Abstract
SCF-type E3-ubiquitin ligases control numerous cellular processes through the ubiquitin-proteasome pathway. However, the regulation of SCF function remains largely uncharacterized. Here, we report a novel SCF complex-interacting protein, Lag2, in Saccharomyces cerevisiae. Lag2 interacts with the SCF complex under physiological conditions. Lag2 negatively controls the ubiquitylation activities of SCF E3 ligase by interrupting the association of Cdc34 to SCF complex. Overexpression of Lag2 increases unrubylated Cdc53, whereas deletion of lag2, together with the deletions of dcn1 and jab1, results in the accumulation of Rub1-modified Cdc53. In vitro rubylation assays show that Lag2 inhibits the conjugation of Rub1 to Cdc53 in competition with Dcn1, which suggest that Lag2 down-regulates the rubylation of Cdc53 rather than promoting derubylation. Furthermore, Dcn1 hinders the association of Lag2 to Cdc53 in vivo. Finally, the deletion of lag2 combined with the deletion of either dcn1 or rub1 suppresses the growth of yeast cells. These observations thus indicate that Lag2 has a significant function in regulating the SCF complex by controlling its ubiquitin ligase activities and its rubylation cycle.
Keywords: E3-ubiquitin ligase, protein degradation, rubylation, SCF complex, ubiquitylation
Introduction
The ubiquitin-proteasome system has a critical function in various cellular processes, including cell-cycle progression, gene transcription, and signal transduction (Hochstrasser, 1996; Hershko and Ciechanover, 1998). Ubiquitylation is achieved through the action of three enzymes: E1-ubiquitin-activating enzymes, E2-ubiquitin-conjugating enzymes, and E3-ubiquitin ligases. First, the C-terminus of ubiquitin is covalently attached to a cysteine residue of E1 through a thioester bond. Ubiquitin is then transferred to a cysteine of E2. Finally, through the action of E3, ubiquitin is transferred from E2 conjugates to a lysine residue of the target protein through an isopeptide bond. There are three main classes of E3-ubiquitin ligases (Hershko and Ciechanover, 1998; Joazeiro and Weissman, 2000; Hatakeyama et al, 2001; Cyr et al, 2002). One class includes an active cysteine in the HECT (homologous to E6-AP COOH terminus) domain, which forms a thioester intermediate with ubiquitin and then transfers the ubiquitin onto substrates. The other two classes contain RING-finger and U-box domains, respectively, which serve to bind ubiquitin-loaded E2s and target proteins to bring them into close proximity for the direct transfer of ubiquitin from E2 to substrate, without forming an E3-ubiquitin intermediate.
One type of cullin-RING E3 ligase (CRL), the SCF complex, consists of Skp1, Cdc53/Cul1, F-box protein, and the RING-finger domain containing Rbx1/Roc1/Hrt1 and regulates a wide variety of protein polyubiquitylation events (Bai et al, 1996; Feldman et al, 1997; Skowyra et al, 1997, 1999; Kamura et al, 1999b; Ohta et al, 1999; Seol et al, 1999; Bosu and Kipreos, 2008). The essential subunit of the SCF complex, Cdc53/Cul1, acts as a scaffold, with its carboxy-terminus binding to Rbx1, which in turn associates with the E2 and facilitates the direct transfer of ubiquitin from the E2 onto the target protein, and with its amino-terminus interacting with the adaptor Skp1, which in turn binds to the substrate recognizing and recruiting F-box proteins. Highly variable F-box proteins carry numerous substrates to be tagged with polyubiquitin and targeted for the subsequent degradation by the 26S proteasome.
The regulatory mechanisms underlying SCF function mainly involve the dimerization, neddylation, and deneddylation of SCF complex (Merlet et al, 2009). F-box protein-mediated dimerization has been shown for the SCF complex (Wolf et al, 1999; Dixon et al, 2003; Hao et al, 2007; Tang et al, 2007). This dimerization may increase the local concentration of substrate and ubiquitin-loaded E2, optimally orient the acceptor lysines of the substrate and E2, or provide distinct catalytic distances that can allow an SCF complex to target substrates of different size and accommodate the changes in the length of the elongating polyubiquitin chain. In addition, distinct F-box proteins recognize and recruit substrates to the SCF complex. They compete with each other to bind the core SCF complex. In yeast, F-box proteins are often unstable because of autoubiquitylation induced by binding to the SCF complex, whereas bound substrates protect F-box proteins from autoubiquitylation and degradation by proteasomes (Zhou and Howley, 1998; Galan and Peter, 1999; Mathias et al, 1999; Wirbelauer et al, 2000; Li et al, 2004). Therefore, the level of F-box proteins influences the activity of the SCF complex.
A small ubiquitin-like protein, Rub1/Nedd8, conjugates to the cullins in a process called rubylation/neddylation, which is essential in all species studied in the literature except for budding yeast (Lammer et al, 1998; Kamura et al, 1999a; Kurz et al, 2005, 2008; Megumi et al, 2005; Merlet et al, 2009). The removal of Nedd8 from cullins mediated by the CSN/COP9 signalosome is called deneddylation/derubylation (Lyapina et al, 2001; Cope et al, 2002). Neddylation is achieved through a ubiquitylation-like enzymatic cascade involving the Nedd8-activating enzyme AppBp1-Uba3, Nedd8-conjugating enzyme Ubc12, and Nedd8 ligases Rbx1 and Dcn1. This Nedd8-conjugating pathway has been shown to stimulate the ubiquitin ligase activity of CRL in vitro and in vivo (Sakata et al, 2007; Wimuttisuk and Singer, 2007; Duda et al, 2008; Saha and Deshaies, 2008). Neddylation on cullins not only stimulates the recruitment of ubiquitin-activated E2s, but also affects the transfer of ubiquitin from SCF-bound E2 to the substrate lysine by inducing a conformational change in the complex that allows for greater structural flexibility to bring ubiquitin-loaded E2 close to the acceptor lysine of the substrate. On the basis of the observation that neddylation enhances CRL ubiquitylation activities, a reduction in deneddylation paradoxically attenuates CRL function (Cope et al, 2002; Doronkin et al, 2003; Liu et al, 2003; Pintard et al, 2003). The current explanation for this paradox proposes that CSN-mediated deneddylation inactivates the CRL complex and protects the CRL subunits from autoubiquitylation and degradation. In addition to preventing the destabilization of CRL components, CSN may also initiate the disassembly of the CRL complex and promote CRL subunit recycling.
Neddylation of cullins is mainly regulated by two proteins: Dcn1 and CAND1 (Liu et al, 2002; Zheng et al, 2002; Goldenberg et al, 2004; Kurz et al, 2005, 2008). Dcn1, a neddylation E3 factor, is required for Cul1 covalent modification by Nedd8. In contrast, CAND1 preferentially binds to unneddylated Cul1-Rbx1 modules, preventing the association of Skp1 to form the SCF complex, and inhibits Nedd8 conjugation to Cul1. How these regulators and the metalloprotease Csn5/Jab1 balance neddylation and deneddylation of cullins and how this balance affects the CRL function remain unresolved questions.
We have identified a novel SCF complex-interacting protein, Lag2, which is specifically associated with unrubylated Cdc53. Lag2 disrupts the ubiquitylation activity of the SCF E3 ligase by interrupting the association of Cdc34 to SCF complex. Lag2 also inhibits the conjugation of Rub1 to Cdc53. Our data indicate that Lag2 acts as a negative modulator in the regulation of the SCF complex function.
