Abstract
Aims/hypothesis
Evidence from candidate gene studies suggests that obesity may modify genetic susceptibility to type 2 diabetes and dyslipidaemia. On an aggregate level, gene–obesity interactions are expected to result in different heritability estimates at different obesity levels. However, this hypothesis has never been tested.
Method
The present study included 2,180 British female twins. BMI was used as an index of general obesity. Outcome measures were insulin sensitivity (indexed by quantitative insulin-sensitivity check index [QUICKI]) and fasting plasma lipid profile. Structural equation modelling was used to test whether BMI interacted with latent genetic and environmental effects to impact on the outcome measures.
Results
Genetic influences on triacylglycerol increased with BMI (p < 0.001) whereas the unique environmental influence on QUICKI decreased with BMI (p < 0.001), resulting in a higher heritability estimate for both measures at higher BMI levels. This was further illustrated by stratified analysis in twin pairs concordant for normal weight and twin pairs concordant for overweight. Heritability was 19 percentage points higher for triacylglycerol (p < 0.001) and 31 percentage points higher for QUICKI (p < 0.01) among twins concordant for overweight than among twins concordant for normal weight. BMI had no moderator effect on the latent genetic and environmental factors for total cholesterol and HDL-cholesterol.
Conclusions/interpretation
Our results suggest that the expression of genes influencing triacylglycerol and insulin sensitivity can vary as a function of obesity status. The substantial increases in the genetic contribution to the total variance in insulin sensitivity and triacylglycerols at higher BMIs may prove extremely valuable in the search for candidate genes.
Keywords: Gene–environment interaction, Heritability, Insulin sensitivity, Lipid, Obesity
Introduction
In the past decade the prevalence of overweight and obesity has increased dramatically [1–4]. Currently in the United States, 65% of adults are overweight and 31% are obese. Obesity is associated with significantly increased risk of dyslipidaemia and type 2 diabetes [1, 2, 5]. Not surprisingly, the prevalence of dyslipidaemia and type 2 diabetes has also increased in the last decade.
Although the association between obesity and dyslipidaemia or type 2 diabetes is well documented [1, 2, 5], the exact nature of this relation remains unclear. The fact that obesity and dyslipidaemia/type 2 diabetes show significant heritability raises the possibility that common genetic vulnerability may account for the association. It is of interest to know whether the association between obesity and dyslipidaemia/type 2 diabetes still exists after excluding common genetic influence. By comparing genetically identical, monozygotic twins discordant for obesity, this hypothesis has been confirmed with significant higher lipid profile and lower insulin sensitivity identified in the obese co-twins [6–9]. On the other hand, it is also possible that obesity modifies genetic susceptibility to dyslipidaemia/type 2 diabetes and that this gene–obesity interaction also partly accounts for the association between obesity and dyslipidaemia/type 2 diabetes. The current evidence is from candidate gene studies. For example, it has been reported that effects of polymorphisms in the apolipoprotein E, apolipoprotein B100 and lipoprotein lipase genes on lipoprotein and lipid levels may depend on obesity [10–13]. Similarly, studies have also shown that BMI or obesity status can modify the associations of interleukin-6, ectonucleotide pyrophosphatase phosphodiesterase 1, angiotensin-converting enzyme and insulin receptor substrate-1 genotypes with insulin resistance or the risk of type 2 diabetes [14–18]. On an aggregate level, gene–obesity interactions are expected to result in different heritability estimates at different BMI levels. However, this hypothesis has never been tested. The large dataset of the Twins UK Registry, which has measures on BMI, lipid profile and insulin sensitivity, provides a unique opportunity to investigate this hypothesis. We therefore investigated, using the data from the Twins UK Registry, the extent to which obesity may modify genetic susceptibility to dyslipidaemia and type 2 diabetes.
