Abstract
Fourteen different genes included in a DNA fragment of 18 kb are involved in the aerobic degradation of phenylacetic acid by Pseudomonas putida U. This catabolic pathway appears to be organized in three contiguous operons that contain the following functional units: (i) a transport system, (ii) a phenylacetic acid activating enzyme, (iii) a ring-hydroxylation complex, (iv) a ring-opening protein, (v) a β-oxidation-like system, and (vi) two regulatory genes. This pathway constitutes the common part (core) of a complex functional unit (catabolon) integrated by several routes that catalyze the transformation of structurally related molecules into a common intermediate (phenylacetyl-CoA).
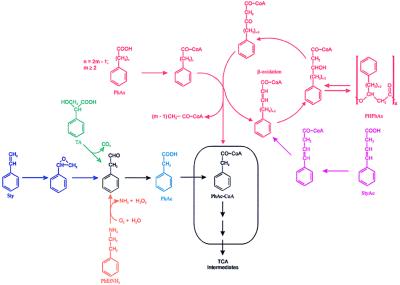
Pseudomonas putida U catabolizes phenylacetic acid (PhAc) aerobically through a catabolic pathway that involves the participation of a phenylacetyl-CoA ligase (PhAc-CoAL) (1, 2). This enzyme has been purified to homogeneity and its gene has been cloned, sequenced, and expressed in different microbes (3). PhAc-CoAL was induced when P. putida U was cultured in chemically minimal defined media (MM) containing as carbon sources PhAc, 2-phenylethylamine (PhEtNH2), phenylacetaldehyde, trans-styrylacetic acid (StyAc), styrene (Sty), or several phenylalkanoic acids (PhAs) with an even number of carbon atoms (4). However, other structurally related compounds, such as hydroxy derivatives of PhAc, l-phenylalanine, l-tyrosine, phenylglycine, or PhAs containing an odd number of carbon atoms, did not induce this enzyme (2, 4). These data suggest that the PhAc catabolic pathway is a convergent route in which other pathways responsible for the catabolism of structurally related compounds end. The ensemble of all them is defined here as a catabolon, and its specific name is given as a function of the common intermediate, PhAc-CoA (see Fig. 1). The genetic organization of the central pathway (catabolon core) and the description of the proteins involved are reported and discussed.
Figure 1.
Biochemical organization of the PhAc-CoA catabolon. PHPhAs, poly(3-hydroxyphenylalkanoic acid)s; TA, tropic acid; TCA, tricarboxylic acid cycle. Box indicates the catabolon core.
MATERIALS AND METHODS
Materials.
Molecular biology products and radioactive molecules were supplied by Amersham. Aromatic compounds were obtained from Lancaster Synthesis. 2-OH-PhAc-CoA was synthesized enzymatically by using 2-hydroxyphenylacetic acid (2-OH-PhAc) as a substrate of PhAc-CoAL (1).
Strains and Vectors.
The P. putida strain U (Colección Española de Cultivos Tipo 4848) was from our collection (1–4). Escherichia coli DH5α′ (Life Technologies, Rockville, MD) was used for plasmid propagation. E. coli strain NM538 (5) was employed for the generation and amplification of a P. putida genomic library. E. coli HB101, carrying the plasmid pGS9, which includes the transposon Tn5 (6), was kindly supplied by J. L. Ramos (Consejo Superior de Investigaciones Cientificas, Granada, Spain). E. coli (pRK600) was used as a helper strain in the triparental filter mating (7). E. coli MC4100 and plasmid pRS551 were specifically used for the identification of promoters (8). Commercial plasmids pUC18, pBSIISK, or KS (Stratagene) were used for subcloning genomic fragments, and the pK18::mob (9) was employed to induce specific gene disruption. DNA manipulations and sequence analysis were carried out as reported previously (10, 11).
Isolation of Mutants.
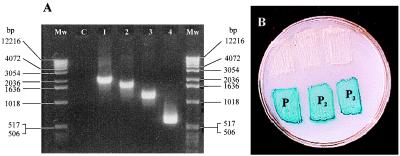
Mutants handicapped in the aerobic catabolism of PhAc (PhAc−) were selected by random insertion of transposon Tn5 (4, 12). The locations of the specific points at which integration of Tn5 occurred were determined by PCR amplification (Fig. 2A).
Figure 2.
(A) PCR amplification of the DNA fragments included between a sequence of Tn5 (5′ → 3′ ACTTGTGTATAAGAGTCAG) and different oligonucleotides present in the genome of P. putida U separated by 400–600 bp. Mw, size markers; C, control without oligonucleotides. (B) Analysis of the P1, P2, and P3 promoters. The existence of a functional promoter implies the expression of the lacZ gene present in the plasmid pRS551. Blue color is generated by the hydrolysis of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal). The upper three patches are E. coli MC4100 transformed with the parental promoter-probe plasmid; the lower three patches are E. coli MC4100 transformed with the recombinant plasmid carrying the indicated promoter.