Results
Identification of Lag2 as an SCF complex-interacting protein
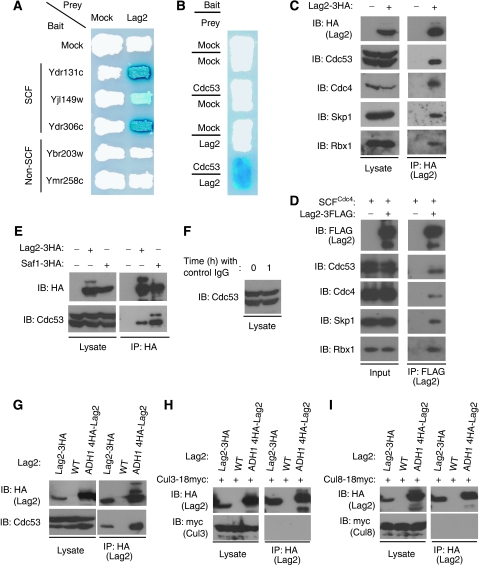
To further investigate the function of the SCF complex in Saccharomyces cerevisiae, we screened a yeast DNA library with the yeast two-hybrid assay using the F-box proteins, Ydr131c, Yjl149w, Ydr306c, Ybr203w, and Ymr258c, as bait, respectively. Interestingly, with several distinct SCF-type F-box proteins, Ydr131c, Yjl149w, and Ydr306c, we identified the same interacting protein, Lag2, which was originally reported to affect cell longevity (Childress et al, 1996). In contrast, the non-SCF-type F-box proteins, Ybr203w and Ymr258c, which bind to Skp1 but not to the Cdc53-Rbx1 module (MK and TK, unpublished data), fail to interact with Lag2 as prey (Figure 1A). These findings raised the possibility that Lag2 might bind to the F-box proteins through the core components of the SCF complex, the Cdc53-Rbx1 module. As anticipated, Lag2 specifically associated with Cdc53 in the β-gal assay (Figure 1B), suggesting that Lag2 directly interacts with Cdc53 rather than with the F-box proteins.
Figure 1.
Lag2 interacts with the SCF complex under physiological conditions. (A, B) The interaction of the indicated F-box proteins (A) and Cdc53 (B) with Lag2 was monitored with a two-hybrid assay by expression of the LacZ-reporter gene. (C) The indicated yeast strains were cultured in YPD medium, collected in the exponential growth phase, and lysed using glass beads and a multibead shocker. Lysates were subjected to immunoprecipitation (IP) with anti-HA antibodies, and the resulting precipitates and total cell lysates were subjected to immunoblot analysis (IB) with anti-HA, anti-Cdc53, anti-Cdc4, anti-Skp1, and anti-Rbx1. (D) The indicated recombinant proteins were mixed and incubated on ice for 1 h. Binding of Lag2 and SCFCdc4 complex was detected by immunoprecipitation (IP) with anti-FLAG, and followed immunoblot analysis (IB) with anti-FLAG, anti-Cdc53, anti-Cdc4, anti-Skp1, and anti-Rbx1. (E) Lysates of the yeast strains expressing Lag2-3HA or Saf1-3HA were prepared as described in (C) and subjected to immunoprecipitation (IP) with anti-HA. The resulting precipitates and total cell lysates were subjected to immunoblot analysis (IB) with anti-HA and anti-Cdc53. (F) Lysates of Lag2-3HA yeast cells before or after incubation with control IgG and protein A beads at 4°C for 1 h were subjected to immunoblot analysis (IB) with anti-Cdc53. (G–I) Lysates of the yeast strains expressing the indicated proteins were prepared as described in (C) and subjected to immunoprecipitation (IP) with anti-HA antibodies. The resulting precipitates and total cell lysates were subjected to immunoblot analysis (IB) with anti-HA (G, H, I), and anti-Cdc53 (G), or anti-myc (G–I).
To explore the possible interactions of Lag2 with the SCF complex, the lag2 gene was tagged with three copies of the hemagglutinin epitope (3HA) at its C-terminus. Cells were cultured in YPD medium, harvested in the exponential growth phase, and lysed with glass beads and a multibeads shocker. Cell lysates were then subjected to immunoprecipitation with anti-HA antibodies (Figure 1C). Immunoblot analysis of the resulting precipitates revealed that Lag2 interacted with native Cdc53, Rbx1, Skp1, and F-box protein Cdc4, indicating that Lag2 physiologically binds to the SCF complex. To investigate the association of Lag2 to the SCF complex in vitro, we mixed recombinant 3FLAG-tagged Lag2 from Escherichia coli with recombinant SCFCdc4 complex purified from insect cells and examined the binding of these proteins by immunoprecipitation analysis with anti-FLAG antibodies. Consistent with in vivo findings, Lag2 bound to SCF complex (Figure 1D).
In experiments investigating the interaction between Lag2 and SCF complex, we noticed that in the physiological condition, Lag2 only bound to one of the two isoforms of Cdc53 seen in the lysate by western blotting. To determine whether Lag2 interacted with rubylated or unrubylated Cdc53, we generated yeast cells, in which SCF-type F-box protein Saf1 was tagged with 3HA at its C-terminus and compared the mobility of Cdc53 in the anti-HA precipitates obtained from Saf1-3HA and Lag2-3HA cells, respectively. As predicted, Saf1 associated with the two isoforms of Cdc53. The mobility of Cdc53 observed in Lag2-3HA cells was similar to that of the unrubylated Cdc53 (Figure 1E). To exclude the possibility that derubylation happened during the process of immunoprecipitation, we examined the rubylation status of Cdc53 in Lag2-3HA cell extracts before and after the incubation with the control antibody and protein A beads, respectively. As shown in Figure 1F, no observable derubylation was detected in the immunoprecipitation process at 4°C. Thus, these findings show that Lag2 preferentially binds to unrubylated Cdc53.
Lag2 specifically interacts with Cdc53
In budding yeast, the cullin protein family includes three members: Cdc53, Cul3, and Cul8/Rtt101 (Michel et al, 2003). Each of these cullin proteins assembles with Rbx1 to reconstitute a module that is capable of stimulating ubiquitylation by E2-ubiquitin-conjugating enzymes. We examined the ability of Lag2 to interact with three cullin molecules, Cdc53, Cul3, and Cul8, under physiological conditions (Figure 1G–I). Consistent with earlier observations, endogenous Lag2 could associate with endogenous Cdc53, but not with endogenous Cul3 or Cul8. Next, we expressed 4HA-tagged Lag2 from the ADH1 promoter and assessed the ability of the overexpressed Lag2 to bind cullin modules. Compared with endogenous Lag2, overexpressed Lag2 bound to a relatively large amount of endogenous Cdc53, whereas neither Cul3 nor Cul8 was immunoprecipitated in detectable quantities with overexpressed Lag2. These results suggest that Lag2 binding to Cdc53 is highly efficient and specific.
Lag2 inhibits Cdc53 rubylation in vivo and in vitro
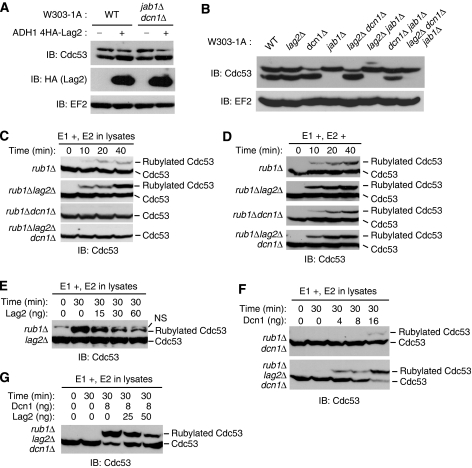
To explore the function of Lag2, we expressed 4HA-tagged Lag2 from the ADH1 promoter and examined the rubylation status of Cdc53. Overexpression of Lag2 increased the ratio of unmodified Cdc53 to rubylated Cdc53 (Figure 2A). As Jab1 and Dcn1 interact with cullins and show derubylating and rubylating activities to Cdc53, respectively (Lyapina et al, 2001; Cope et al, 2002), we tested whether Lag2 functioned independently of Jab1 and Dcn1 or not. We generated dcn1Δ jab1Δ double mutant cells and examined the effect of Lag2 overexpression on Rub1 conjugation to Cdc53. An increased level of unrubylated Cdc53 was clearly observed in dcn1Δ jab1Δ double mutant cells with overexpressed Lag2. We further verified this effect of Lag2 on Cdc53 rubylation in lag2Δ cells (Figure 2B). Neither the lag2Δ single mutant nor the dcn1Δ lag2Δ double mutant produced any detectable effect on Cdc53 rubylation. However, compared with the dcn1Δ jab1Δ double mutant, the lag2Δ dcn1Δ jab1Δ triple mutant resulted in significantly increased levels of rubylated Cdc53. These findings imply that Lag2 functions to reduce Rub1 conjugation to Cdc53 independently of Dcn1 and Jab1.
