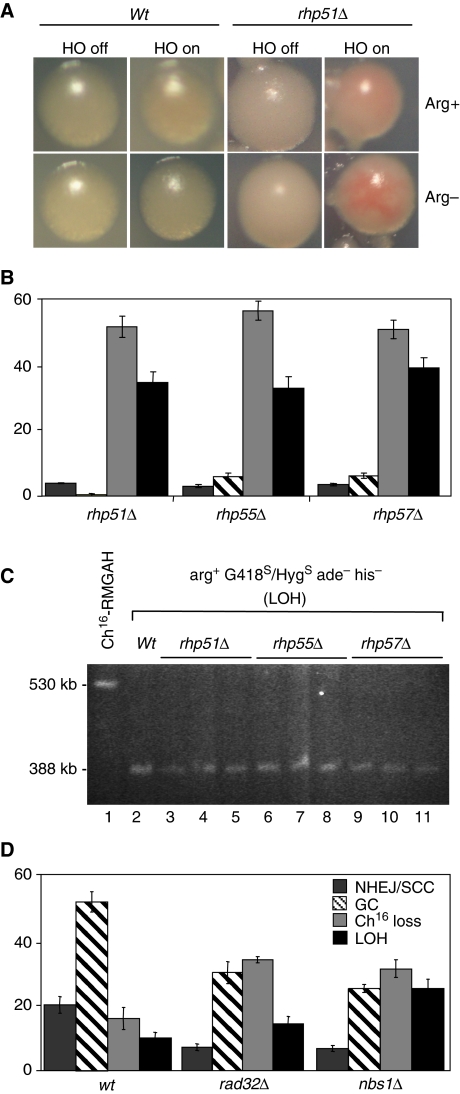
To identify suppressors of break-induced LOH, we screened for mutants that exhibited elevated levels of break-induced LOH, thus resulting in colony sectoring on low adenine plates (described in Supplementary Data). One candidate, loh4-1, exhibited reproducible break-induced sectoring, and showed a marked increase in both minichromosome loss (46% P=0) and extensive LOH (25% P=0.01), which was consistent with isochromosome formation (Supplementary Figure 1). Further analysis indicated that loh4-1 encoded a mutation in the rad51 gene, in which the glutamic acid residue at position 344 was substituted by a lysine in the Rad51 domain (E344K; Supplementary Figure 1). This indicated that mutating rhp51 resulted in a significant increase in break-induced LOH. To test this further, sectoring and marker loss in an rhp51∷ura4 deletion background was examined. Break-induced sectoring was observed in an rhp51Δ background either in the presence or absence of arginine (Figure 6A). High levels of break-induced LOH (35% P=0.01), minichromosome loss (52% P=0) and striking reduction in gene conversion (0.5% P=0 against wild type) were observed in the rhp51Δ mutant, thus confirming a role for rhp51+ in suppressing break-induced LOH (Figure 6B). Subsequent analysis of 21 rhp51Δ arg+ G418S ade− his− colonies indicated that they each carried truncated minichromosomes of a size identical to that of a known isochromosome (Figure 6C, compare lanes 2 with 3–5). CGH analysis of genomic DNA from an rhp51Δ arg+ G418S ade− his− strain confirmed the loss of the right arm and duplication of the entire left arm, again consistent with isochromosome formation (our unpublished results).
Figure 6.
HR genes suppress break-induced isochromosome formation. (A) Colony colouration of arg+ G418S/HygS his− ade− wild-type (wt) (TH2130) or rhp51Δ colonies (TH2945) grown on EMM plus uracil, histidine and low adenine (5 mg/l) with (arg+) or without (arg−) arginine, in the presence (HO off) or absence (HO on) of thiamine (see Supplementary Data). (B) Percentage DSB-induced marker loss in rhp51Δ (TH2945-7, TH2942-4), rhp55Δ (TH3407, TH3847-8) and rhp57Δ (TH3243, TH3869-70). The levels of non-homologous end joining/sister chromatid conversion (NHEJ/SCC), gene conversion (GC), minichromosome loss (Ch16 loss), and LOH are shown. s.e.m. values are indicated. (C) High-resolution PFGE analysis of individual arg+ G418S/HygS his− ade− (LOH) colonies from wild-type (lane 2) rhp51Δ (lane 3–5), rhp55Δ (lane 6–8) and rhp57Δ (lane 9–11) backgrounds after DSB induction are shown. (D) Percentage DSB-induced marker loss in wild-type (TH2130-3 and TH2357) (wt), rad32Δ (TH3966-9, TH3971-2) and nbs1Δ (TH3918-21, TH3922) with outcome classifications as above. s.e.m. values are indicated.