Methods
Participants
The present study included 430 monozygotic (215 pairs) and 1,750 dizygotic (875 pairs) British female twins (mean ± SD age 45.9 ± 12.1; range 18.4–76.3 years) from the Twins UK Registry, which comprises unselected, mostly women volunteers from the general population, recruited through national media campaigns in the UK [19]. Means and ranges of quantitative phenotypes in Twins UK are similar to the age-matched general population in the UK [20]. Patients with diabetes and on lipid-lowering medications were excluded from the current study. Zygosity was determined by standardised questionnaire and confirmed by DNA fingerprinting. Written informed consent was obtained from all participants before they entered the study, which was approved by the local Research Ethics Committee.
Measurements
Height was measured to the nearest 0.5 cm using a wall-mounted stadiometer. Weight (light clothing only) was measured to the nearest 0.1 kg using digital scales. BMI was calculated as weight divided by height squared (kg/m2). A venous blood sample was taken between 08:00 and 10:00 hours after an overnight fast. Plasma levels of total cholesterol, HDL-cholesterol and triacylglycerol were measured using an analysing device (Cobas Fara; Roche Diagnostics, Lewes, UK) [21]. Fasting insulin was measured by immunoassay (Abbott Laboratories, Maidenhead, UK) and fasting glucose was measured on a multichannel analyser using an enzymatic colorimetric slide assay (Ektachem 700; Johnson and Johnson Clinical Diagnostic Systems, Amersham, UK). Based on fasting glucose and insulin, quantitative insulin sensitivity check index (QUICKI) was calculated to index insulin sensitivity [22]. QUICKI is among the most thoroughly evaluated and validated surrogate indices for insulin sensitivity. Studies have demonstrated that as an index of insulin sensitivity, QUICKI is reliable, reproducible, accurate and has excellent predictive power [22]. In our cohort, the correlation between QUICKI and HOMA index of insulin resistance (HOMA-IR) was −0.99 [22].
Statistical analysis
Structural equation modelling was the primary method of analysis. It is based on comparison of the variance–covariance matrices in monozygotic and dizygotic twin pairs and allows separation of the observed phenotypic variance into its genetic and environmental components, i.e. additive (A) or dominant (D) genetic components, and common (C) and unique (E) environmental components [23]. We focused on additive genetic, common and unique environmental effects as previously published twin literature [24–26] had found little evidence for dominant genetic effects on dyslipidaemia and type 2 diabetes.
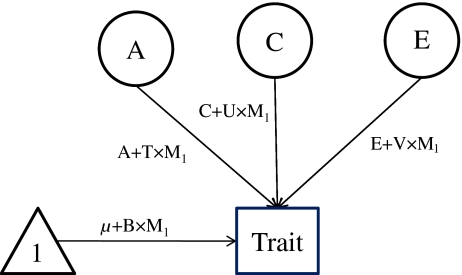
We first fit the basic gene–environment interaction models as described by Purcell [27], using BMI as a continuous moderator incorporating all the available twin pairs. In this gene–environment interaction model (Fig. 1), the phenotypic variance of the outcome variables (e.g. total cholesterol, HDL-cholesterol, triacylglycerol and QUICKI) was portioned into additive, common and unique components (A, C, E as above), with the path coefficients associated with each variable expressed as linear functions of the moderator, e.g. A + T × M1, C + U × M1, E + V × M1, where T represents the effects of the moderator on additive genetic variance, and U and V represent the effects of the moderator on common and unique environmental variance, respectively. In addition, the effect of the moderate on the mean was also modelled, e.g. µ + B × M1, where µ represents the constant term, M1 represents the value of the moderator and B represents linear effects on the outcome. The mean structure encompasses any phenotypic correlation between BMI and the outcome variables, permitting analyses of gene–BMI interaction independent of any gene–BMI correlation. As we were primarily interested in the effects of BMI on variance components, we forced B in the model to guard against the influence of gene–BMI correlation. On the other hand, a significant compromise of model fit when variables T, U or V are fixed to zero reflects evidence of significant moderation of additive genetic, common environmental or unique environmental variance by BMI, respectively.
Fig. 1.