Figure 2.
Lag2 inhibits the Rub1 conjugation to Cdc53 in vivo and in vitro. (A, B) The indicated yeast cells were cultured in YPD medium, collected in the exponential growth phase, and lysed by the TCA lysis method. Lysates were then subjected to immunoblot analysis (IB) with anti-Cdc53 (A, B), anti-HA (A), and anti-EF2 (A, B). (C) Deletion of Lag2 stimulates the rubylation of Cdc53. The indicated yeast strains were cultured in YPD medium, collected in the exponential growth phase, and lysed using glass beads and a multibeads shocker. For the Dcn1-dependent in vitro Rub1 conjugation assay, the lysates were mixed with Ula1, Uba3, and His6-Rub1 in a 6-μl reaction in the presence of ATP. Reaction mixtures were incubated at 26°C for the indicated times. Rubylated native Cdc53 was detected by immunoblot analysis (IB) using anti-Cdc53. (D) The addition of recombinant Ubc12 activates the Rub1 conjugation to Cdc53. The extracts were prepared as described in (C). For the Dcn1-independent in vitro Rub1 conjugation assay, recombinant Ubc12 was added to the reactions shown in (C). (E) Recombinant Lag2 inhibits Rub1 conjugation to Cdc53. The lysate from the rub1Δ lag2Δ strain was subjected to the Dcn1-dependent in vitro Rub1 conjugation assay as in (C) with the indicated amount of recombinant Lag2. NS: non-specific band. (F, G) Lag2 inhibits Dcn1-dependent Rub1 conjugation to Cdc53. Lysates from the indicated strains were subjected to the Dcn1-dependent in vitro Rub1 conjugation assay as in (C) with the indicated amounts of Dcn1 (F) and Dcn1 and Lag2 (G).
As we earlier observed that Lag2 only bound to unrubylated Cdc53 (Figure 1C, E, and G) in vivo, we reasoned that Lag2 might inhibit Rub1 conjugation to Cdc53. To test this hypothesis, yeast cells were cultured in YPD medium, collected in the exponential growth phase, and lysed with glass beads and a multibeads shocker. We compared the lysates obtained from four yeast strains, the rub1Δ single mutant, rub1Δ lag2Δ double mutant, rub1Δ dcn1Δ double mutant, and rub1Δ lag2Δ dcn1Δ triple mutant, for their ability to catalyse the rubylation of Cdc53 in vitro (Figure 2C). In this experiment, by deleting rub1, we used endogenous Cdc53 as a substrate and endogenous Ubc12 as an E2 for Rub1 conjugation to reduce non-specific reactions. In the presence of ATP, recombinant His6-tagged Ula1, recombinant His6-tagged Uba3, recombinant His6-tagged Rub1, and native E2 (low concentration) in lysates, we observed a faint rubylation of native Cdc53 in the rub1Δ mutant, whereas abundant rubylated Cdc53 was detected in the rub1Δ lag2Δ double mutant in a time-dependent manner. Consistent with earlier reports (Kurz et al, 2008), in cell extracts from the dcn1Δ mutant, no rubylation of Cdc53 was achieved in this low-E2-concentration lysate. Next, we examined the effect of recombinant Ubc12 on the rubylation reaction in vitro (Figure 2D). In high concentration of E2, Cdc53 rubylation was markedly greater in the rub1Δ lag2Δ double mutant than that in the rub1Δ mutant, which was similar to that in low-E2-concentration lysate. Meanwhile, Rub1 conjugation to Cdc53 in the rub1Δdcn1Δ and rub1Δdcn1Δlag2Δ mutants was clearly detected, even if at lower abundance than those in rub1Δ and rub1Δ lag2Δ mutants, respectively. These findings indicate that Lag2 and Dcn1 modulate the rubylation of Cdc53 in E2-concentration-independent and -dependent manners, respectively. On the basis of these observations, we speculated that Lag2 might function to inhibit the rubylation of Cdc53. To explore this possibility, we tested the effects of recombinant Lag2 on rubylation in low-E2-concentration lysate (Figure 2E). As predicted, Lag2 inhibited Rub1 conjugation to Cdc53 in a dose-dependent manner in the rub1Δ lag2Δ double mutant. We next checked the function of Dcn1 with a similar assay, as shown in Figure 2C. Using the lysate from rub1Δ dcn1Δ double mutant and recombinant His6-tagged Dcn1 from E. coli, we observed that Dcn1 slightly enhanced the accumulation of rubylated Cdc53 (Figure 2F). The reaction activity changed with Dcn1 in a dose-dependent manner. In contrast, deletion of lag2 in the rub1Δ dcn1Δ double mutant promoted a notable rise in Rub1-conjugated Cdc53. To further investigate the effects of Lag2 and Dcn1 on Cdc53 rubylation, we added bacterially expressed recombinant Dcn1 to the rubylation reaction, using the rub1Δ lag2Δ dcn1Δ triple mutant, and obtained higher level of rubylated Cdc53 (Figure 2G). However, additional recombinant Lag2 inhibited the rubylation of Cdc53 in a dose-dependent manner. Together, these data indicate that Lag2 negatively regulates Rub1 conjugation to Cdc53 by counteracting the activity of Dcn1.
