Abstract
The epidermal growth factor receptor (EGFR) has been shown to be able to translocate to the nucleus where it is involved in many cellular process including transcriptional regulation and DNA repair. Recently, it has been shown that the DNA damage-inducing agents ionizing radiation (IR) and cisplatin are able to induce EGFR nuclear localization, and this nuclear localization is correlated with increased DNA-PK activity, which plays an essential role in DNA double stranded repair. Here we sought to determine if there is a causal relationship between nuclear EGFR and DNA repair activity. We found that mutation in the nuclear localization signal (NLS) of EGFR (mNLS), known to be unable to translocate to the nucleus, released EGFR induced resistance to cisplatin. Re-introduction of an NLS in the C-terminal allowed EGFR to re-enter the nucleus and the cells regained resistance to cisplatin. In addition, we show that the re-expression of a functional nuclear localization sequence in EGFR (mNLS-R) is not only able to restore its resistance to cisplatin, but also reduced the DNA damage caused by cisplatin, and restored DNA repair activity. Thus, we demonstrate here that nuclear EGFR is required for DNA repair and resistance to cisplatin treatment.
Keywords: Epidermal growth factor receptor, nuclear localization, dna damage, DNA repair, cisplatin
Introduction
Traditionally, the epidermal growth factor receptor (EGFR) is known to function at the cell membrane to induce the activation of several different signaling pathways involved in many different cellular functions such as cell proliferation and survival. Overexpression of EGFR occurs in many cancer types and is frequently a poor prognostic factor [1]. In addition to the traditional signaling pathway, EGFR has been shown to translocate to the nucleus upon EGF stimulation. Once in the nucleus, EGFR is involved in many cellular process, such as the transcription of genes associated with cell proliferation, tumor growth and metastasis [2–4], DNA synthesis [5], and DNA repair [5–7]. Although the function and mechanisms of nuclear EGFR are still continuing to be elucidated, the importance of nuclear EGFR is demonstrated by its association with poor prognosis in breast cancer [8], oranpharygeal cancer [9], esophageal squamous cell carcinoma [10] and ovarian cancer [11].
The involvement of nuclear EGFR in DNA repair occurs through its interactions with DNA proliferating cell nuclear antigen (PCNA) [5] and DNA dependent kinase (DNA-PK), which is required for DNA repair [7, 12, 13]. Dittmann et. al. has shown that after ionizing radiation or cisplatin treatment, both known to induce DNA damage, EGFR translocates to the nucleus. The translocation of EGFR to the nucleus was associated with an increase in DNA-PK activity and was inhibited by treatment with the C225 monoclonal antibody against EGFR [7]. Although a correlation between nuclear EGFR and DNA repair activity was seen, no causal effect was shown. Therefore, in this study, we sought to determine if EGFR nuclear localization is required for DNA repair induced by cisplatin treatment. Here we found that wildtype EGFR is resistant to cisplatin treatment and is able to induce DNA repair, both of which requires nuclear EGFR.
Materials and methods
Cell culture and transfection
The human HeLa epidermal carcinoma cells, HEK293 (human embryonic kidney cell), Chinese hamster ovary (CHO) and MCF7 breast cancer cells were purchased from American Tissue Type Collection (ATCC, Manassas, VA) and tested negative for Mycoplasma contamination in periodical evaluations. All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM)/F-12 with 10% fetal bovine serum and of 100 units/ml of penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2. Transfection was done with the cationic liposome SN (Stabilized Non-viral) or lipofectamine 2000 (Invitrogen, Carlsbad, CA) as previously described [14, 15]. Briefly, cells were grown on petri dishes or slides overnight and incubated with plasmid/liposome complex in Opti-MEM medium for 4 hours, after which the medium was replaced by complete medium and the cells were incubated at 37°C for 24–48 hours. MCF7 cells stably transfected with an empty vector, the wild-type or mutant EGFR were selected and maintained in 10 μg/ml blasticidin (Invivogen, San Diego, CA).
Plasmid constructs
The pcDNA6A-EGFR plasmid, expressing full-length human EGFR with a carboxyl-terminal myc-6His tag, was constructed and described previously [14]. Briefly, the DNA fragments amplified by PCR with a forward primer 5-ATTAAGCTTCGGGGAGCAGCGATG-3' and a reverse primer 5'-CCTTCTAGATGCTCCAATAA-ATTCACTG-3' were digested with HindIII and XbaI and cloned into the corresponding sites of the pcDNA6A/myc-His vector (Invitrogen). The EGFR mNLS (previously named NLSm12; 645AAAHIVAAA653) has a mutation in the first two basic amino acid clusters of EGFR tripartite NLS [14]. The EGFR mNLS-R was constructed by insertion of a short peptide encoding NLS of SV40 large T antigen (PKKRKV) at the C-terminus of EGFR mNLS. Primers 5'-CTAGACCAAAGAAGAAGAGAAAGGT GT T-3'and 5'-CGAACACCTTTCTCTTCTTCTTTGG T-3' were annealed and ligated to the plasmid EGFR mNLS digested with XbaI and BstBI.