These findings prompted us to test the possible roles of Rhp55 and Rhp57 in suppressing break-induced LOH. Break-induced LOH (33% P=0.01) and minichromosome loss (56% P=0) were significantly increased in an rhp55Δ background, and gene conversion significantly reduced (6% P=0) compared with wild type (Figure 6B). Similarly, break-induced LOH (39% P=0) and minichromosome loss (51% P=0) were significantly increased, and gene conversion significantly reduced (6% P=0) in an rhp57Δ background compared with wild type (Figure 6B). Further, analysis of approximately 20 arg+ HygS ade− his− colonies obtained from either rhp55Δ or rhp57Δ backgrounds indicated that the minichromosomes had been truncated (Figure 6C), and CGH analysis of representative arg+ HygS ade− his− colonies confirmed that these contained isochromosomes (our unpublished data). These data together identify a role for homologous recombination genes rhp51+, rhp55+ and rhp57+ in efficiently suppressing both break-induced minichromosome loss and break-induced LOH.
The MRN complex suppresses break-induced isochromosome formation
To examine the role of HR further, levels of extensive LOH were also examined in strains in which the MRN complex was disrupted. The level of break-induced minichromosome loss was increased in a rad32Δ background (37% P=0.01) compared with wild type, as was break-induced LOH (16% P=0.13). This corresponded with a significant reduction in the levels of gene conversion (32% P=0.02) compared with wild type (53%; Figure 6D). Similarly, in an nbs1Δ background, levels of both LOH (27% P=0.02) and minichromosome loss (34% P=0.01) were significantly increased, compared with those of wild type, whereas levels of gene conversion were significantly reduced (27% P=0; Figure 6D). The break-induced marker-loss profile in nbs1Δ was not significantly different from rad32Δ. Further analysis of each of these mutant backgrounds indicated that arg+ G418S ade− his− (LOH) colonies had truncated minichromosomes consistent with isochromosome formation (our unpublished results). Thus the MRN complex suppresses both break-induced isochromosome formation and minichromosome loss.
Isochromosome formation results from extensive end processing
We wished to test whether the loss of the right arm of the minichromosome during isochromosome formation arose through extensive end processing associated with failed gene conversion following break induction.
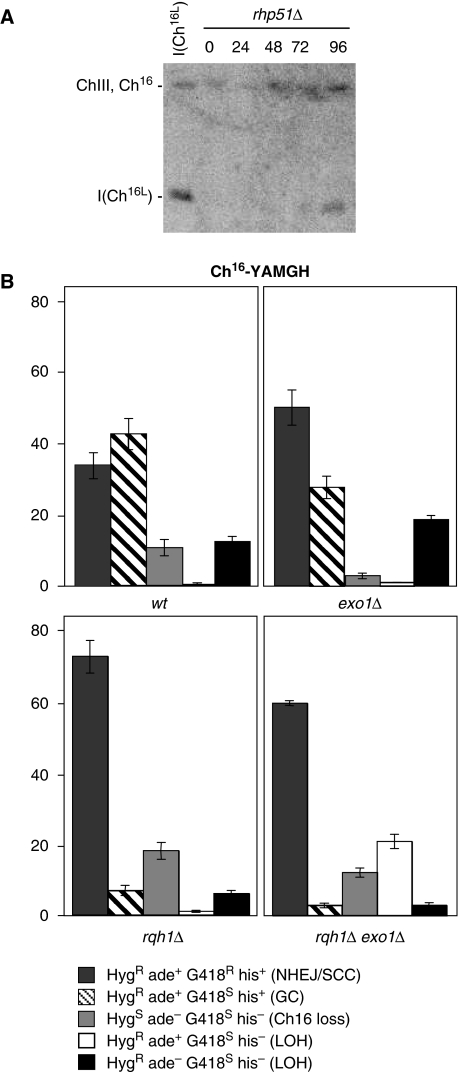
The. resection speed in S. cerevisiae has been estimated to be 4.4 kb per h (Zhu et al, 2008). Although the resection speed has not yet been established in S. pombe, resection from the DSB site in Ch16-RMGAH to the centromere ∼160 kb away would be predicted to take ∼36 h following break induction that occurs ∼18–24 h after thiamine removal (Prudden et al, 2003). Consistent with this, a time course of isochromosome formation in an rhp51Δ background revealed that isochromosomes could be weakly detected 48 h after thiamine removal, but were more strongly observed at later times (Figure 7A). Thus the timing of isochromosome formation is consistent with extensive end processing from the MATa break site.
Figure 7.
Break-induced isochromosome formation requires extensive end processing. (A) PFGE analysis of samples taken from rhp51Δ cells carrying Ch16-RMGAH grown in absence of thiamine for the indicated time periods (see Materials and methods). Bands corresponding to Chromosome III, (ChIII), minichromosome, (Ch16), and isochromosome, I(Ch16L), are indicated. (B) Percentage DSB-induced marker loss in wt (TH3315-20), exo1Δ (TH4422,24; TH3319,20), rqh1Δ (TH3896-8, TH3899-01), or rqh1Δ exo1Δ, (TH4344-7, TH4348-50) backgrounds carrying Ch16-YAMGH. The levels of non-homologous end joining/sister chromatid conversion (NHEJ/SCC), gene conversion (GC), minichromosome loss (Ch16 loss), and LOH types are shown. s.e.m. values are indicated.