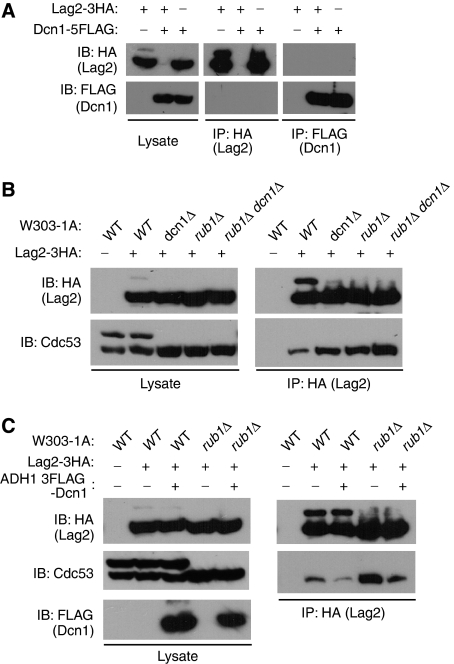
Dcn1 interrupts the interaction of Lag2 to unrubylated Cdc53
To investigate how Lag2 competes with Dcn1 in Rub1 conjugation to Cdc53, we generated yeast cells, in which endogenous Lag2 and Dcn1 were tagged with 3HA and 5FLAG, respectively, and then examined the physiological interaction between these two proteins. As shown in Figure 3A, we failed to observe the binding between Lag2 and Dcn1. We next tested the ability of endogenous or overexpressed Dcn1 to interact with endogenous Cdc53. Unfortunately, we could not detect the association of endogenous or overexpressed Dcn1 with Cdc53 (data not shown). However, in earlier reports (Kurz et al, 2005, 2008) interaction between Dcn1 and Cdc53 was shown in a yeast two-hybrid assay and this direct interaction was further confirmed with in vitro binding assay using recombinant proteins. So, we predicted that Dcn1 might interact with Cdc53 with a weak affinity in vivo. Then, we examined the effect of deletion of dcn1 on the binding of Lag2 to Cdc53 (Figure 3B). Coimmunoprecipitation analysis revealed that Lag2 in dcn1Δ cells bound to a relatively large amount of unrubylated Cdc53 compared to that in wild-type cells. To clarify whether this effect resulted from the loss of binding competition or the increased amount of unrubylated Cdc53 executed by deletion of dcn1, we tested the ability of Lag2 on binding to Cdc53 in rub1Δ and rub1Δ dcn1Δ mutants, respectively. Deletion of rub1 induced a similar up-regulation of binding ability of Lag2 to Cdc53 compared with that in dcn1Δ cells, whereas the association of Lag2 with Cdc53 was dominantly increased by the additional dcn1 deletion. The importance of Dcn1 on the interaction of Lag2 to Cdc53 was further examined by the Dcn1 overexpression experiments. As shown in Figure 3C, overexpression of Dcn1 in wild-type or rub1Δ cells induced a marked decrease in the binding between Lag2 and Cdc53 compared with that apparent in wild-type or rub1Δ cells. These data suggest that Dcn1 modulates the association between Lag2 and Cdc53 by competing the binding of Lag2 to Cdc53 as well as regulating the rubylation status of Cdc53.
Figure 3.
Dcn1 interrupts the association of Lag2 to Cdc53. (A) Lysates of the indicated yeast cells were prepared as described in Figure 1C and subjected to immunoprecipitation (IP) with anti-HA or anti-FLAG. The resulting precipitates and total cell lysates were subjected to immunoblot analysis (IB) with anti-HA and anti-FLAG. (B, C) Indicated yeast cell lysates were prepared as described in Figure 1C and subjected to immunoprecipitation (IP) with anti-HA. The resulting precipitates and total cell lysates were subjected to immunoblot analysis (IB) with anti-HA (B, C), anti-Cdc53 (B, C), and anti-FLAG (C).
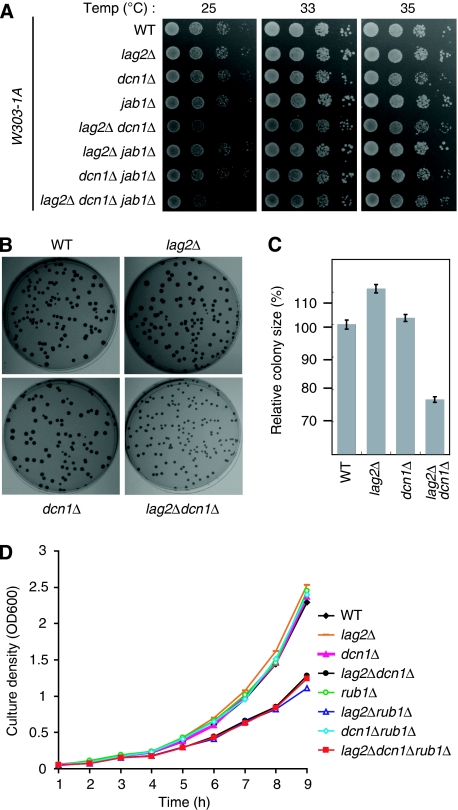
Deletion of lag2 combined with the deletion of either dcn1 or rub1 inhibits the growth of W303-1A cells
As Lag2 was originally reported to affect cell longevity (Childress et al, 1996), we further tested the possible effects exerted by lag2 on cell growth. We examined the genetically combined functions of lag2Δ, dcn1Δ, and jab1Δ on cell growth in the W303-1A strain (Figure 4A). No clear growth difference was observed for any single gene deletion mutant, whereas the lag2Δ dcn1Δ double mutant obviously hindered cell growth at 25, 33, and 35°C. Additional jab1 gene deletion in lag2Δ dcn1Δ double mutant did not show any synthetic effect on cell viability at 25°C and slightly rescued cell growth at 33 and 35°C. On the basis of these findings, we predicted that this growth defect was mainly caused by the deficiency of both Lag2 and Dcn1 functions. We continued to compare the growth of single colonies in each mutant (Figure 4B and C). Compared with W303-1A wild-type cells, mildly increased growth was observed in the lag2Δ mutant, whereas a marked growth defect was detected in lag2Δ dcn1Δ mutant. Additional deletion of the jab1 gene did not affect the growth defect in the lag2Δ dcn1Δ double mutant (data not shown). To further confirm the growth defect, we measured their growth rates in YPD medium (Figure 4D). Consistent with the earlier results in Figure 4A–C, double deletions of lag2 and dcn1 inhibit the cell growth. As loss of Dcn1 function suppresses the rubylation of Cdc53, we wondered how the combinational deletions of rub1, lag2, and dcn1 influenced the cell growth. Single deletion of rub1 did not show clear growth effect. However, double deletion of lag2 and rub1 displayed similar growth defect to that of double deletion of lag2 and dcn1. Triple deletion of lag2, dcn1, and rub1 did not show further growth effect, compared with the double deletion of lag2 and rub1. These results imply that the loss of Lag2 function, together with the defect of rubylation of Cdc53, results in the retardation of cell growth.
Figure 4.
Deletion of lag2 combined with the deletion of either dcn1 or rub1 inhibits the growth of W303-1A cells. (A) The indicated yeast cells were grown to log phase in YPD medium. Cells were harvested and resuspended in YPD medium to an OD600 of 0.4; 3 μl of 10-fold serial dilutions was spotted onto YPD plates, incubated for 24 h at the indicated temperature, and photographed. (B) The indicated cells were plated onto the YPD plates, grown for 6 days at 30°C, and photographed. (C) The diameter of the colonies shown in (B) was measured. Data are expressed as a percentage of the size of W303-1A wild-type cells and are the means±s.d. from 60 independent colonies. (D) Overnight cultures of the indicated cells were inoculated into YPD medium at a density of an OD600=0.05. Samples were taken every 1 h to measure the cell density.
Genetic phenotype exerted by the combination of lag2, dcn1, and jab1 deletion
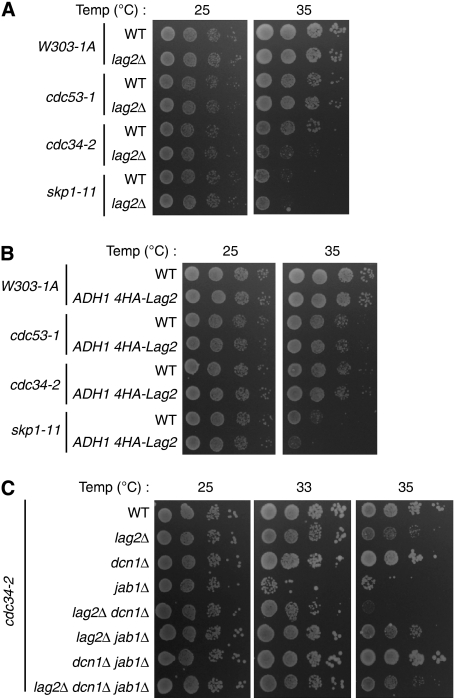
To clarify whether Lag2 affects the functions of the SCF E3-ubiquitin ligase, we tested the synthetic interaction between Lag2 and SCF temperature-sensitive (ts) mutants, cdc53-1, cdc34-2, and skp1-11. We observed that the lag2Δ mutation enhanced the phenotype of cdc34-2 ts cell (Figure 5A), whereas in cdc53-1 and skp1-11, no growth change was detectable after the deletion of lag2. In addition, overexpression of lag2 slightly inhibited the growth of skp1-11, but failed to affect the viability of cdc53-1 and cdc34-2 mutants (Figure 5B).