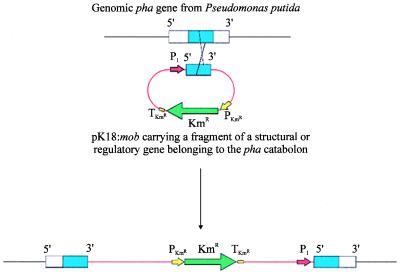
To analyze the function of a particular gene, it was disrupted by homologous recombination. Thus, an internal fragment (300–900 bp) belonging to the gene to be mutated was cloned in pK18:mob (9), and the resulting construct was introduced into P. putida U by triparental mating (7). To ensure the expression of the genes located downstream from the disrupted gene, a P. putida promoter was placed upstream of the internal fragment cloned in pK18:mob (Fig. 3). After 2 days of incubation at 30°C, single transconjugant colonies were isolated and replicated in LB (10) or in the required medium. PhAc− mutants were classified according to the Tn5 insertion point or as a function of the accumulated catabolite. Samples of either culture broth or cell-free extracts (1 ml) were analyzed by HPLC.
Figure 3.
Gene disruption by homologous recombination. P1, P. putida promoter belonging to the pha catabolic pathway; KmR, kanamycin-resistance gene; PKmR, kanamycin-resistance promoter; TKmR kanamycin-resistance terminator.
HPLC Analyses.
The rate of utilization of carbon sources and the identification of the intermediates accumulated by each mutant were analyzed by HPLC (4) using as mobile phase 0.2 M KH2PO4, pH 4.5 (component A), and CH3CN (component B) in a linear gradient varying from 95% A/5% B (vol/vol) to 50% A/50% B over 1 h (1 ml⋅min−1), and the absorbance of the eluate was monitored at 254 nm. The retention times (Rts) for 2,5-diOH-PhAc, 3,4-diOH-PhAc, 4-OH-PhAc, 3-OH-PhAc, 2-OH-PhAc, PhAc, StyAc, 4-phenylbutyric acid (PhB), 6-phenylhexanoic acid (PhH), and 8-phenyloctanoic acid (PhO) were 8, 11, 19, 22, 25, 30, 37, 39, 45, and 56 min, respectively. When other phenyl derivatives were analyzed the mobile phase was 0.2 M KH2PO4, pH 4.5/2-propanol (92:8, vol/vol); the Rts for 2-OH-PhAc, PhAc, 2-OH-PhAc-CoA, and PhAc-CoA were 8, 12, 26, and 27 min, respectively. OH-PhAc derivatives were also monitored at 280 nm.
In other experiments a refractometer (Waters 410) and a Aminex HPX-87 H (Bio-Rad) column were used and the mobile phase was 4 mM H2SO4 (0.6 ml⋅min−1). The Rts for trans-trans-1,3-butadiene-1,4-dicarboxylic acid (muconic acid), 2-OH-PhAc, 4-OH-PhAc, and PhAc were 29, 35, 40, and 57 min, respectively.
[1-14C]PhAc Uptake.
The uptake of labeled PhAc, sodium octanoate, l-arginine, l-proline, l-ornithine, l-histidine, and PhEtNH2 by P. putida U (wild type or mutants) was carried out as reported (2, 13).
RESULTS AND DISCUSSION
Classification of PhAc− Mutants and Metabolic Studies.
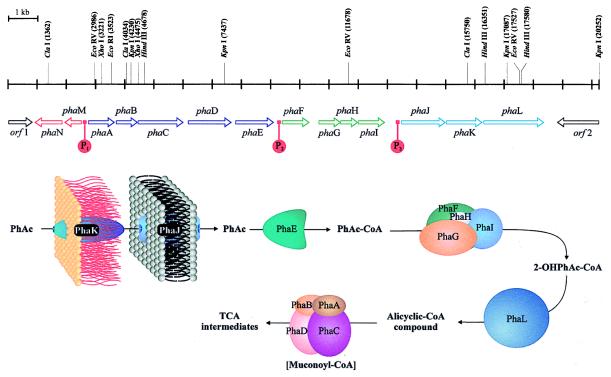
Mutagenesis of P. putida U with the transposon Tn5 allowed the isolation of different mutants unable to catabolize PhAc (PhAc−). Analysis of these strains revealed that the insertion of the transposon occurred in an 18-kb fragment of DNA (G+C content of about 65%) containing 14 ORFs, phaA to phaM (Fig. 4).
Figure 4.
Genetic map of the pha catabolic pathway and hypothetical function of the different proteins. A putative intermediate is shown in brackets.
We classified all PhAc− mutants in PhAc-CoAL+ or PhAc-CoAL− as a function of the presence or absence of PhAc-CoAL activity. The mutants were cultured in MM containing as the carbon source 4-OH-PhAc, which does not induce PhAc-CoAL but does support bacterial growth (4), and PhAc, which, although it cannot be assimilated, is able to induce PhAc-CoAL. Whereas the mutants in which Tn5 was inserted upstream from phaE (Fig. 4) did not show PhAc-CoAL activity, those mutants in which Tn5 was inserted downstream of this gene had high PhAc-CoAL activity. These data suggest that phaE, as well as the other catabolic genes located upstream (phaA, phaB, phaC, and phaD), would be controlled by a single promoter (P1 promoter, see later) and their activities would be blocked by the polar effect of Tn5 (12).