Partial path diagram for the basic gene–environment interaction model. A, additive genetic effects; C, common environmental effects; E, unique environmental effects; M, moderator; T, moderated component of A; U, moderated component of C; V, moderated component of E; B, linear effects of moderator on mean (forced entry)
Next, for the outcome variables in which a significant gene–BMI interaction was identified in the above model, we further stratified the sample into normal weight (BMI < 25 kg/m2) and overweight (BMI ≥ 25 mg/m2) to explore differences in additive, common and unique components (A, C, E as above). In this round of analyses, twin pairs who were either concordant for normal weight or concordant for overweight were included in the analysis.
Prior to analysis, QUICKI and lipid profile measures were log-transformed to obtain a better approximation of normal distribution. Effect of age was regressed out for the above outcome variables before using the residuals in model fitting. The significance of variance components T, U, V, A and C (as above) was assessed by testing the deterioration in model fit after each component was dropped from the full model. Standard hierarchic χ 2 tests were used to select the best fitting models in combination with Akaike's information criterion (AIC) (AIC = χ 2 − 2df). The model with the lowest AIC reflects the best balance of goodness of fit and parsimony [23]. Preliminary analyses were done using STATA 8.0 (Stata, College Station, TX, USA). Genetic modelling was carried out with Mx, a computer program specifically designed for the analysis of twin and family data [28]. To avoid the potential influences of participants with extreme obesity, we repeated all the analyses after excluding 14 participants with BMI ≥40 kg/m2. The results were virtually identical (data not shown).
Results
Table 1 presents the general characteristics of participants by zygosity. The average age, BMI, mean QUICKI and mean concentrations of total cholesterol, HDL-cholesterol and triacylglycerol were very similar in these two zygosity groups.
Table 1.
General characteristics of study participants by zygosity
| Variables | Monozygotic twins | Dizygotic twins |
|---|---|---|
| n | 430 | 1,750 |
| Age (years) | 46.1 ± 13.7 | 45.9 ± 11.6 |
| BMI (kg/m2) | 24.1 ± 3.8 | 25.0 ± 4.6 |
| QUICKIa | 0.24 ± 0.03 | 0.24 ± 0.03 |
| Triacylglycerol (mmol/l)b | 1.16 ± 0.66 | 1.20 ± 0.73 |
| Total cholesterol (mmol/l)b | 5.54 ± 1.27 | 5.45 ± 1.23 |
| HDL-cholesterol (mmol/l)b | 1.53 ± 0.37 | 1.53 ± 0.39 |
Values are means ± SD
aData available on 135 monozygotic and 685 dizygotic pairs
bData available on 193 monozygotic and 826 dizygotic pairs
Comparative model fits testing the extent to which BMI served as a moderator of QUICKI and lipid profile are presented in Table 2. Each model includes the variable B, representing linear effects of BMI on the mean of respective outcome variables. The full model (ACETUVB), including additive genetic (A), shared environmental (C), unique environment (E) effects, as well as terms for the moderation of each by BMI (T, U and V, respectively), is presented first. Next, a backwards stepwise elimination procedure is followed for the individual variables, testing the extent to which the variable associated with the smallest change in log-likelihood ratio contributes significantly to the model. Moderation terms are tested first, followed by variance components with no evidence of moderation by BMI. In the final model, each variable contributed significantly to model fit.
Table 2.