Exo1 and Sgs1 have recently been shown to function in extensive end processing in S. cerevisiae (Zhu et al, 2008; Mimitou and Symington, 2009). Therefore to test whether isochromosome formation resulted from extensive end processing, levels of break-induced marker loss were examined in strains containing Ch16-YAMGH in which the homologous S. pombe rqh1+ and exo1+ genes were deleted. Deletion of exo1+ in this context was found to result in significantly reduced levels of gene conversion (27% P=0.02) and chromosome loss (3% P=0.02), whereas levels of HygR ade− G418S his− LOH colonies were found to be increased (19% P=0.01) compared with wild type (Figure 7B). Deletion of rqh1+ resulted in a significant increase in NHEJ/SCC, (68% P=0) consistent with other studies (Hope et al, 2006). This was associated with a significant reduction in gene conversion (6% P=0), and reduced levels of both HygR ade+ G418S his− LOH colonies (1% P=0.07) and HygR ade− G418S his− LOH colonies (6% P=0). Similarly, DSB induction in an exo1Δ rqh1Δ background resulted in high levels of NHEJ/SCC (58% P=0), and significantly reduced levels of gene conversion (3%; P=0 Figure 7B). However, LOH resulting in HygR ade+ G418S his− colonies in an exo1Δ rqh1Δ background was increased significantly compared with wild type (20%; P=0), rqh1Δ (1%; P=0) and exo1Δ (1%; P=0) mutants (Figure 7B). In contrast, levels of LOH resulting in HygR ade− G418S his− colonies were significantly reduced (3%; P=0) compared with wild type, exo1Δ (P=0) and rqh1Δ (P=0.04) backgrounds (Figure 7B). PFGE analysis indicated that LOH was associated with truncations in the HygR ade+ G418S his− colonies, whereas LOH resulted from isochromosome formation in HygR ade− G418S his− LOH colonies (our unpublished results). As the ade6-M216 marker, 30-kb centromere-proximal to the MATa break site in Ch16-YAMGH, is retained to a significantly greater degree in an rqh1Δ exo1Δ background, these findings are consistent with Rqh1 and Exo1 together being required for extensive end processing and efficient isochromosome formation.
Isochromosome formation results from replication of the intact arm
Break-induced isochromosome formation could have arisen from replication of the intact left arm of the chromosome following extensive end processing. If the intact arm was replicated, both daughter cells would be expected to inherit an isochromosome. Alternatively, isochromosome formation could have arisen through break-induced isochromatid fusion in which two broken sister chromatids fuse following loss of their right arms. Such a mechanism would result in co-segregation of the fused two left arms of the sister chromatids into one of the daughter cells at mitosis. To distinguish between these possibilities, pedigree analysis was carried out using a strain carrying Ch16-RMGAH, in which elongated cells, indicative of DSB-induced checkpoint activation, were allowed to divide and were separated. Marker loss and chromosome sizes were subsequently determined for colonies formed from each separated daughter cell. Of 800 daughter cells separated, 578 (72%) grew, of which 135 (23%) formed arg+ G418S ade− his− (LOH) colonies. Of these, 93 (69%) had a viable sister. The sisters of 25 (27%) of these also formed arg+ G418S ade− his− (LOH) colonies, and which were found to be consistent with isochromosome formation by PFGE analysis. Broken sister chromatids were also repaired predominantly by gene conversion (27%). In addition, the sisters of 40 LOH colonies (43%) were arg− G418S ade− his−, consistent with either failed repair, or isochromosome formation by ligation of truncated sister chromatids and co-segregation into one of the two daughters (Supplementary Table 2). Similar results were obtained from pedigree analysis of a wild-type strain carrying the Ch16-YAMGH minichromosome and an rhp51Δ strain carrying Ch16-RMGAH (Supplementary Tables 3 and 4). For both daughter cells to contain an isochromosome, or for one to contain an isochromosome and the other to contain an intact minichromosome repaired by either NHEJ or gene conversion (48% of all the LOH colonies), duplication of the left arm of the minichromosome must have arisen through replication rather than co-segregation event.