Figure 5.
Genetic interactions of lag2, dcn1, and jab1 in SCF ts cells. (A) Deletion of lag2 lowers the restrictive temperature of cdc34-2 cells. The growth of the indicated yeast cells was compared as described in Figure 3A at the indicated temperature for 48 h. (B) Overexpression of Lag2 lowers the permissive temperature of skp1-11 cells. The indicated yeast strains were spotted onto YPD plates as shown in Figure 3A, incubated for 48 h at the indicated temperature, and photographed. (C) The combination of deletions of lag2, dcn1, and jab1 affects the ts phenotype of cdc34-2 cells. The indicated cells were spotted onto YPD plates as described in Figure 3A at the indicated temperature for 48 h.
Deletion of lag2 induced a growth defect only in cdc34-2 ts cells, so we further tested the effects of the combination of lag2Δ, dcn1Δ, and jab1Δ in cdc34-2 (Figure 5C). Lag2Δ and jab1Δ lowered the permissive temperature mildly and severely, respectively. No discernible effects were detected in the dcn1Δ mutant. The lag2Δ dcn1Δ mutant dramatically lowered the restrictive temperature. Lag2Δ jab1Δ moderately suppressed cell viability. Dcn1Δ jab1Δ displayed a growth phenotype similar to that of cdc34-2 cells. The lag2Δ dcn1Δ jab1Δ triple mutant showed mild growth defects. These observations suggest that these three rubylation modulators, Lag2, Dcn1, and Jab1, affect cdc34-2 cell growth by regulating the activities of SCF complex through the rubylation cycle.
Lag2 negatively regulates SCF complex function in vivo and in vitro
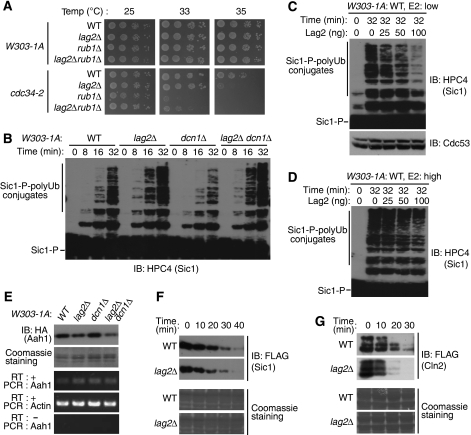
As reported, without rub1, cdc34-2 cells could not grow, even at 33°C (Lammer et al, 1998). Surprisingly, the double deletion of rub1 and lag2 partially rescued rub1Δ cdc34-2 cell viability (Figure 6A). This finding, together with our observations in growth curve check (Figure 4D), raised the possibility that Lag2 not only affects the rubylation of Cdc53, but also regulates other SCF functions. We hypothesized that Lag2 might affect ubiquitylation reactions executed by the SCF complex. To test this, we examined the ubiquitylation of Sic1 in vitro. Yeast cells were cultured in YPD medium, collected in the exponential growth phase, and lysed with glass beads and a multibeads shocker. We compared the abilities of the lysates obtained from four yeast strains: wild type, lag2Δ, dcn1Δ, and the lag2Δ dcn1Δ double mutant to catalyse the ubiquitylation of Sic1 in vitro (Figure 6B). In the presence of ATP, recombinant Uba1, recombinant Cdc34 (low concentration), and recombinant His6-tagged ubiquitin, we observed that with extracts from lag2Δ and lag2Δ dcn1Δ double mutant cells, polyubiquitylation of Sic1 was clearly increased compared with that of the wild-type cells, whereas in the dcn1Δ strain, polyubiquitylation of Sic1 was faintly inhibited. Consistent with these findings, the addition of recombinant Lag2 obviously inhibited polyubiquitin chain conjugation to Sic1 in a dose-dependent manner (Figure 6C). To rule out the possibility that Lag2 suppresses the ubiquitylation of Sic1 by altering the rubylation of Cdc53, we detected the level of Cdc53. The constant level of rubylated Cdc53 suggests that Lag2 does not reduce the rubylation of Cdc53. To further investigate whether the Cdc34 concentration influences the effect of Lag2 on ubiquitylation of Sic1 or not, we conducted the in vitro ubiquitylation assay with five times larger amount of recombinant Cdc34 (Figure 6D). In this high E2 condition, the inhibitory effect of Lag2 on Sic1 ubiquitylation clearly decreased.
Figure 6.
Lag2 inhibits the activity of the SCF E3 ligase complex. (A) Deletion of lag2 partially rescues the viability of rub1Δ cdc34-2 cells. The indicated yeast cells were spotted onto YPD plates and cultured as described in Figure 4A at the indicated temperature for 48 h. (B–D) Lag2 negatively regulates polyubiquitin conjugation to phosphorylated Sic1. The indicated yeast strains were cultured in YPD medium, harvested in the exponential growth phase, and lysed using glass beads and a multibeads shocker. The cell extracts were incubated with phosphorylated Sic1 (Sic1-P), Uba1, Cdc34, and His6-ubiquitin for the indicated times in the absence (B) of endogenous Lag2 or presence (C, D) of recombinant Lag2. Ubiquitylation of Sic1 was detected by immunoblot analysis (IB) using anti-HPC4 (B–D) and anti-Cdc53 (C). (E) Deletion of Lag2 decreases the abundance of Aah1 in vivo. The indicated cells were cultured in YPD medium to an OD600=6 and harvested. One half of the cells were lysed by the TCA lysis method. Then, extracts were subjected to immunoblot analysis (IB) with anti-HA, and the membrane was stained with Coomassie as a loading control. Total RNA was isolated from the other half of the cells. The abundance of mRNAs for Aah1 and Actin (internal standard) was determined by RT–PCR. PCR of Aah1 without RT reaction was used for negative control. (F) Deletion of Lag2 decreases the stability of Sic1 in vivo. The indicated cells were cultured in YPD medium to an OD600=0.5, and then in YP raffinose medium for 1 h. Cells were arrested by 2 μl/ml α factor for 2 h, and then the expression of Sic1 was induced by YPG for 2 h. The medium was changed back to YPD. Yeast cells were harvested and lysed by the TCA lysis method as the indicated time interval. Extracts were subjected to immunoblot analysis (IB) with anti-FLAG, and the membrane was stained with Coomassie as a loading control. (G) Deletion of Lag2 decreases the stability of Cln2 in vivo. The indicated cells were cultured in YPD medium to an OD600=0.4, and then the expression of Cln2 was inducted in YPG medium for 5 h. The medium was changed back to YPD. Turnover of Cln2 was examined with the same method as shown in (F).
Next, we examined the effects of Lag2 on SCF E3 ligase function in vivo. Adenine deaminase, Aah1, is targeted and degraded through SCFSaf1 (Escusa et al, 2006, 2007). We generated yeast cells with C-terminal 3HA-tagged Aah1, cultured with YPD medium, harvested to an OD600=6, lysed with trichloroacetic acid (TCA) precipitation method, and checked the expression of Aah1 (Figure 6E). Compared with wild-type yeast cells, no discernable change in Aah1 expression was observed in dcn1Δ cells. In contrast, deleting lag2 or both lag2 and dcn1 simultaneously results in an obvious decrease of Aah1. At the same time, we proved that mRNA of Aah1 in detected yeast stains did not show clear difference. As we tested the expression of Aah1 in the condition of an OD600=6, we could not measure Aah1 turnover. Therefore, to further investigate the effects of Lag2 on SCF function, we generated yeast cells that expressed N-terminally 3FLAG-tagged Sic1 controlled by regulatory Gal1 promoter in wild-type and lag2Δ cells (Figure 6F). Cells were arrested in G1 phase by α factor, and then the expression of Sic1 was induced by YPG. We changed medium back to YPD to shut off the expression of Sic1 and took samples in 10 min interval for detecting the turnover of Sic1. The deletion of lag2 induced a decrease of the stability of Sic1, compared with that in wild-type cells. Next, we detected the stability of Cln2 in asynchoronized cells (Figure 6G). Similarly, a reduced stability of Cln2 was observed in the lag2 deleted cells.