Comparative models fits for BMI as a continuous moderator of insulin sensitivity and lipid profile
| Variables | Comparative model fit | ||||
|---|---|---|---|---|---|
| Model | Δ-2LL | df | p value | Test | |
| QUICKI | |||||
| 1. Full | ACETUVB | ||||
| 2. U = 0 | ACETVB | 1.52 | 1 | 0.22 | 2 vs 1 |
| 3. T = 0 U = 0 | ACEVB | 0.44 | 1 | 0.51 | 3 vs 2 |
| 4. V = 0 T = U = 0 | ACEB | 25.2 | 1 | <0.001 | 4 vs 3 |
| 5. C = 0 T = U = 0a | AEVB | 0.42 | 1 | 0.52 | 5 vs 3 |
| Triacylglycerol | |||||
| 1. Full | ACETUVB | ||||
| 2. U = 0 | ACETVB | 0.01 | 1 | 0.96 | 2 vs 1 |
| 3. V = 0 U = 0 | ACETB | 0.04 | 1 | 0.82 | 3 vs 2 |
| 4. T = 0 U = V = 0 | ACEB | 14.7 | 1 | <0.001 | 4 vs 3 |
| 5. C = 0 U = V = 0a | AETB | 0.00 | 1 | 1.00 | 5 vs 3 |
| Total cholesterol | |||||
| 1. Full | ACETUVB | ||||
| 2. U = 0 | ACETVB | 2.27 | 1 | 0.13 | 2 vs 1 |
| 3. V = 0 U = 0 | ACETB | 0.01 | 1 | 0.96 | 3 vs 2 |
| 4. T = 0 U = V = 0a | ACEB | 3.59 | 1 | 0.06 | 4 vs 3 |
| 5. C = 0 T = U = V = 0 | AEB | 4.40 | 1 | 0.04 | 5 vs 4 |
| HDL-cholesterol | |||||
| 1. Full | ACETUVB | ||||
| 2. U = 0 | ACETVB | 0.12 | 1 | 0.73 | 2 vs 1 |
| 3. V = 0 U = 0 | ACETB | 0.18 | 1 | 0.67 | 3 vs 2 |
| 4. T = 0 U = V = 0a | ACEB | 3.09 | 1 | 0.08 | 4 vs 3 |
| 5. C = 0 T = U = V = 0 | AEB | 9.60 | 1 | 0.002 | 5 vs 4 |
aThe best fitting model
Δ-2LL, log likelihood; A, additive genetic variance; B, linear effects of BMI on means of the outcome variables; C, shared environmental variance; df, degrees of freedom; E, unique environmental variance; T, moderation of additive genetic variance by BMI; U, moderation of shared environmental variance by BMI; V, moderation of unique environmental variance by BMI
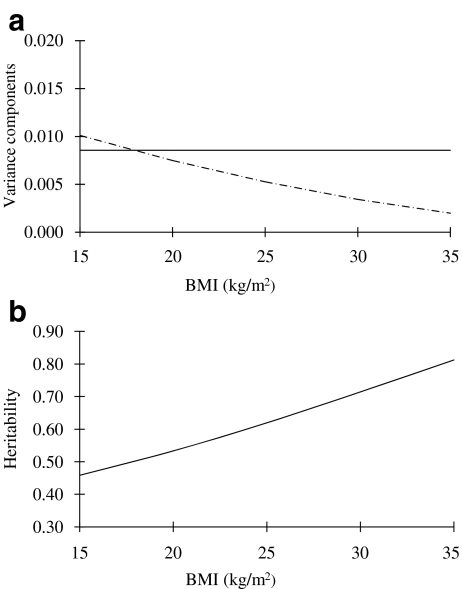
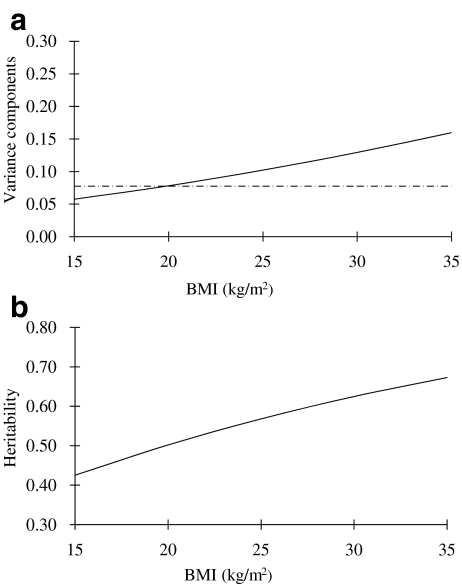
The best fitting model for QUICKI was AEVB, in which moderation of the unique environment component by BMI (V) contributed significantly to the model (p < 0.001). As shown in Fig. 2a, the unique environmental influence on QUICKI decreased with BMI. This resulted in a higher heritability estimate for QUICKI at higher BMI levels (Fig. 2b). At the mean BMI level (25 kg/m2), the heritability of QUICKI was estimated to be 0.61 (95% CI 0.54–0.68). With regard to the lipid profile, the best fitting model for triacylglycerol was AETB, in which moderation of the additive genetic component by BMI contributed significantly to the model (p < 0.001). As shown in Fig. 3a, the genetic influence on triacylglycerol increased with BMI, also resulting in a higher heritability estimate for triacylglycerol at higher BMI levels (Fig. 3b). At the mean BMI level (25 kg/m2), heritability of triacylglycerol was estimated to be 0.57 (95% CI 0.49–0.64). The best fitting model for total cholesterol and HDL-cholesterol was ACEB, which showed that BMI had no moderator effect on the latent genetic and environmental factors for these two outcome variables. The estimated heritability from the best fitting model was 0.48 (95% CI 0.28–0.66) for total cholesterol and 0.50 (95% CI 0.33–0.65) for HDL-cholesterol, respectively.