PhAc− mutants, with the exception of those mutated in the PhAc-CoA ligase or in the regulatory genes (see below), grow very poorly in minimal media containing PhAs with an even number of carbon atoms (C2n+2PhAs) (Fig. 5). Because the catabolism of PhAs proceeds via β-oxidation of the side chain (14), the utilization of acetyl-CoA residues released by such β-oxidation can explain the residual growth observed. Therefore, the fact that Tn5 insertion in phaE (PhAc-CoAL) allows the assimilation of such compounds, suggests that: (i) this ligase is not required for the activation of C2n+2PhAs to their CoA thioesters, and (ii) a promoter (P2, see later) that ensures the expression of the genes located downstream of phaE exists. Furthermore, all the mutants affected in the structural genes were also unable to catabolize PhEtNH2 and Sty, whereas they grew well in MM containing 4-OH-PhAc, tyrosine, phenylalanine, phenylglycine, or PhAs with an odd number of carbon atoms (C2n+1PhAs) as carbon sources.
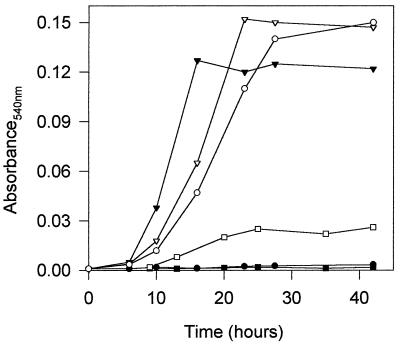
Figure 5.
Bacterial growth (OD540) of P. putida U (▾, ▿) or mutants in which the genes encoding: PhAc-CoAL (phaE) (•, ○) or the enoyl-CoA hydratase I (phaA) had been disrupted (▪, □) were cultured in MM containing 5 mM PhAc (▾, •, ▪) or in MM containing 5 mM PhH (▿, ○, □). Disruption of phaA, phaB, phaC, or phaD genes led to similar results.
Analysis of the products released to the broths by these PhAc− mutants, which grow poorly in C2n+2PhAs, revealed that when they were cultured in MM containing PhH or PhO and 4-OH-PhAc these strains accumulated PhAc. Therefore, these results agree with previous observations that C2n+2PhAs are transformed into PhAc-CoA through a β-oxidation pathway (14). This thioester (a multipathway convergent catabolite) is further catabolized by a specific route that constitutes the core of the PhAc catabolon (Fig. 1). Thus, when this pathway is blocked, the PhAc-CoA originated by convergent routes is hydrolyzed to PhAc. Therefore, the PhAc moiety, which cannot be further catabolized, is excreted and accumulated in the culture broth. Interestingly, PhAc accumulation was also observed when these mutants were incubated in MM containing 4-OH-PhAc (which supports bacterial growth) and PhEtNH2, Sty, or StyAc (used as source of catabolites), indicating that the three compounds are catabolized through the PhAc catabolic pathway (Fig. 1). Furthermore, PhAc-CoAL activity was induced by some PhAc precursors (PhEtNH2, Sty, StyAc, PhB, PhH, and PhO), suggesting that a common catabolite (probably PhAc-CoA) is the true inducer.
Functional Analysis of the PhAc Catabolic Pathway.
Two different set of genes were identified in the PhAc catabolic pathway: (i) regulatory regions and (ii) catabolic genes (phaA to phaL). The regulatory regions include two genes (phaN and phaM, encoding a transcriptional repressor and a protein of unknown function, respectively) and three promoters (P1, P2, and P3) located upstream from phaA, phaF, and phaJ, respectively. The structural catabolic genes were grouped into five functional units: (i) a transport system (encoded by phaJ and phaK); (ii) a PhAc activating enzyme (phaE product) (1, 3); (iii) a ring-hydroxylation system (encoded by phaF, phaG, phaH, and phaI); (iv) a protein that could catalyze the opening of the aromatic ring (phaL product); and, finally, (v) a β-oxidation-like system (encoded by phaA to phaD). Functional analyses of these proteins were performed by directed disruption of the 14 genes through homologous recombination (Fig. 3).
An overall database comparative sequence analysis of the 14 deduced proteins revealed that all them were homologous to several proteins of unknown function encoded by a gene cluster recently sequenced in E. coli K-12 (D90778) and whose analogous proteins are involved in the metabolism of PhAc in E. coli W (A.F., unpublished results).
In the following we will provide evidence for the specific function of each of these 14 genes.
Transport System.
The genes phaJ and phaK encode a permease and a specific-channel-forming protein (15, 16). Disruption of either of these two genes resulted in the inability of these mutants to grow in chemically defined media containing PhAc as the sole carbon source. However, they grew well in media in which PhAc had been replaced by 4-OH-PhAc, phenylalanine, tyrosine, or various C2n+2PhAs or C2n+1PhAs. This observation indicated that: (i) both proteins are essential for the uptake of PhAc and (ii) they are not required for the uptake of other related compounds.