Lag2 inhibits the association of Cdc34 to SCF complex in vitro
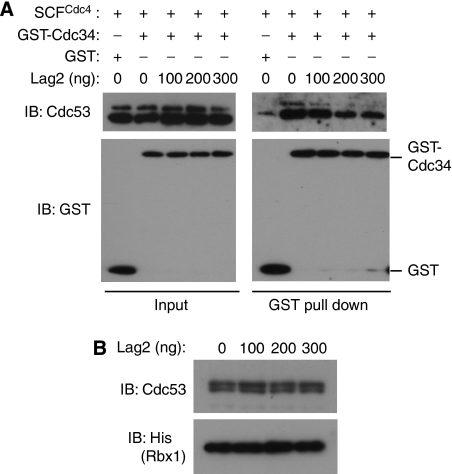
On the basis of this negative regulation on SCF E3 ligase exerted by Lag2, we hypothesized that binding of Lag2 to SCF complex might interrupt the association of E2 to E3. To test this notion, we mixed recombinant GST-tagged Cdc34 from E. coli with recombinant SCFCdc4 complex purified from insect cells in vitro and examined the binding of these proteins (Figure 7A). GST-tagged Cdc34 bound to a relatively large amount of SCF complex, compared with that of GST alone. We further checked the effects of recombinant His6-tagged Lag2 from E. coli on this interaction. As observed, Lag2 competed with Cdc34 to bind SCFCdc4 complex in a concentration-dependent manner. In our study, Lag2 reproducibly suppressed the binding of Cdc34 to SCFcdc4, even though the inhibiting efficiency displayed some kind of variability. We noticed that Lag2 inhibited Cdc34 binding to both rubylated and unrubylated Cdc53. This result might contradict with the in vivo data (Figure 1C, E, and G), which showed that Lag2 only interacted with unrubylated Cdc53. However, in the binding experiment in vitro (Figure 1D), Lag2 bound to small fraction of rubylated Cdc53, in addition to large amount of unrubylated Cdc53, which indicated that Lag2 had weak binding affinity with rubylated Cdc53 in vitro. This observation suggests that Lag2 inhibits Cdc34 binding to both rubylated and unrubylated Cdc53 in vitro. Similarly, we examined the effects of Lag2 to neddylation-conjugating enzyme Ubc12. But we failed to detect the interaction of Ubc12 with SCF complex (data not shown). Finally, we examined whether Lag2 dissociated Rbx1 from Cdc53 in vitro. As shown in Figure 7B, the increasing amount of recombinant Lag2 did not induce the dissociation of Rbx1 from Cdc53. Together, these findings suggest that Lag2 suppresses the function of the SCF E3 ligase by interrupting the association of E2 to SCF complex and negatively modulating rubylation cycling.
Figure 7.
Lag2 dose dependently inhibits the binding of Cdc34 to SCFCdc4 complex in vitro. (A) The indicated recombinant proteins were mixed and incubated on ice for 1 h. Binding of Cdc34 and SCFCdc4 complex was detected by GST pull-down analysis and followed immunoblot analysis (IB) with anti-Cdc53 and anti-GST. (B) Ni2+-agarose beads containing Cdc53/His6-Rbx1 complex was mixed with the indicated amount of His6-Lag2 at 4°C for 1 h. After washing, bound proteins were separated by SDS/PAGE and subjected to immunoblot analysis (IB) with anti-Cdc53 and anti-His.
Discussion
Protein degradation by the ubiquitin-proteasome pathway has a fundamental function in controlling the abundance of important regulatory proteins. Diverse ubiquitin E3 ligases determine the substrate specificity in this pathway. However, how the E3 ligase function is regulated remains largely unknown. In this report, we identified a novel protein, Lag2, which interacts with the SCF complex under physiological conditions. Lag2 inhibits Cdc34 association to SCF complex, in turn suppresses the ubiquitylation activity of SCF E3 ligase. And in addition, Lag2 negatively regulates Rub1 conjugation to Cdc53.
Although the high-speed progress in the study of SCF E3 ligase, as far as we know, no regulator was reported to block the association of E2-ubiquitin-conjugating enzyme to SCF complex. Our in vitro binding competition assay shows that Lag2 inhibits Cdc34 interaction with SCF complex. There are two underlying mechanisms to explain this inhibition. One is that Lag2 may directly bind to RING-finger domain of Rbx1, which blocks Cdc34 association. The other possibility is that the interaction of Lag2 to SCF complex induces the conformational change, which interrupts Cdc34 access to SCF complex. The exact mechanism needs to clarify with further structural analysis.
We show the molecular mechanism underlying the inhibition of SCF activity executed by Lag2 in two different direct and indirect pathways. First, as we mentioned above, Lag2 directly inhibits the activity of SCF E3 ligase by blocking E2 association to SCF. This notion is also supported by our finding that in rub1Δ cdc34-2 cell viability was partially rescued by the further loss of Lag2 function. Second, Lag2 interrupts rubylation of Cdc53, resulting in the down-regulation of SCF activity, which consists of two mechanisms. One is that Lag2 competes with Dcn1 in binding to Cdc53, which interrupts Rub1 conjugation to Cdc53 activated by Dcn1. The other is that Lag2 itself directly suppresses the rubylation of Cdc53, which is supported by our in vivo and in vitro experiments that even in the absence of Dcn1, deletion of lag2 stimulates rubylation activity.
In genetic experiments, we observed that single deletion of lag2, dcn1, or rub1 gene did not display obvious effects on W303-1A cell growth, whereas the double deletion of lag2 and dcn1 or lag2 and rub1 inhibited cell growth in a synthetic manner. It suggests that this growth defect is caused by the loss of both Lag2 function and rubylation of Cdc53, rather than rubylation cycle alone. In the absence of Rub1 conjugation to Cdc53, loss of Lag2 function may strongly affect SCF E3 activity, which disturbs the balance of cell-cycle regulators, in turn results in the retardation of cell growth. This imbalance of SCF E3 function may also explain the growth discrepancy that is observed in cdc34-2 lag2Δ mutant in the presence or absence of Rub1.
During further examination of Lag2 function, consistent with an earlier report, we observed that Dcn1 promoted the transfer of Rub1 onto Cdc53. However, with recombinant E2, Rub1 was conjugated to Cdc53 independently of Dcn1. One explanation is that high concentrations of E2 may be partially responsible for this Dcn1-independent in vitro rubylation. We also found that in dcn1Δ yeast cells, about 15% of Cdc53 was in the rubylated form, whereas in the triple mutant lag2Δ dcn1Δ jab1Δ, almost all Cdc53 was in rubylated form. Combined with in vitro observations, these data suggest that in living cells, the local concentration of E2 around the Cdc53-Rbx1 module could be high, so that Rub1 can conjugate to Cdc53, even in the absence of Dcn1. There still remains a minor possibility that some other unknown Dcn1-independent rubylation pathways may exist. In addition, in cdc34-2 ts cells, rub1 gene deletion resulted in cell inviability. However, dcn1 deletion in the cdc34-2 ts cell did not show any growth deficiency. Is there other unknown function of Dcn1 unrelated to the rubylation of Cdc53? The further mechanical analysis of Dcn1 remains to be clarified.