Fig. 2.
Change of (a) additive genetic (continuous line) and unique environmental (dashed line) variance of QUICKI with increasing level of BMI, and (b) heritability of QUICKI with increasing level of BMI
Fig. 3.
Change of (a) additive genetic (continuous line) and unique environmental (dashed line) variance of triacylglycerol with increasing level of BMI, and (b) of heritability of triacylglycerol with increasing level of BMI
The increased heritability of QUICKI and triacylglycerol with increased BMI was further illustrated by stratified analysis in twin pairs concordant for normal weight (BMI < 25 kg/m2) and twin pairs concordant for overweight (BMI ≥ 25 kg/m2). As shown in Table 3, heritability was 19 percentage points higher for triacylglycerol (p < 0.001) among twins concordant for overweight (h2 = 0.67; 95% CI 0.52–0.78) relative to twins concordant for normal weight (h2 = 0.48; 95% CI 0.36–0.60). Similarly, heritability was 31 percentage points higher for QUICKI (p < 0.001) among twins concordant for overweight (h2 = 0.81; 95% CI 0.65–0.90) relative to twins concordant for normal weight (h2 = 0.50; 95% CI 0.37–0.60).
Table 3.
Twin pair correlations and univariate twin structural equation model variable estimates for QUICKI and triacylglycerol for normal weight and overweight participants
| Variables | Number of pairs | Correlations | h2 (95% CI) | e 2 (95% CI) | ||
|---|---|---|---|---|---|---|
| MZ | DZ | r mz | r dz | |||
| QUICKI | ||||||
| Normal weight | 73 | 282 | 0.48 | 0.31 | 0.50 (0.37–0.60) | 0.50 (0.40–0.63) |
| Overweight | 26 | 154 | 0.90 | 0.23 | 0.81 (0.65–0.90) | 0.19 (0.10–0.35) |
| Triacylglycerol | ||||||
| Normal weight | 99 | 342 | 0.52 | 0.21 | 0.48 (0.36–0.60) | 0.52 (0.40–0.64) |
| Overweight | 42 | 186 | 0.66 | 0.35 | 0.67 (0.52–0.78) | 0.33 (0.22–0.48) |
DZ, dizygotic twins; e2, variance explained by unique environment; h2, variance explained by additive genetics (heritability); MZ, monozygotic twins
Discussion
The important findings in this study are that the additive genetic influence on triacylglycerol levels and the unique environmental influence on QUICKI vary as a function of obesity. The influence of genetic factors on variation of triacylglycerol was more important and the influence of unique environmental factors on variation of QUICKI was less important in obese participants, resulting in a higher heritability estimate for triacylglycerol and QUICKI in participants with higher BMI levels.