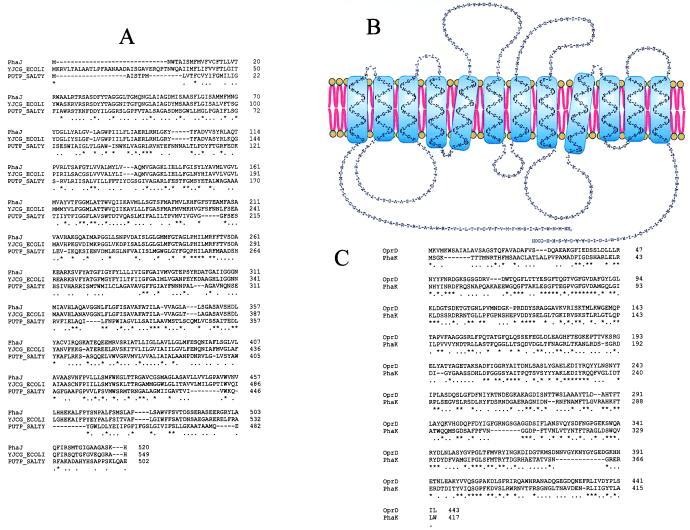
The gene phaJ encodes a protein of 520 amino acids (Mr 55,066) that has 12 putative transmembrane regions (16) (Fig. 6 A and B). This protein seems to be related to the permeases of E. coli and Salmonella typhimurium (65% and 25% identity, respectively) involved in the uptake of proline (17). However, the disruption of phaJ did not affect the rate of [14C]proline transport in P. putida U (data not shown), indicating that PhaJ has a different function in this bacterium. In this sense, we have observed that the uptake of [1-14C]PhAc in phaJ mutants decreases (Fig. 7). This observation, together with the fact that these mutants were unable to grow on PhAc, indicated that (i) PhAc uptake is mediated by a protein, and (ii) the residual uptake observed in the phaJ mutants (Fig. 7) is insufficient to ensure bacterial growth. Similar results have been reported in a study of the uptake of fatty acids in Saccharomyces cerevisiae (18). Although PhAc uptake has been studied in different microbes (2, 19, 20), the sequence of this permease has not been described previously.
Figure 6.
(A) Multiple alignment of the amino acid sequence of PhaJ, the proline permease from Salmonella typhimurium (PUTP-SALTY) and a sodium/solute symport protein from E. coli (YJCG-ECOLI). (B) Putative transmembrane regions of PhaJ. (C) Pairwise alignment of the amino acid sequence of PhaK from P. putida U and the D2 protein from Pseudomonas aeruginosa (OprD). ∗ indicates that a position in the alignment is perfectly conserved; ., that it is well conserved.
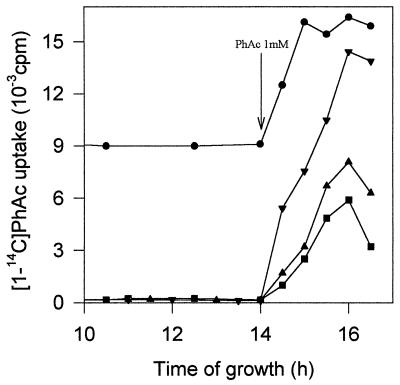
Figure 7.
Analysis of the PhAc transport system when P. putida U and various mutants were cultured in MM containing glucose (5 mM), and once the sugar had been exhausted 1 mM PhAc (inducer) was added to the bacterial population. Shown is [1-14C]PhAc uptake by P. putida U (▾) and by mutants in which the genes encoding permease (▪), the specific channel (▴), or the repressor protein (•) have been disrupted. Arrow indicates addition of PhAc.
A second gene, phaK, encodes a protein of 417 amino acids (Mr 46,078) that shows similarity (37%) with the D2 protein of P. aeruginosa (Fig. 6C), a paradigm of specific channels (porins) (21, 22). Protein D2 facilitates the permeation of imipenem across the outer membrane barrier (19) and is also involved in the uptake of basic amino acids as well as in the incorporation of peptides containing these amino acids (21). Although a disruption of the phaK gene handicapped these mutants for growing in chemically defined media containing PhAc, they were still able to assimilate other related compounds (4-OH-PhAc, PhAs, phenylalanine, tyrosine and PhEtNH2), suggesting that the channel contains specific ligand-binding sites that facilitate the diffusion of PhAc (but not of other analogues) from the external part of the outer membrane to the periplasmic space. Furthermore, a mutation in phaK reduced the uptake of [1-14C]PhAc (Fig. 7), suggesting that this channel is necessary to accelerate PhAc diffusion. Moreover, the uptake of l-lysine, l-arginine, l-ornithine, and l-histidine in phaK mutants remained unaffected and the incorporation of PhAc was not modified when basic amino acids were added to the transport assay system, suggesting that (i) PhaK and D2 have different substrate specificities and (ii) the specificity of the channel would be defined not only by the presence of ligand-binding sites but also by the catabolic proteins linked to the transporter(s).