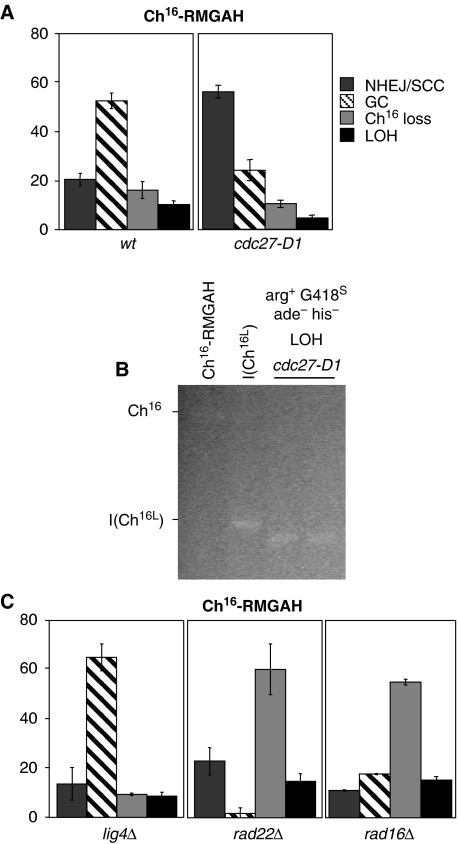
Pol32 has been shown to be required for BIR in S. cerevisiae (Lydeard et al, 2007). To test the possible role of BIR in isochromosome formation, marker loss was examined in a strain containing a cold-sensitive mutation in the fission yeast homologue, Cdc27 (Tanaka et al, 2004). DSB induction in a cdc27-D1 mutant within Ch16-RMGAH at 30°C resulted in significantly increased levels of NHEJ/SCC (59%; P=0), markedly reduced levels of gene conversion (25%; P=0.02), and reduced minichromosome loss (11% P=0.17). Furthermore, levels of break-induced LOH were significantly reduced (5%; P=0.03) compared with those of wild type (Figure 8A). PFGE analysis of 20 LOH colonies revealed that these had resulted from truncations and not from isochromosome formation (Figure 8B). Thus the Pol32 homologue, Cdc27 is required for isochromosome formation in S. pombe, which is strongly suggestive of BIR as the mechanism by which the left arm is replicated during isochromosome formation.
Figure 8.
Genetic determinants of break-induced isochromosome formation. (A) Percentage DSB-induced marker loss of strains carrying Ch16-RMGAH in wild-type (wt) (TH2130-3, TH2357) and cdc27-D1 (TH4460-1, TH4463-4) backgrounds at 30°C. The levels of non-homologous end joining/sister chromatid conversion (NHEJ/SCC), gene conversion (GC), minichromosome loss (Ch16 loss), and LOH are shown. s.e.m. values are indicated. (B) High-resolution PFGE analysis of parental wild-type (lane 1) and individual arg+G418S his−ade− (LOH) colonies from wild-type (lane 2) and cdc27-D1 background (lane 3–4) after DSB induction. (C) Percentage DSB-induced marker loss in lig4Δ (TH3569-70, TH3575-7), rad22Δ (TH4135-7, TH4138-9) and rad16Δ (TH4156-9, TH4161-2) backgrounds with outcome classification as above. s.e.m. values are indicated.
In contrast, levels of LOH in a lig4Δ background (9%), in which ligation through NHEJ is abrogated (Manolis et al, 2001), were similar to those of wild type (Figure 8C), and arose through isochromosome formation (our unpublished results). Thus isochromosome formation is Lig4 independent, and unlikely to have resulted from ligation of two resected sister chromatids. In S. cerevisiae, single-strand annealing requires Rad52 (Rad22Sp) and Rad1 (Rad16Sp), but is Rad51 independent (Haber, 2006). Although levels of break-induced minichromosome loss were significantly increased in a rad22 null mutant background (60% P=0.04), levels of LOH (15%) were similar to those of wild type (Figure 8C), and arose through isochromosome formation (our unpublished results). Similarly, in a rad16Δ background, although levels of break-induced minichromosome loss were significantly increased (55.1% P=0), presumably as a result of being unable to remove the non-homologous MATa–KanMX tails during repair (Prudden et al, 2003), levels of LOH (15%) were similar to those of wild type, and arose through isochromosome formation (our unpublished results). Thus single-strand annealing seems not to be required for isochromosome formation.
Discussion
A new model for isochromosome formation
Isochromosome formation is associated with particular genetic disorders, including Turner syndrome (Palmer and Reichmann, 1976) and Pallister–Killian syndrome (Reynolds et al, 1987); malignant diseases such as therapy-related myelodysplastic syndrome (Andersen and Pedersen-Bjergaard, 2000); and is frequently observed in specific types of human cancers, including testicular germ cell tumours, in which it is used as a characteristic genetic marker (Atkin and Baker, 1982; Putnam et al, 2005; Mitelman et al, 2008). Further, gene duplication through isochromosome formation is associated with diverse forms of drug resistance such as azole resistance in the fungal opportunist Candida albicans (Selmecki et al, 2008).