These findings are in line with most previous candidate association studies [10–13, 15–18], which report that obese individuals in particular display the greatest genetic vulnerability to risk of dyslipidaemia and type 2 diabetes. For example, Gerdes et al. [12] showed that only in participants with BMI above the mean of the sample (26 kg/m2) carriers of the N9 allele of the lipoprotein lipase gene D9N variant had significantly higher plasma triacylglycerol levels than non-carriers. A study by Muthumala et al. [16] showed that only in obese participants did carriers of the deletion allele of the angiotensin-converting enzyme gene insertion/deletion polymorphism have a significantly higher risk of developing type 2 diabetes than non-carriers. Stolerman et al. [15] showed that the effect of the Q allele of the ectonucleotide pyrophosphatase phosphodiesterase 1 gene K12Q polymorphism on HOMA-IR was stronger in individuals with a higher BMI. Using a twin design, the current study not only for the first time confirmed the existence of gene–obesity interaction on triacylglycerol and QUICKI on an aggregate level, but also demonstrated that the mechanism behind this interaction might be different for these two traits. For triacylglycerol, expression of genes influencing triacylglycerol levels can vary as a function of obesity status, which indicates that new genes may be expressed at different BMI levels. These genes can only be detected by testing gene–BMI interactions in genetic association studies. On the other hand, our model suggests that no new genes are expressed in connection with QUICKI levels, because the increased heritability at higher BMI values is solely due to decreased unique environmental influences. In this case, although the genetic variants responsible for QUICKI can be detected in normal weight participants, the power to detect these variants will be larger if obese participants are used instead. These findings may prove extremely helpful in efforts to look for genes. Recently, the hypothesis-free genome-wide association (GWA) approach has successfully located many genes for dyslipidaemia and type 2 diabetes [29, 30]. However, these new discoveries only represent a small proportion (5–10%) of the genetic variation underlying susceptibility to these two disorders [29, 30]. Scrutiny of the most recent crop of GWA studies shows only a modest increment in new loci over the 2008 GWA for lipids and type 2 diabetes, indicating that the ‘low-hanging fruit’ has been harvested [29, 30]. Investigators have begun to realise the importance of gene–environment interactions. The identified gene–obesity interaction for triacylglycerol and QUICKI on an aggregate level shows strong support for gene–obesity interaction to be fully considered in the next wave of GWA studies on dyslipidaemia and type 2 diabetes.
We failed to detect gene–obesity interactions for total cholesterol and HDL-cholesterol. There could be several explanations for this. One explanation is gene–obesity correlation, which could be induced by pleiotropic genes influencing both BMI and the outcome of interest, i.e. total cholesterol or HDL-cholesterol. In our analysis, BMI (the moderator) was entered into the model to allow for a main effect on total cholesterol or HDL-cholesterol. However, this effectively removes all genetic effects that are shared between trait and moderator from the covariance model. Thus, any interactions detected cannot be due to gene–BMI correlation. Such an approach would fail to detect gene–BMI interaction in the presence of gene–BMI correlation, which could be the case here. However, after additionally fitting the extended gene–environment interaction model suggested by Purcell et al. [27] to allow for gene–environment correlation, we still did not observe gene–BMI interactions for total cholesterol and HDL-cholesterol (data not shown). A second explanation is that interaction effects are generally small and hence difficult to identify [31], suggesting that a larger sample might be required. In our study, the moderation effect of BMI (T) on the genetic component of total cholesterol and HDL-cholesterol was borderline significant (p = 0.06 for total cholesterol, p = 0.08 for HDL-cholesterol) (Table 2). By forcing T (effects of the moderator on additive genetic variance) into the model, we observed that the moderation effect of BMI on the genetic influence of total cholesterol and HDL-cholesterol followed the same pattern as that for triacylglycerol, i.e. the genetic influence on total cholesterol and HDL-cholesterol increased with BMI. Studies with larger sample sizes might be able to confirm this finding. Finally, it is also possible that there were no gene–BMI interaction effects for total cholesterol and HDL-cholesterol, at least not large enough to detect on an aggregate level.
Some limitations of the study warrant discussion. First, the model used to test for interaction including a continuously measured environmental variable is relatively new. However, the same pattern was found when the sample was split into overweight and normal weight groups, further supporting our conclusion. Second, our twin sample included only women and only whites; the generalisability of these results to men and to other ethnic populations remains to be determined. Third, BMI may affect the expression of different sets of genetic factors. To explore this possibility, we fitted another gene–environment interaction model as suggested by other investigators (M. C. Neal, M. C. Keller, unpublished results) for triacylglycerol and QUICKI. In this extended model, two sets of genetic factors are modelled. One represents those that contribute to constant heritability, while the second changes linearly with effect of the moderator. These two components are allowed to correlate freely. Using this model, we did not find any evidence to support the above possibility. However, with the current sample size, we might not have had sufficient power to detect this complex interaction.