PhAc-CoAL.
The phaE gene encodes a PhAc-CoAL (3). Specific disruption of this gene prevents P. putida U from growing in MM containing PhAc, PhEtNH2, or Sty, indicating that the pathway involved in the degradation of these compounds generates PhAc as an intermediate (Fig. 1). However, these mutants grew well in minimal medium containing 4-OH-PhAc, phenylalanine, tyrosine, StyAc, and C2n+2PhAs or C2n+1PhAs, suggesting that these compounds are not catabolized through the same pathway or that PhAc-CoAL is not required. However, most PhAc− mutants were unable to degrade C2n+2PhAs completely, suggesting that these compounds are catabolized to PhAc-CoA by β-oxidation, and later assimilated through the PhAc pathway. The fact that mutants disrupted in the phaE gene were not impaired in the degradation of C2n+2PhAs allowed us to conclude that the activation of these acids is not carried out by PhAc-CoAL. In fact, we have isolated a mutant lacking the acyl-CoA synthetase that activates octanoic acid to octanoyl-CoA (23), and we found that it did not grow in MM containing hexanoic acid, octanoic acid, PhH, or PhO, whereas it grew well in the same medium containing only PhAc. In sum, the phaE mutants do not activate PhAc to PhAc-CoA but can completely catabolize all molecules that synthesize PhAc-CoA through other routes (Fig. 1).
Hydroxylation System.
Downstream from phaE four different ORFs, (genes phaF to phaI) were identified (Fig. 4). Mutation with Tn5 or specific disruption of any of these genes prevented these mutants from growing on MM containing PhAc, PhEtNH2, Sty, or StyAc and allowed them to grow only very poorly when the carbon sources were C2n+2PhAs. Analysis of the products accumulated by these mutants revealed that when they were cultured in MM containing PhAc (as a source of intermediates) and 4-OH-PhAc (to support bacterial growth), PhAc remained unaltered in the broth. However, when the source of intermediates was PhH or PhO, PhAc was accumulated extracellularly (Table 1). These data agree with the hypothesis that PhH and PhO are catabolized to PhAc-CoA through the β-oxidation pathway. The existence of a blockade in the PhAc catabolic pathway appears to prevent any further catabolism of PhAc-CoA, which is therefore hydrolyzed, and the PhAc moiety is released to the broth. When [1-14C]PhAc was added to the cultures of these mutants this acid remained unaltered in the broth. Interestingly, when the same experiment was performed with phaL mutants, radioactive 2-OH-PhAc was accumulated. These data suggest that the first two reactions of the PhAc catabolic pathway involve (i) activation of PhAc to PhAc-CoA (catalyzed by PhaE) and (ii) hydroxylation of PhAc-CoA to 2-OH-PhAc-CoA (performed by the enzymes PhaF, PhaG, PhaH, and PhaI). This hypothesis is supported by the fact that the expression of the equivalent phaE, phaF, phaG, phaH, and phaI genes of E. coli W led to the production of 2-OH-PhAc, whereas when these genes were lacking or the phaL gene was simultaneously expressed, 2-OH-PhAc was not found (A.F., unpublished results).
Table 1.
Compounds accumulated in the culture broths when mutants of P. putida U, specifically disrupted in a PhAc catabolic gene, were cultured in MM containing 4-OH-PhAc + PhAc or 4-OH-PhAc + PhH
| Additions to MM | Compound accumulated when gene disrupted
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| phaA | phaB | phaC | phaD | phaE | phaF | phaG | phaH | phaI | phaL | |
| 4-OH-PhAc (5 mM) + PhAc (5 mM) | PhAc | PhAc | PhAc | PhAc | PhAc | PhAc | PhAc | PhAc | PhAc | 2-OH-PhAc |
| 4-OH-PhAc (5 mM) + PhH (5 mM) | PhAc + 2-OH-PhAc | PhAc + 2-OH-PhAc | PhAc | PhAc + muconic acid (traces) | None | PhAc | PhAc | PhAc | PhAc | 2-OH-PhAc |
Muconic acid is trans-trans-1,3-butadiene-1,4-dicarboxylic acid. Results similar to those when 4-OH-PhAc and PhH were added were obtained when P. putida U was cultured in MM + 4-OH-PhAc (5 mM) + PhO (5 mM).
Comparative study of the deduced amino acid sequences of these four enzymes revealed that they show similarities with oxidases or enzymes involved in electron transfer processes. Thus, PhaG has a small identity with certain hemoproteins (15%); PhaF seems to be related to a subunit of the toluene 4-monooxygenase system (14% identity); and PhaI conserves a ferredoxin consensus sequence (CKAGVCSTC, positions 257–265) (24–27). However, although these results seem to be consistent with the hydroxylative model proposed, it does not escape us that much more work is needed to delineate the functions of these proteins.
Ring-Opening Enzyme.