It is interesting to note that, functionally, Lag2 is partially similar to CAND1, which inhibits neddylation of cullins, interferes with ubiquitylation in vitro, and regulates SCF E3-ubiquitin ligase function (Liu et al, 2002; Zheng et al, 2002). A CAND1-like protein has not been identified in S. cerevisiae until now. Lag2 binds to Cdc53 and inhibits the rubylation of Cdc53, which indicates that Lag2 might function as a counterpart of CAND1 in budding yeast. CAND1 associates to unneddylated cullins and the modification of Nedd8 to cullins kicks off CAND1 from cullins. Similarly, Lag2 specifically binds to unrubylated Cdc53, which suggests that the rubylation of Cdc53 may block the association of Lag2 to Cdc53. However, two important differences between CAND1 and Lag2 attract our attention. One difference is that CAND1 binds to all scaffold cullins, but Lag2 only detectably interacts with Cdc53. The other difference is that CAND1 interacts with cullins and Rbx1 in an exclusively competitive reaction to Skp1. In other words, CAND1 interrupts the assembly of the SCF complex. In contrast, Lag2 binds SCF complex formed by Cdc53, Rbx1, Skp1, and F-box protein. We propose that Lag2 functions in a different manner from CAND1.
Lag2 was originally reported to be involved in determining cell longevity. On the basis of our observations, Lag2 inhibits the ubiquitylation and rubylation activities of the SCF complex in budding yeast. However, we failed to confirm the ubiquitylation and rubylation inhibition executed by Lag2 in in vitro reactions with baculovirus or bacterially expressed recombinant proteins. This implies that additional components in yeast may be involved in these reactions. Regardless of the exact mechanism of Lag2, our findings show an expanded function for Lag2 in regulating cellular longevity by controlling SCF function.
Materials and methods
Yeast strains and growth conditions
Yeast strains used in this study are listed in Supplementary Table I. All strains except for L40 were derived from W303-1A. The strains were constructed with the use of standard genetic techniques and grown in rich medium, YPD (1% yeast extract, 1% peptone, and 2% glucose), YPG (1% yeast extract, 1% peptone, and 2% galactose), or standard minimal medium, SD (0.67% Yeast nitrogen base without amino acids and 2% glucose) lacking the appropriate amino acid (Sherman et al, 1986).
Antibodies
Mouse monoclonal antibodies to HA (12CA5) and c-myc (9E10) were obtained from Roche. The mouse monoclonal antibody to FLAG (M2) was from Sigma. The mouse monoclonal antibody to GST (4C10) was from Covance. The mouse monoclonal antibody to Protein C (HPC4) was from Roche. Rabbit polyclonal antibodies to Cdc53 (y-300) and goat polyclonal antibodies to Skp1 (yC-20), Cdc4 (yN-19) and Rbx1 (yC-14) were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies to EF2 were described earlier (Inada and Aiba, 2005).
Plasmid construction
The pLexA-KM bait vector was constructed by replacing the GAL4 (AD) gene and the ampicillin resistance gene of pACT2 (BD Biosciences) with the LexA (BD) gene and the kanamycin resistance gene, respectively. DNAs encoding S. cerevisiae Ydr131c, S. cerevisiae Yjl149w, S. cerevisiae Ydr306c, S. cerevisiae Ybr203w, and S. cerevisiae Ymr258c were subcloned into the pLexA-KM vector. The pRS306ADH vector was generated by subcloning the SacI/KpnI fragment from p416ADH into the SacI/KpnI sites of pRS306. A DNA encoding S. cerevisiae Lag2 N-terminally tagged with four copies of the HA epitope (4HA) was subcloned into the pRS306ADH vector. The pRS304GAL vector was generated by subcloning the PCR fragment of Gal1 promoter into the EcoRI/BamHI sites of pRS304. DNAs encoding S. cerevisiae Sic1 and Cln2 tagged at their N-termini with a His6 epitope and 3FLAG were subcloned into the pRS304GAL vector. DNAs for S. cerevisiae Lag2, S. cerevisiae Lag2 C-terminally tagged with 3FLAG, S. cerevisiae Dcn1, S. cerevisiae Rub1, and S. cerevisiae Sic1 tagged at its N-terminus with an HPC4 epitope were subcloned into the pRSETB vector (Invitrogen). A DNA for S. cerevisiae Ubc12 was subcloned into the pGEX-6P-2 vector (GE Healthcare). DNAs for S. cerevisiae Cdc4 tagged at its N-terminus with a His6 epitope and S. cerevisiae Skp1, S. cerevisiae Cdc53, and S. cerevisiae Rbx1 N-terminally tagged with a His6 epitope were subcloned into the pFastBac Dual vector (Invitrogen). Expression vectors for mouse ubiquitin, S. cerevisiae Ula1, S. cerevisiae Uba3, S. cerevisiae Ubc12, S. cerevisiae Cdc34, S. cerevisiae Cln2, and S. cerevisiae Cdc28 were described earlier (Kamura et al, 1999a, 2004).
Yeast two-hybrid system
Yeast strain L40 was transformed both with the plasmid pLexA-KM encoding the F-box proteins Ydr131c, Yjl149w, Ydr306c, Ybr203w, or Ymr258c and with a budding yeast DNA library in the pJG4-5 vector (Origene). The cells were then streaked on plates of medium lacking histidine to detect interaction-dependent activation of HIS3.
Yeast extracts
Yeast cells grown in YPD were collected in the exponential growth phase. Total cell extracts were prepared by the TCA lysis method (Avaro et al, 2002).
Immunoprecipitation and immunoblot analysis
Cells were harvested and lysed with a solution containing 40 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, 0.5% Triton X-100, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin using glass beads and a multibeads shocker (Yasui Kikai, Japan). Lysates were incubated with anti-HA (12CA5) and protein A Sepharose for 1 h at 4°C. The beads were then washed three times with a solution containing 40 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, and 0.5% Triton X-100. Immunoprecipitated proteins were separated by SDS–PAGE, transferred to Hybond-P membranes (GE Healthcare), and subjected to immunoblot analysis. Immune complexes were detected with Supersignal West Pico or West Dura chemiluminescence reagents (Pierce).
In vitro Rub1 conjugation assay
His6-myc-Ula1 and His6-FLAG-Uba3 were coexpressed in Sf21 cells and purified by Ni2+-agarose affinity chromatography, as described earlier (Kamura et al, 1999a). His6-FLAG-Ubc12, His6-Rub1, His6-Lag2, and His6-Dcn1 were expressed in E. coli BL21 (DE3) and purified by Ni2+-agarose affinity chromatography. Yeast cells were harvested in the exponential growth phase and lysed with a solution containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, 0.2% Triton X-100, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin using glass beads and a multibead shocker. For the Dcn1-dependent in vitro Rub1 conjugation assay, 15 μg of protein lysate from the yeast cells was mixed with 30 ng of Ula1, 20 ng of Uba3, and 600 ng of His6-Rub1 in a 6-μl reaction containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 mM EDTA (pH 7.5), 10% (vol/vol) glycerol, and 1.5 mM ATP. In addition, recombinant His6-Lag2 and His6-Dcn1 were added to the reactions as shown in Figure 2E–G. For the Dcn1-independent in vitro Rub1 conjugation assay, 60 ng of Ubc12 was added to the reactions. Reaction mixtures were incubated at 26°C for the times indicated in the figures. Rubylated native Cdc53 was detected by immunoblotting with anti-Cdc53 antibodies.