In conclusion, the current twin study confirms the existence of gene–obesity interaction for triacylglycerol and QUICKI on an aggregate level. The substantial increases in the genetic contribution to the total variance in QUICKI and triacylglycerol may prove extremely helpful in gene-finding efforts.
Acknowledgements
The Twins UK (TUK) study was funded by: the Wellcome Trust; the European Community’s Seventh Framework Programme (FP7/2007–2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and FP7/2007–2013; the ENGAGE project grant agreement HEALTH-F4-2007-201413; and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also received support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London. T. D. Spector is a NIHR senior investigator. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (G20234).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AIC
Akaike's information criterion
- GWA
Genome-wide association
- HOMA-IR
HOMA index of insulin resistance
- QUICKI
Quantitative insulin-sensitivity check index
References
- 1.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 3.Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world—a growing challenge. N Engl J Med. 2007;356:213–215. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- 4.Caterson ID, Hubbard V, Bray GA, et al. Prevention Conference VII: obesity, a worldwide epidemic related to heart disease and stroke: Group III: worldwide comorbidities of obesity. Circulation. 2004;110:e476–e483. doi: 10.1161/01.CIR.0000140114.83145.59. [DOI] [PubMed] [Google Scholar]
- 5.Poirier P, Giles TD, Bray GA, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on Obesity and Heart Disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2006;113:898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 6.Pietilainen KH, Sysi-Aho M, Rissanen A, et al. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects—a monozygotic twin study. PLoS One. 2007;2:e218. doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronnemaa T, Marniemi J, Savolainen MJ, et al. Serum lipids, lipoproteins, and lipid metabolizing enzymes in identical twins discordant for obesity. J Clin Endocrinol Metab. 1998;83:2792–2799. doi: 10.1210/jc.83.8.2792. [DOI] [PubMed] [Google Scholar]
- 8.Mayer EJ, Newman B, Austin MA, et al. Genetic and environmental influences on insulin levels and the insulin resistance syndrome: an analysis of women twins. Am J Epidemiol. 1996;143:323–332. doi: 10.1093/oxfordjournals.aje.a008746. [DOI] [PubMed] [Google Scholar]
- 9.Newman B, Selby JV, Quesenberry CP, Jr, King MC, Friedman GD, Fabsitz RR. Nongenetic influences of obesity on other cardiovascular disease risk factors: an analysis of identical twins. Am J Public Health. 1990;80:675–678. doi: 10.2105/AJPH.80.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jemaa R, Elasmi M, Naouali C, et al. Apolipoprotein E polymorphism in the Tunisian population: frequency and effect on lipid parameters. Clin Biochem. 2006;39:816–820. doi: 10.1016/j.clinbiochem.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Turner PR, Talmud PJ, Visvikis S, Ehnholm C, Tiret L. DNA polymorphisms of the apoprotein B gene are associated with altered plasma lipoprotein concentrations but not with perceived risk of cardiovascular disease: European Atherosclerosis Research Study. Atherosclerosis. 1995;116:221–234. doi: 10.1016/0021-9150(94)05550-3. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes C, Fisher RM, Nicaud V, et al. Lipoprotein lipase variants D9N and N291S are associated with increased plasma triglyceride and lower high-density lipoprotein cholesterol concentrations: studies in the fasting and postprandial states: the European Atherosclerosis Research Studies. Circulation. 1997;96:733–740. doi: 10.1161/01.cir.96.3.733. [DOI] [PubMed] [Google Scholar]