At the 3′ end of the 18-kb DNA fragment is the phaL gene (Fig. 4), which encodes a protein of 688 amino acids (Mr 73,559). Specific disruption of this gene prevented these mutants from growing in MM containing PhAc (or PhEtNH2) and allowed only poor growth when the carbon source was PhH or PhO. Analysis of the product accumulated revealed that when the carbon source was PhEtNH2, PhH, or PhO, only 2-OH-PhAc was detected in the broth (see Table 1), suggesting that PhAc-CoA is hydroxylated to 2-OH-PhAc-CoA and later transformed by PhaL into an alicyclic compound (Fig. 4). Thus, mutants blocked in phaL should accumulate 2-OH-PhAc-CoA intracellularly when grown in MM containing PhH. As expected, when ethanolic extracts (2) of the phaL mutants were analyzed by HPLC, a product with the same retention time as a standard of 2-OH-PhAc-CoA was found, allowing us to conclude that 2-OH-PhAc-CoA is a true catabolic intermediate (Fig. 3). Furthermore, the absence of 2-OH-PhAc from the broth as well as the lack of 2-OH-PhAc-CoA in the ethanolic extracts when PhaL is fully functional might indicate that 2-OH-PhAc-CoA is the substrate for this enzyme.
The amino acid sequence of PhaL revealed that it has two domains: the N-terminal, which shows certain similarity (29% identity) with aldehyde dehydrogenases, and the C-terminal, which has 53% identity with the maoC gene product, a protein that seems to be involved in the catabolism of catecholamine in Klebsiella aerogenes (28).
β-Oxidation Unit.
The catabolic genes located at the 5′ end of the DNA fragment (phaA to phaD) encode a β-oxidation-like pathway. Thus, PhaA (292 amino acids, Mr 31,353) and PhaB (263 amino acids, Mr 28,648) show significant similarity with enoyl-CoA hydratases (46% and 47% identity, respectively) (29), suggesting that these enzymes could be involved in the hydroxylation of double bonds once the aromatic nucleus has been opened. PhaC (505 amino acids, Mr 53,265), seems to be a 3-hydroxyacyl-CoA dehydrogenase (42% identity) (30) and PhaD (497 amino acids, Mr 52,248), has been identified as a ketothiolase (45% identity) (31). These genes together with phaE form a single operon under the control of the P1 promoter. Thus, insertion of Tn5 in any of them results directly in the lack of PhAc-CoAL activity. Moreover, a mutation in phaA, phaB, phaC, or phaD renders P. putida U unable to catabolize PhAc, StyAc, or PhEtNH2. Furthermore, these mutants could not completely degrade C2n+2PhAs, whereas they grow well when C2n+1PhAs or aliphatic compounds were used as the carbon sources. The slow growth observed when these mutants were cultured in MM containing PhH (Fig. 5) could be explained because they can catabolize the two molecules of acetyl-CoA released only through β-oxidation of the side chain, whereas the final product, PhAc-CoA, cannot be assimilated owing to the existence of a later interruption in the pathway. Although different intermediates would be expected to be accumulated by these mutants, we found only PhAc and 2-OH-PhAc (Table 1), suggesting that the metabolic flux is halted immediately when some intermediates are accumulated. The same results were obtained when PhEtNH2 was the carbon source. Nevertheless, very low quantities of a compound identified as trans-trans-butadiene-1,4-dicarboxylic acid (muconic acid) were found in the broth of phaD mutants (Table 1), suggesting that it could be a catabolic intermediate (Fig. 4).
These data point to the existence of two β-oxidation systems in P. putida U: one corresponding to the enzymes involved in the β-oxidation of aliphatic compounds that are also involved in the degradation of the aliphatic moieties of PhAs giving PhAc-CoA or benzoyl-CoA, and the other (products of phaABCD) that catalyzes the degradation of an alicyclic PhAc-CoA derivative (Fig. 4).
Regulatory Elements.
Three promoter regions (P1, P2, and P3) have been identified in the catabolic pathway (Fig. 4). The first one (P1) is located before phaA and controls the expression of the genes phaABCDE. The second one (P2) is placed after phaE and controls the genes phaFGHI. The third promoter (P3) is located before phaJ and controls the expression of the genes phaJKL. Once cloned in E. coli MC4100 (8), their promoter activities have been tested by monitoring the expression of the lacZ gene (Fig. 2B).
Furthermore, two additional genes (phaN and phaM) that can be involved in the transcriptional regulation of this catabolic pathway were identified. The phaN gene encodes a protein of 307 amino acids (Mr 35,100) that acts as a transcriptional repressor, because a blockade in this gene implies the constitutive expression of the PhAc-CoAL and the PhAc-transport system (both enzymatic systems are inducible in the wild type) (Fig. 7). Surprisingly, we have also observed that a blockade of the phaN gene avoids the catabolic repression of this pathway caused by glucose. Thus, in the presence of 5 mM glucose, neither PhAc-CoAL activity nor [1-14C]PhAc uptake was detected, even in the presence of PhAc (inducer). These results could lead to important biotechnological and environmental applications, as the strains mutated at phaN gene could be employed for the degradation of PhAc (or compounds that generate PhAc through other catabolic pathways; see Fig. 1) in natural media, even in the presence of repressive carbon sources.