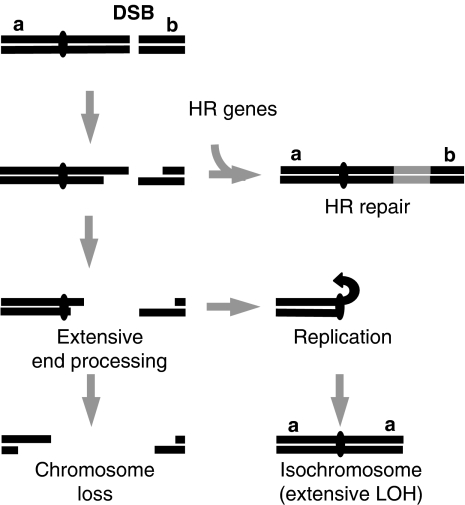
Several mechanisms of isochromosome formation have been proposed which can lead to either a classically defined isochromosome, in which a single functional centromere separates two inverted identical chromosome arms, or a broader class of chromosome rearrangements, including isodicentrics or duplicated non-identical arms. These mechanisms include centromere misdivision (Darlington, 1939), homologous chromosomal interchanges (Lorda-Sanchez et al, 1991), sister chromatid breakage and reunion (Phelan et al, 1988) or crossing over within an inversion loop (Therman et al, 1974). Partial isochromosomes have also previously been shown to result from BIR in S. cerevisiae in which chromosome ends are duplicated (Bosco and Haber, 1998). The data presented in this study support a new ‘break chew and copy' mechanism of isochromosome formation, in which after a DSB, extensive end processing associated with failed gene conversion leads to the loss of the broken chromosome arm. The processed end then initiates replication from within the centromere, resulting in duplication of the intact chromosome arm (Figure 9).
Figure 9.
Model for break-induced isochromosome formation. Extensive end processing arising from failed or inefficient HR repair results in loss of the broken chromosome arm and replication of the intact arm, leading to chromosome loss, isochromosome formation, or potentially other types of chromosomal rearrangement. Distant break-induced chromosomal rearrangements are expected to be more frequent in HR or other mutants that facilitate extensive end processing resulting from failed or inefficient HR repair. See text for details.
The fact that isochromosomes arise through extensive end processing from a break is supported by the following findings: first, isochromosome formation was DSB dependent, but break-site independent, consistent with extensive 5′–3′ degradation arising from un-repaired breaks leading to the loss of the broken chromosome arm. Second, the timing of isochromosome formation was in accordance with the predicted time required for such extensive end processing and subsequent replication. Third, both efficient isochromosome formation and efficient ade6-M216 marker loss 30-kb centromere-proximal from the break-site required Rqh1 and Exo1, whose homologues in S. cerevisiae are required for extensive end processing (Zhu et al, 2008; Mimitou and Symington, 2009). The observation that duplication of the intact arm is initiated by replication as opposed to isochromatid fusion is supported by pedigree analysis, in which isochromosomes were detected in both daughter cells; an outcome that would not be possible if they resulted from isochromatid fusion. In addition, disruption of genes required for NHEJ or SSA, which could have facilitated isochromatid fusion, had little effect on the efficiency of isochromosome formation.
The replication of the intact chromosome arm could have arisen through BIR initiated by strand invasion of the extensively processed end-invading homologous sequences within the centromere. Alternatively, exonucleolytic resection could have exposed single-strand inverted repeat regions, resulting in a hairpin loop close to the centromere, which after a subsequent S phase, might lead to isochromosome formation (Lobachev et al, 2002). Our data support a role for BIR in duplication of the intact arm during isochromosome formation. First, isochromosome formation requires Cdc27, a homologue of Pol32, which is required for BIR in S. cerevisiae (Lydeard et al, 2007). Second, sequence gaps were observed within the centromere which are unlikely to have resulted from normal S phase replication. Third, subtelomeric regions of the endogenous chromosomes were also duplicated on some isochromosomes, which also cannot be easily explained by the hairpin model. This pattern may reflect a subpopulation of isochromosomes whose formation resulted from a number of short ectopic DNA synthesis and dissociation events before telomere capture and chromosome stabilization by the replicated arm. These isochromosomes would be of heterogeneous lengths and thus not readily detectable by PFGE. Such a duplication pattern is consistent with template switching associated with BIR (Smith et al, 2007). These findings are consistent with BIR being mutagenic (Llorente et al, 2008), and further suggest BIR to be a potent mechanism for global copy number variation.
Sequence analysis revealed that that although the left arm of the centromere and minichromosome had been duplicated during break-induced isochromosome formation, the central centromeric region of the isochromosome was not. This central centromeric region consists of two very large (∼13 kb) inverted repeats, each encompassing imr3L and imr3R, respectively, and separated by ∼4.8 kb encompassing cnt3. The absence of the irc3R and otr3R–imr3R junction from the isochromosome and retention of the cnt3–imr3R junction support a model in which end processing initiated at a distant break continues into the right inverted repeat in which the 3′ end is subsequently able to initiate a complex pattern of DNA replication using the left inverted repeat as a replication template. This would result in the duplication of the left arm of the centromere and minichromosome, but not the central centromeric region of the minichromosome encompassing imr3L, cnt3 and imr3R, as observed. It is possible that these inverted repeats form a large hairpin loop structure, resembling the intramolecular loop conformation of pericentric chromatin observed in S. cerevisiae (Yeh et al, 2008). Such a structure could potentially further facilitate BIR. An alternative model in which exonucleolytic resection promotes annealing of the single-strand inverted repeat regions would result in a large single-stranded hairpin loop containing cnt3 that would be potentially unstable, and probably non-functional during the subsequent mitosis.