- 13.Mailly F, Fisher RM, Nicaud V, et al. Association between the LPL-D9N mutation in the lipoprotein lipase gene and plasma lipid traits in myocardial infarction survivors from the ECTIM Study. Atherosclerosis. 1996;122:21–28. doi: 10.1016/0021-9150(95)05736-6. [DOI] [PubMed] [Google Scholar]
- 14.Herbert A, Liu C, Karamohamed S, et al. BMI modifies associations of IL-6 genotypes with insulin resistance: the Framingham Study. Obesity (Silver Spring) 2006;14:1454–1461. doi: 10.1038/oby.2006.165. [DOI] [PubMed] [Google Scholar]
- 15.Stolerman ES, Manning AK, McAteer JB, et al. Haplotype structure of the ENPP1 gene and nominal association of the K121Q missense single nucleotide polymorphism with glycemic traits in the Framingham Heart Study. Diabetes. 2008;57:1971–1977. doi: 10.2337/db08-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muthumala A, Gable DR, Palmen J, et al. Is the influence of variation in the ACE gene on the prospective risk of type 2 diabetes in middle-aged men modified by obesity? Clin Sci (Lond) 2007;113:467–472. doi: 10.1042/CS20070158. [DOI] [PubMed] [Google Scholar]
- 17.Baroni MG, Arca M, Sentinelli F, et al. The G972R variant of the insulin receptor substrate-1 (IRS-1) gene, body fat distribution and insulin-resistance. Diabetologia. 2001;44:367–372. doi: 10.1007/s001250051628. [DOI] [PubMed] [Google Scholar]
- 18.Clausen JO, Hansen T, Bjorbaek C, et al. Insulin resistance: interactions between obesity and a common variant of insulin receptor substrate-1. Lancet. 1995;346:397–402. doi: 10.1016/S0140-6736(95)92779-4. [DOI] [PubMed] [Google Scholar]
- 19.Spector TD, Williams FM. The UK Adult Twin Registry (TwinsUK) Twin Res Hum Genet. 2006;9:899–906. doi: 10.1375/twin.9.6.899. [DOI] [PubMed] [Google Scholar]
- 20.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–477. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 21.Middelberg RP, Spector TD, Swaminathan R, Snieder H. Genetic and environmental influences on lipids, lipoproteins, and apolipoproteins: effects of menopause. Arterioscler Thromb Vasc Biol. 2002;22:1142–1147. doi: 10.1161/01.ATV.0000022889.85440.79. [DOI] [PubMed] [Google Scholar]
- 22.Muniyappa R, Lee S, Chen H, Quon MJ. Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. Am J Physiol. 2008;294:E15–E26. doi: 10.1152/ajpendo.00645.2007. [DOI] [PubMed] [Google Scholar]
- 23.Neale MC, Cardon LR. Methodologies for genetic studies of twins and families. Dordrecht: Kluwer; 1992. [Google Scholar]
- 24.Beekman M, Heijmans BT, Martin NG, et al. Heritabilities of apolipoprotein and lipid levels in three countries. Twin Res. 2002;5:87–97. doi: 10.1375/1369052022956. [DOI] [PubMed] [Google Scholar]
- 25.Schousboe K, Visscher PM, Henriksen JE, Hopper JL, Sorensen TI, Kyvik KO. Twin study of genetic and environmental influences on glucose tolerance and indices of insulin sensitivity and secretion. Diabetologia. 2003;46:1276–1283. doi: 10.1007/s00125-003-1165-x. [DOI] [PubMed] [Google Scholar]
- 26.Snieder H, van Doornen LJ, Boomsma DI. Dissecting the genetic architecture of lipids, lipoproteins, and apolipoproteins: lessons from twin studies. Arterioscler Thromb Vasc Biol. 1999;19:2826–2834. doi: 10.1161/01.atv.19.12.2826. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S. Variance components models for gene–environment interaction in twin analysis. Twin Res. 2002;5:554–571. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- 28.Neale MC, Boker SM, Xie G, Maes HH (1999) Mx: statistical modeling. Virginia Commonwealth University, Richmond. Available from www.vcu.edu/mx/, accessed 1 March 2004
- 29.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 30.Florez JC. Clinical review: the genetics of type 2 diabetes: a realistic appraisal in 2008. J Clin Endocrinol Metab. 2008;93:4633–4642. doi: 10.1210/jc.2008-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silberg J, Rutter M, Neale M, Eaves L. Genetic moderation of environmental risk for depression and anxiety in adolescent girls. Br J Psychiatry. 2001;179:116–121. doi: 10.1192/bjp.179.2.116. [DOI] [PubMed] [Google Scholar]