The role of the phaM gene, which encodes a protein of 199 amino acids (Mr 21,076), is unknown. Thus, its disruption does not affect the catabolism of PhAc, PhH, PhO, or PhEtNH2. However, PhaM shares high identity (59%) with the protein encoded by the caiE gene of E. coli, suggesting that it could be, as this protein, a regulator (32).
In summary, all the above data point to the existence of a previously unknown pathway involved in the aerobic catabolism of PhAc that seems to be a central route in which other peripheral catabolic pathways, leading to PhAc or PhAc-CoA, converge (Fig. 1). Taking this pathway as a model, we have introduced the term catabolon to define a complex catabolic unit integrated by several degradative routes which catalyze the transformation of structurally related molecules (in this case PhAc, Sty, StAc, PhEtNH2, C2n+2PhAs, tropic acid; see Fig. 1) into a common intermediate (convergent point) and which are, or could be, cross-link regulated. The genetic characterization of the PhAc catabolic pathway (core of the PhAc-CoA catabolon) contributes not only an increase in our basic knowledge about the bacterial assimilation of aromatic compounds but also possibilities of new biotechnological and environmental applications. Thus, (i) the expression of phaE, phaJ, and phaK genes in Penicillium chrysogenum or in Aspergillus nidulans could help to increase the rate of benzylpenicillin intermediate biosynthesis (3) or to produce modified β-lactam antibiotics; (ii) mutants specifically altered in the regulatory mechanisms (such as those mutated in the phaN gene) could be used for the removal of PhAc and some toxic related compounds—e.g., Sty and StyAc—under a wide range of environmental and metabolic conditions; and (iii) the synthesis of new bacterial plastics such as the poly(3-hydroxyphenylalkanoate)s (33) could be approached.
Of special interest will be the study of this catabolic pathway from an evolutionary point of view. It is worth noting that the pha gene cluster is particularly rich in short repeat sequences. Although their role remains unclear, it has been proposed that they could be involved in gene expression and in mRNA stability (34). However, the presence of these sequences could also contribute to the evolution of the catabolic pathways by allowing the exchange of either a single gene or a set of genes that catalyze a particular function between different operons. Thus, by means of a single translocation, a new function could be introduced into a preexisting pathway, thereby improving the rate of assimilation of a particular compound or enlarging the number of structurally related molecules that can be catabolized along the pathway. The high catabolic versatility of P. putida U and its impressive rate of adaptation to new metabolic conditions could be due, at least in part, to a mechanism of gene mobilization that would permit the reorganization of certain catabolic pathways by including cassettes of genes that encode specific functions. Thus, preliminary studies have shown that the organization of the different functional units that integrate the catabolon core described here is not the same in all microbes analyzed (A.F., unpublished results). Accordingly, comparative study of this route in E. coli and in different Pseudomonas species might help to clarify how the different operons integrated in the PhAc pathway originated, and also to determine how and why different activities (CoA activation, ring-hydroxylation, β-oxidation) evolved together in a particular catabolon.
The organization of the enzymes in catabolons could represent an important adaptive advantage because, as occurs in Pseudomonas putida U, they seem to contribute to the rapid biochemical evolution of the catabolic pathways. Thus, inclusion of a new gene (or function) in an operon belonging to a particular catabolon could increase the number of enzymatic modifications that can be introduced into a molecule (or in structurally related compounds), broadening the range of products that can be assimilated or transformed by the microbe. In sum, catabolons, which probably arose in response to a need to adapt to special habitats, could be considered as an important evolutionary tool, because such entities facilitate the catabolic polarization of a particular strain, thus increasing its survival possibilities in the biosphere (even among the members of the same species). Furthermore, study of the changes undergone by a catabolon under controlled conditions of selective pressure could yield basic models useful for understanding the evolution of catabolic pathways.
Acknowledgments
This work is dedicated to the memory of Dr. Clemente Luengo. This work was supported by Grant AMB97–0603-C02–01 (Comision Interministerial de Ciencia y Tecnología, Madrid) and Grant LE 42/96 (Junta de Castilla y León). A.F. and B.G. have predoctoral fellowships from the Plan Nacional de Formación de Personal Investigador, Ministerio de Educación y Ciencia and the Fundación Ramón Areces (Madrid), respectively.
ABBREVIATIONS
- PhAc
phenylacetic acid
- PhAc-CoAL
phenylacetyl-CoA ligase
- PhEtNH2
2-phenylethylamine
- Sty
styrene
- StyAc
trans-styrylacetic acid
- PhAs
phenylalkanoic acids
- 2-OH-PhAc and 4-OH-PhAc
2- and 4-hydroxyphenylacetic acid
- PhB
4-phenylbutyric acid
- PhH
6-phenylhexanoic acid
- PhO
8-phenyloctanoic acid
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF029714 for pha gene cluster).