BIR in this context seems to be Rhp51 independent. This may reflect the context in which strand invasion is predicted to occur. A minor Rad51-independent pathway for strand invasion has also been described in S. cerevisiae, which requires Rad52, but not Rad51, Rad55 or Rad57 (Bartsch et al, 2000; Pohl and Nickoloff, 2008). It is therefore possible that a similar pathway in S. pombe triggers efficient BIR in rhp51Δ, rhp55Δ or rhp57Δ backgrounds in this context. The fact that isochromosome formation was still observed in a rad22 null background, albeit without enhancing isochromosome formation, was unexpected as RAD52, the homologue of rad22+, is required for BIR in S. cerevisiae (Malkova et al, 1996; Bosco and Haber, 1998). Furthermore, the reduced requirement for rad22+ may reflect the highly repetitive centromeric context in which BIR is predicted to be initiated in our assay. It is also possible that the S. pombe homologue, rti1+, could be partially redundant with rad22+, thus facilitating BIR in its absence (Suto et al, 1999; van den Bosch et al, 2001). Alternatively, rhp51+ may facilitate BIR in a rad22 null background, but less efficiently than that facilitated by rad22+, in rhp51Δ, rhp55Δ or rhp57Δ backgrounds. Unfortunately, we were unable to generate viable rad22Δ rti1Δ or rad22Δ rhp51Δ cells containing a minichromosome to test these hypotheses.
Suppressing chromosomal rearrangements
The ‘break, chew and copy' model for isochromosome formation shares features in common with the mechanisms proposed to account for other types of chromosomal rearrangements (Bosco and Haber, 1998; Maringele and Lydall, 2004; VanHulle et al, 2007). In this respect, it is possible that had end-processing exposed different regions of homology or inverted repeats before reaching the centromere, then partial isochromosomes or other chromosomal rearrangements, similar to those previously described may also have arisen. We have previously found extensive LOH to arise from large translocations associated with Rad51-dependent gene conversion in a wild-type background, or through de novo telomere addition at or near the break site, which was prevented by HR through competition for resected ends (Cullen et al, 2007). In these cases, LOH was associated with events occurring at or near the break site, resulting in retention of a selectable ade6-M216 marker proximal to the DSB, and occurred in approximately 2% of the total population (Cullen et al, 2007). In this study, using a minichromosome with markers on both arms of the minichromosome, isochromosome formation was identified as a distinct mechanism of break-induced LOH. This mechanism contrasts with those previously observed in that it is associated with events occurring at a considerable distance from the break site, resulting in the loss of the selectable ade6-M216 marker proximal to the DSB, and indeed loss of the entire broken chromosome arm. These events were not previously detected using Ch16-MGH due to the loss of the selectable ade6-M216 marker proximal to the DSB, but were detected in this study using Ch16-RMGAH, as the selectable arg+ marker was present on the intact left arm of the minichromosome. This mechanism of LOH was more frequently observed in our assays compared with those previously identified, and is likely to have arisen through extensive end processing as a result of failed gene conversion. Consistent with this, isochromosome formation was found to occur significantly more frequently in rhp51Δ, rhp55Δ, rhp57Δ or nbs1Δ backgrounds, in which gene conversion was either abrogated or inefficient. Break induction in a wild-type background led to 10% LOH through isochromosome formation and an additional 16% chromosomal loss, presumably as a result of continued end processing beyond the centromere. Similar ratios of LOH to chromosome loss were observed in rhp51Δ, rhp55Δ, rhp57Δ and nbs1Δ backgrounds. The fact that extensive LOH was associated with minichromosome loss is in accordance with both events having resulted from failed gene conversion.
HR genes have been found to suppress spontaneous and DSB-induced chromosomal rearrangements in S. cerevisiae, (Myung et al, 2001; Myung and Kolodner, 2003; Deem et al, 2008) and S. pombe (Nakamura et al, 2008). Similarly, the role of HR genes, in particular, BRCA2, in suppressing chromosomal rearrangements and tumourigenesis in mammalian cells is well established (Christ et al, 2007). Thus, extensive end processing resulting from failed gene conversion may be a general mechanism driving chromosomal rearrangements. We anticipate that although the precise nature of the rearrangement would be influenced by the sequence of the single-stranded DNA exposed and the location of homologous sequences, the frequency of such events will be determined, in part, by the stability of such rearranged chromosomes and also by the efficiency of HR repair. In this respect other classes of genes required for efficient HR repair may also be predicted to suppress extensive LOH and chromosomal rearrangements, and thus to maintain genome stability.
Materials and methods
Yeast strains, media and genetic methods
All S. pombe strains were cultured, manipulated and stored as previously described by Prudden et al (2003). All strain genotypes are listed in Supplementary Table 5 and details of their construction are described in Supplementary Data.