In vitro ubiquitylation assay
Myc-Uba1-His6, His6-Cdc34, His6-Ubiquitin, His6-Lag2, and His6-HPC4-Sic1 were expressed in E. coli BL21 (DE3) and purified by Ni2+-agarose affinity chromatography (Invitrogen). The complex of His6-HA-Cln2 and His6-myc-Cdc28 was purified by Ni2+-agarose chromatography from lysates of Sf21 cells that had been coinfected with baculoviruses encoding Cln2 and Cdc28. Sic1 was phosphorylated by incubation with the Cln2-Cdc28 complex in a solution containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 mM EDTA (pH 7.5), 10% (vol/vol) glycerol, and 1.5 mM ATP at 26°C for 1 h. Yeast cells were harvested in the exponential growth phase and lysed with a solution containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, 0.2% Triton X-100, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin using glass beads and a multibeads shocker. For the in vitro Sic1 ubiquitylation assay, 30 μg of the lysates from yeast cells were mixed with 30 ng of phosphorylated Sic1, 30 ng of Uba1, 30 ng of Cdc34, and 1 μg of His6-ubiquitin in a 8-μl reaction containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 mM EDTA (pH 7.5), 10% (vol/vol) glycerol, and 1.5 mM ATP. In addition, His6-Lag2 was added to the reactions as shown in Figure 6C and D. Reaction mixtures were incubated at 26°C for the times indicated in Figure 6.
In vitro binding assay
SCFCdc4 complex was purified by Ni2+-agarose chromatography from lysates of Sf21 cells that had been coinfected with baculoviruses encoding His6-Cdc4, Cdc53, Skp1, and His6-Rbx1. GST-Cdc34, His6-Lag2, and His6-Lag2-3FLAG were expressed in E. coli BL21 (DE3) and purified by glutathione sepharose (GE Healthcare) or Ni2+-agarose affinity chromatography. For the in vitro binding experiments between Lag2 and SCFCdc4 complex, 250 ng of His6-Lag2-3FLAG was mixed with 1 μg of SCFCdc4 complex in a 12-μl reaction containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, and 10% (vol/vol) glycerol at 4°C for 1 h, and then subjected to immunoprecipitaion analysis with anti-FLAG (M2). To examine the ability of Lag2 to inhibit the association of Cdc34 to SCF complex, 250 ng of GST-Cdc34 was mixed with 200 ng of SCFCdc4 complex and His6-Lag2 as shown in Figure 7A in a 20-μl reaction containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, and 10% (vol/vol) glycerol at 4°C for 1 h, and then subjected to glutathione-sepharose affinity purification. To test the ability of Lag2 to inhibit the association of Cdc53 to Rbx1, Cdc53/His6-Rbx1 complex was expressed in Sf21 cells and immobilized on Ni2+-agarose beads; 5 μl of Ni2+-agarose beads containing 50 ng of Cdc53/His6-Rbx1 complex was mixed with His6-Lag2 as shown in Figure 7B in a 20-μl reaction containing 40 mM Tris–HCl (pH 7.5), 60 mM NaCl, 1 mM dithiothreitol, and 10% (vol/vol) glycerol at 4°C for 1 h. The beads were then washed three times with a solution containing 40 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, and 0.5% Triton X-100. Bound proteins were separated by SDS–PAGE and subjected to immunoblot analysis.
RT–PCR
Total RNA (0.5 μg) isolated from yeast cells with the use of an ISOGEN kit (Nippongene) was subjected to reverse transcription (RT) with a RevaTra Ace kit (Toyobo). The resulting cDNA was subjected to PCR with a Phusion High-Fidelity DNA Polymerase (New England BioLabs).
Supplementary Material
Supplementary Table I
Review Process File
Acknowledgments
We thank Mike Tyers and Yoshiko Kikuchi for yeast strains, Yasushi Saeki for GST-Cdc34 vector, and Mie Hirano for help in preparation of the paper. This work was supported in part by a grant from the Ministry of Education, Science, Sports, and Culture of Japan, the Sumitomo Foundation, the Uehara Foundation, and the Saibou Kagaku Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Avaro S, Belgareh-Touze N, Sibella-Arguelles C, Volland C, Haguenauer-Tsapis R (2002) Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast 19: 351–371 [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET (2008) Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress AM, Franklin DS, Pinswasdi C, Kale S (1996) LAG2, a gene that determines yeast longevity. Microbiology 142 (Part 8): 2289–2297 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Cyr DM, Hohfeld J, Patterson C (2002) Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci 27: 368–375 [DOI] [PubMed] [Google Scholar]
- Dixon C, Brunson LE, Roy MM, Smothers D, Sehorn MG, Mathias N (2003) Overproduction of polypeptides corresponding to the amino terminus of the F-box proteins Cdc4p and Met30p inhibits ubiquitin ligase activities of their SCF complexes. Eukaryot Cell 2: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronkin S, Djagaeva I, Beckendorf SK (2003) The COP9 signalosome promotes degradation of cyclin E during early Drosophila oogenesis. Dev Cell 4: 699–710 [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa S, Camblong J, Galan JM, Pinson B, Daignan-Fornier B (2006) Proteasome- and SCF-dependent degradation of yeast adenine deaminase upon transition from proliferation to quiescence requires a new F-box protein named Saf1p. Mol Microbiol 60: 1014–1025 [DOI] [PubMed] [Google Scholar]
- Escusa S, Laporte D, Massoni A, Boucherie H, Dautant A, Daignan-Fornier B (2007) Skp1-Cullin-F-box-dependent degradation of Aah1p requires its interaction with the F-box protein Saf1p. J Biol Chem 282: 20097–20103 [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ (1997) A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- Galan JM, Peter M (1999) Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119: 517–528 [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP (2007) Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111–33120 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Inada T, Aiba H (2005) Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J 24: 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW (1999a) The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev 13: 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI (2004) Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol 6: 1229–1235 [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG Jr, Elledge SJ, Conaway RC, Harper JW, Conaway JW (1999b) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284: 657–661 [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F (2008) Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell 29: 23–35 [DOI] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O′Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M (2005) The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435: 1257–1261 [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev 12: 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gazdoiu S, Pan ZQ, Fuchs SY (2004) Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J Biol Chem 279: 11074–11080 [DOI] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T (2003) Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10: 1511–1518 [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson S, Byers B, Goebl M (1999) The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol Cell Biol 19: 1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megumi Y, Miyauchi Y, Sakurai H, Nobeyama H, Lorick K, Nakamura E, Chiba T, Tanaka K, Weissman AM, Kirisako T, Ogawa O, Iwai K (2005) Multiple roles of Rbx1 in the VBC-Cul2 ubiquitin ligase complex. Genes Cells 10: 679–691 [DOI] [PubMed] [Google Scholar]
- Merlet J, Burger J, Gomes JE, Pintard L (2009) Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66: 1924–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel JJ, McCarville JF, Xiong Y (2003) A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J Biol Chem 278: 22828–22837 [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell 3: 535–541 [DOI] [PubMed] [Google Scholar]
- Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B (2003) Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol 13: 911–921 [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Miyauchi Y, Iwai K, Chiba T, Saeki Y, Matsuda N, Tanaka K, Kato K (2007) Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol 14: 167–168 [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev 13: 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB (eds) (1986) Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284: 662–665 [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M (2007) Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Wimuttisuk W, Singer JD (2007) The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell 18: 899–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C, Sutterluty H, Blondel M, Gstaiger M, Peter M, Reymond F, Krek W (2000) The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J 19: 5362–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA, McKeon F, Jackson PK (1999) F-box/WD-repeat proteins pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of cdc18p. Curr Biol 9: 373–376 [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H (2002) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 10: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Zhou P, Howley PM (1998) Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol Cell 2: 571–580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I
Review Process File