References
- 1.Martínez-Blanco H, Reglero A, Rodríguez-Aparicio L B, Luengo J M. J Biol Chem. 1990;265:7084–7090. [PubMed] [Google Scholar]
- 2.Schleissner C, Olivera E R, Fernández-Valverde M, Luengo J M. J Bacteriol. 1994;176:7667–7676. doi: 10.1128/jb.176.24.7667-7676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miñambres B, Martínez-Blanco H, Olivera E R, García B, Díez B, Barredo J L, Moreno M A, Schleissner C, Salto F, Luengo J M. J Biol Chem. 1996;271:33531–33538. doi: 10.1074/jbc.271.52.33531. [DOI] [PubMed] [Google Scholar]
- 4.Olivera E R, Reglero A, Martínez-Blanco H, Fernández-Medarde A, Moreno M A, Luengo J M. Eur J Biochem. 1994;221:375–381. doi: 10.1111/j.1432-1033.1994.tb18749.x. [DOI] [PubMed] [Google Scholar]
- 5.Frischauf A M, Lehrach H, Poustka A, Murray N. J Mol Biol. 1983;170:827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- 6.Selvaraj G, Iyer V N. J Bacteriol. 1983;156:1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrero M, de Lorenzo V, Timmis K N. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadaban M J, Martínez-Arias A, Shapira S K, Chou J. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 9.Schäfer A, Tauch A, Jäger W, Kalinowski J, Tierbach G, Pühler A. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 11.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg D E. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 185–210. [Google Scholar]
- 13.Carnicero D, Fernández-Valverde M, Cañedo L M, Schleissner C, Luengo J M. FEMS Microbiol Lett. 1997;149:51–58. [Google Scholar]
- 14.Smith M R. Biodegradation. 1990;1:191–206. doi: 10.1007/BF00058836. [DOI] [PubMed] [Google Scholar]
- 15.Nikaido H. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 16.Henderson P J F. Curr Opin Cell Biol. 1993;5:708–721. doi: 10.1016/0955-0674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 17.Nelson K, Selander R K. J Bacteriol. 1992;174:6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faergeman N J, DiRusso C C, Elberger A, Knudsen J, Black P N. J Biol Chem. 1997;272:8531–8538. doi: 10.1074/jbc.272.13.8531. [DOI] [PubMed] [Google Scholar]
- 19.Fernández-Cañón J M, Reglero A, Martínez-Blanco H, Luengo J M. J Antibiot. 1989;42:1398–1409. doi: 10.7164/antibiotics.42.1398. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-Cañón J M, Luengo J M. J Antibiot. 1997;50:45–52. doi: 10.7164/antibiotics.50.45. [DOI] [PubMed] [Google Scholar]
- 21.Trias J, Nikaido H. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 22.Nikaido H. In: Pseudomonas: Molecular Biology and Biotechnology. Galli E, Silver S, Witholt B, editors. Washington, DC: Am. Soc. Microbiol.; 1992. pp. 146–153. [Google Scholar]
- 23.Fernández-Valverde M, Reglero A, Martínez-Blanco H, Luengo J M. Appl Environ Microbiol. 1993;59:1149–1154. doi: 10.1128/aem.59.4.1149-1154.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaac P G, Jones V P, Leaver C J. EMBO J. 1985;4:1617–1623. doi: 10.1002/j.1460-2075.1985.tb03828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen K M, Karl M R, Blatt L M, Simon M J, Winter R B, Fausset P R, Lu H S, Harcourt A A, Chen K K. J Bacteriol. 1991;173:5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harayama S, Rekik M, Bairoch A, Neidle E L, Ornston L N. J Bacteriol. 1991;173:7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harayama S, Polissi A, Rekik M. FEBS Lett. 1991;285:85–88. doi: 10.1016/0014-5793(91)80730-q. [DOI] [PubMed] [Google Scholar]
- 28.Sugino H, Sasaki M, Azakami H, Yamashita M, Murooka Y. J Bacteriol. 1992;174:2485–2492. doi: 10.1128/jb.174.8.2485-2492.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanazawa M, Ohtake A, Abe H, Yamamoto S, Satoh Y, Takayanagi M, Niimi H, Mori M, Hashimoto T. Enzyme Protein. 1993;47:9–13. doi: 10.1159/000468650. [DOI] [PubMed] [Google Scholar]
- 30.Boynton Z L, Bennet G N, Rudolph F B. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebergesell M, Steinbüchel A. Eur J Biochem. 1992;209:135–150. doi: 10.1111/j.1432-1033.1992.tb17270.x. [DOI] [PubMed] [Google Scholar]
- 32.Eichler K, Bourgis F, Buchet A, Kleber H-P, Mandrand-Berthelot M-A. Mol Microbiol. 1994;13:775–786. doi: 10.1111/j.1365-2958.1994.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Valverde, M., Cañedo L. M. & Luengo, J. M. (1997) Spanish Patent P9700356.
- 34.Stern M J, Ames G F-L, Smith N J, Robinson E C, Higgins C F. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]