Site-specific DSB assay
Assay was carried out as described previously by Prudden et al (2003). The percentage of colonies undergoing NHEJ/SCC (arg+ G418R/HygR ade+ his+), gene conversion (arg+ G418S/HygS ade+ his+), minichromosome loss (arg+ G418R/HygR ade+ his+), or LOH (arg+ G418S/HygS ade− his−; HygR ade− G418S his− for Ch16-YAMGH) were calculated. To determine the levels of break-induced minichromosome loss, background minichromosome loss at 48h-T in blank vector assays was subtracted from break-induced minichromosome loss at 48h-T in cells transformed with pREP81X-HO. More than 1000 colonies were scored for each time point and each experiment was carried out three times using three independently derived strains for all mutants tested. Southern blots were carried out as previously described by Prudden et al (2003). For the time course experiment, cells were inoculated onto EMM+U+A+H plates, and collected in 0.05 M EDTA at times indicated, and fixed using 5% formaldehyde before PFGE analysis.
Pedigree analysis
The procedures used in this study for pedigree analysis have been previously described by Cullen et al (2007).
Pulsed field gel electrophoresis
The procedures used in this study for PFGE analysis have been described previously (Cullen et al, 2007). For separation of ApaI-digested Ch16, a 1% chromosomal grade agarose gel was used with the following conditions: 6 V/cm, 120° angle with an initial switch time of 2.16 s and a final switch time of 51.17 s. Samples were separated for 27 h in 0.5 × TBE at 14°C.
Chromosome purification
Isochromosomes and minichromosomes were separated by PFGE, electroeluted from an excised gel fragment and ethanol precipitated. DNA was re-suspended in 50 μl 10 mM Tris, 1 mM EDTA (pH 8.0) and subjected to further analysis as indicated.
Microarray design
The microarrays used were ORF arrays covering the coding regions and intergenic arrays covering all non-coding regions. ORF microarrays were designed as described by Lyne et al (2003). The intergenic microarrays were developed using similar approaches as described by (Heichinger et al, 2006). The inter-probe distance was 1.3 kb on average. The microarrays do not cover the 1.2-Mb rDNA repeats proximal to the telomeres on ChIII and centromere core regions.
Genomic DNA labelling, microarray hybridization, data acquisition and normalization
A total of 0.6 μg of genomic DNA from all experimental and reference samples was labelled using a Bioprime labelling kit (Invitrogen) with either fluorescent Cy3- or Cy5-dCTP (GE Healthcare), and were hybridized to S. pombe cDNA microarrays as previously described (Heichinger et al, 2006). Microarrays were subsequently scanned using a GenePix 4000B laser scanner (Axon Instruments) and fluorescence intensity ratios calculated with GenePix Pro (Axon Instruments). The data were normalized using an in house script (Lyne et al, 2003).
Sequence analysis
Illumina GAII 36-bp reads from the minichromosome and isochromosome were mapped to the S. pombe reference genome using MAQ (Li et al, 2008).
Supplementary Material
Supplementary Data
Acknowledgments
We thank the laboratories of Tony Carr, Hiroshi Iwasaki, Jürg Kohli and Paul Russell for strains and reagents. We further thank Lorne Lonie and Lorna Gregory for Illumina GA II sequencing production, and Sarah Trewick for helpful discussions. This study was funded by the Medical Research Council and Cancer Research UK.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allshire R (2004) Centromere and kinetochore structure and function. In The molecular biology of Schizosaccharomyces pombe, Egel R (ed), Vol. 10, pp 150–169. Heidelberg: Springer-Verlag [Google Scholar]
- Andersen MK, Pedersen-Bjergaard J (2000) Increased frequency of dicentric chromosomes in therapy-related MDS and AML compared to de novo disease is significantly related to previous treatment with alkylating agents and suggests a specific susceptibility to chromosome breakage at the centromere. Leukemia 14: 105–111 [DOI] [PubMed] [Google Scholar]
- Atkin NB, Baker MC (1982) Specific chromosome change, i(12p), in testicular tumours? Lancet 2: 1349. [DOI] [PubMed] [Google Scholar]
- Bartsch S, Kang LE, Symington LS (2000) RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol 20: 1194–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Haber JE (1998) Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150: 1037–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ N, Moynahan M, Jasin M (2007) BRCA2: safeguarding the genome through homologous recombination. In Molecular genetics of recombination, Aguilera A, Rothstein R (eds), pp 363–380. Berlin-Heidelberg: Springer-Verlag [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280: 38631–38638 [DOI] [PubMed] [Google Scholar]
- Cullen JK, Hussey SP, Walker C, Prudden J, Wee BY, Dave A, Findlay JS, Savory AP, Humphrey TC (2007) Break-induced loss of heterozygosity in fission yeast: dual roles for homologous recombination in promoting translocations and preventing de novo telomere addition. Mol Cell Biol 27: 7745–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington C (1939) Misdivision and the genetics of the centromere. J Genet 37: 341–364 [Google Scholar]
- Davis AP, Symington LS (2004) RAD51-dependent break-induced replication in yeast. Mol Cell Biol 24: 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem AK, Barker K, Vanhulle K, Downing BD, Vayl A, Malkova A (2008) Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics 179: 1845–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE (2006) Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amst) 5: 998–1009 [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P (2006) Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope JC, Mense SM, Jalakas M, Mitsumoto J, Freyer GA (2006) Rqh1 blocks recombination between sister chromatids during double strand break repair, independent of its helicase activity. Proc Natl Acad Sci USA 103: 5875–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M (2000) LOH and mitotic recombination. In DNA alterations in Cancer, Ehrlich M (ed), pp 191–209. Natick, MA, USA: Eaton Publishing [Google Scholar]
- Knudson AG (1993) Antioncogenes and human cancer. Proc Natl Acad Sci USA 90: 10914–10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS (2004) Recombination proteins in yeast. Annu Rev Genet 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Lasko D, Cavenee W, Nordenskjold M (1991) Loss of constitutional heterozygosity in human cancer. Annu Rev Genet 25: 281–314 [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS (2008) Break-induced replication: what is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′–3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183–193 [DOI] [PubMed] [Google Scholar]
- Lorda-Sanchez I, Binkert F, Maechler M, Schinzel A (1991) A molecular study of X isochromosomes: parental origin, centromeric structure, and mechanisms of formation. Am J Hum Genet 49: 1034–1040 [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bahler J (2003) Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA 93: 7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE (2005) RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol 25: 933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolis KG, Nimmo ER, Hartsuiker E, Carr AM, Jeggo PA, Allshire RC (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J 20: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2004) Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev 18: 2663–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS (2009) Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci 34: 264–272 [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F (2008) Mitelman Database of Chromosome Aberrations in Cancer
- Myung K, Chen C, Kolodner RD (2001) Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076 [DOI] [PubMed] [Google Scholar]
- Myung K, Kolodner RD (2003) Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2: 243–258 [DOI] [PubMed] [Google Scholar]
- Nakamura K, Okamoto A, Katou Y, Yadani C, Shitanda T, Kaweeteerawat C, Takahashi TS, Itoh T, Shirahige K, Masukata H, Nakagawa T (2008) Rad51 suppresses gross chromosomal rearrangement at centromere in Schizosaccharomyces pombe. EMBO J 27: 3036–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M (1986) Construction of a minichromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Gen Genet 203: 397–405 [Google Scholar]
- Palmer CG, Reichmann A (1976) Chromosomal and clinical findings in 110 females with Turner syndrome. Hum Genet 35: 35–49 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P, Goedecke W, Obe G (2000) Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15: 289–302 [DOI] [PubMed] [Google Scholar]
- Phelan MC, Prouty LA, Stevenson RE, Howard-Peebles PN, Page DC, Schwartz CE (1988) The parental origin and mechanism of formation of three dicentric X chromosomes. Hum Genet 80: 81–84 [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12: 521–534 [DOI] [PubMed] [Google Scholar]
- Pohl TJ, Nickoloff JA (2008) Rad51-independent interchromosomal double-strand break repair by gene conversion requires Rad52 but not Rad55, Rad57, or Dmc1. Mol Cell Biol 28: 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Evans JS, Hussey SP, Deans B, O'Neill P, Thacker J, Humphrey T (2003) Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J 22: 1419–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Pennaneach V, Kolodner RD (2005) Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol Cell Biol 25: 7226–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JF, Daniel A, Kelly TE, Gollin SM, Stephan MJ, Carey J, Adkins WN, Webb MJ, Char F, Jimenez JF, Opitz JM (1987) Isochromosome 12p mosaicism (Pallister mosaic aneuploidy or Pallister–Killian syndrome): report of 11 cases. Am J Med Genet 27: 257–274 [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J (2008) An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol 68: 624–641 [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS (2007) Template switching during break-induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE (2003) In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sung P (1997a) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197 [DOI] [PubMed] [Google Scholar]
- Sung P (1997b) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Suto K, Nagata A, Murakami H, Okayama H (1999) A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol Biol Cell 10: 3331–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW (1983) The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Ryu GH, Seo YS, MacNeill SA (2004) Genetics of lagging strand DNA synthesis and maturation in fission yeast: suppression analysis links the Dna2–Cdc24 complex to DNA polymerase delta. Nucleic Acids Res 32: 6367–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therman E, Sarto GE, Patau K (1974) Apparently isodicentric but functionally monocentric X chromosome in man. Am J Hum Genet 26: 83–92 [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol 23: 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H (2000) Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell 11: 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M, Vreeken K, Zonneveld JB, Brandsma JA, Lombaerts M, Murray JM, Lohman PH, Pastink A (2001) Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat Res 461: 311–323 [DOI] [PubMed] [Google Scholar]
- VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A (2007) Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol 27: 2601–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner B, van Komen S, Sung P, Peterson CL (2003) Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol Cell 12: 221–232 [DOI] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS (2008) Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol 18: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data