Cellular fractionation and western blotting analyses
Cellular fractionation was performed as described previously [14]. Briefly, cells were washed twice with ice-cold phosphate-buffered saline, harvested by scraping with a rubber policeman, and lysed in a hypotonic lysis buffer (20 mM HEPES, pH 7.0, 10 mM KCl, 2 mM MgCl2, 0.5% NP-40, 1 mM Na3VO4, 10 mM NaF, 1 mM phenylmethanesulfonyl fluoride, 2 μg/ml aprotinin). After a 10 minute incubation on ice, cells were homogenized by 15–20 strokes in a tightly fitting Dounce homogenizer. To to confirm that more than 98% of cells had been lysed an aliquot of cells was checked for cell lysis by light microscopy. The homogenate was centrifuged 5 minutes at 1,500 × g to sediment the nuclei. The supernatant was then centrifuged at 16,100 × g for 20 minutes, and the resulting supernatant formed the non-nuclear fraction. To remove contamination from cytoplasmic membranes the nuclear pellet was washed three times with hypotonic lysis buffer, and the purity of the nuclei was confirmed by light microscopy. To extract nuclear proteins, the isolated nuclei were resuspended in NETN buffer (150 mM NaCl, 1 mM EDTA, 20 mM Tris-Cl [pH 8.0], 0.5% NP-40, 1 mM Na3VO4, 10 mM NaF, 1 mM phenylmethanesulfonyl fluoride, and 2 μg/ml aprotinin) and the mixture was sonicated briefly to aid nuclear lysis. After centrifugation at 16,100 × g for 20 minutes at 4°C the nuclear lysates were collected. To obtain whole-cell lysates, cells were lysed in NETN buffer with sonication; lysates were then centrifuged at 16,100 × g for 20 minutes. Protein concentrations were determined by using the BCA protein assay kit (Pierce, Rockford, IL) and mixing the samples with Laemmli sample buffer and heating them at 95°C for 5 minutes. Samples were then subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to nitrocellulose membranes. Prestained molecular mass standards for electrophoresis were obtained from Bio-Rad. Membranes were probed with monoclonal or polyclonal antibodies that were followed by horseradish peroxidase-labeled secondary antibodies.
Immunoreactive protein bands were detected with an enhanced chemiluminescence reagent (Pierce, Rockford, IL or Amersham Pharmacia Biotech, Piscataway, NJ). When required, the intensity of the expression proteins was quantified with a computer-assisted scanning densitometer equipped with Quantity One software (Bio-Rad, Hercules, CA). The antibodies used in this study were as follows: anti-EGFR (Santa Cruz, Santa Cruz, CA.; #sc-03); anti-phosphotyrosine, 4G10 (Millipore, Billerica, MA; #05–321); anti-phospho-EGFR (Tyr1173) (Cell Signaling Danvers, MA; #4407); anti-lamin B (Calbiochem, San Diego, CA; #NA12); anti-α-tubulin (Sigma, St. Louis, MO; #T5168); anti-c-myc (Roche, Indianapolis, IN; 9E10); anti-AKT (Cell Signaling; #9272); anti-phospho-AKT (Ser473) (Cell Signaling; #9271); anti-Erk1/2 (Cell Signaling; #9102); anti-phospho-Erk1/2 (Ser202/ Tyr204) (Cell Signaling; #9101); anti-His (Santa Cruz; #sc-8036); anti-γH2A (Millipore; #05-636); anti-actin (Sigma; #A2066) . All secondary antibodies were obtained from Vector Laboratories (Burlingame, CA) and Jackson Immuno Research (West Grove, PA).
Confocal analyses
Cultured cells were washed three times with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.2% Triton X-100 for 5 minutes, and incubated with 2% bovine serum albumin for 1 hour. Cells were then incubated with the anti-EGFR antibodies (Lab Vision, Fremont, CA; Ab13) overnight in 4°C, washed, and further incubated with the appropriate secondary antibody, tagged with fluorescein isothiocyanate (FITC). Slides were mounted by Prolong Gold antifade reagent with DAPI (Invitrogen). Confocal fluorescence images were captured with Olympus FV300 confocal laser scanning biological microscope. The images were captured in the middle sections of nuclei.
EGFR immuno-complex kinase assay
HEK293 cells were transfected with indicated constructs at 70% confluence. Thirty-six hours later, cells were harvested and lysed in NETN buffer. EGFR proteins were immunoprecipitated from 500 μg of whole cell lysate using anti-EGFR antibodies (Lab Vision; Ab13). The EGFR immunoprecipitates were washed three times of NETN buffer and one time with kinase buffer (15 mM HEPES [pH 7.5], 15 mM MgCl2, 6 mM MnCl2, 150 uM Na3VO4, 60 μM ATP). Immunoprecipitates were then resuspended in 30 μl kinase buffer containing 5 μCi [γ-32P] ATP and 1 μg of recombinant of His-tag γ-enolase (Calbiochem, #524811) and incubated at 30°C for 30 min with occasional flip. The reaction was stopped by adding Laemmli sample buffer and samples were boiled, loaded onto an SDS-PAGE, electro-transferred onto nitrocellulose membrane and subjected to autoradiography. Following autoradiography, the membranes were subjected to Western Blotting.
MTT cell proliferation and survival assay
MTT assays were performed as described [16]. For cell proliferation assay, 2000 cells were seeded in 96-well plates in quadruplicate. After incubation at 37°C for the indicated time, the colorimetric reaction was initiated by adding 25 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution (5 mg/ml in PBS) and the plates were incubated at 37°C for 2 hour. The insoluble colored formazan produced by viable cells was solubilized by adding 100 μl lysis buffer (20% SDS, 50% DMF, 2.5% acetic acid, 2.5% HCl, pH 4.7) to each well and incubated overnight at 37°C. The solution was assayed colorimetrically by measuring the absorbance at 570 nm on a microplate reader (Labsystems, Waltham, MA). For cell survival assay, 5000 cells were seeded in 96-well plate for overnight then 25 μM cisplatin (final concentration) was added for an additional 2 days. Addition of MTT solution and cell lysis buffer was performed as previously described.
DNA repair/End-Joining Assay
pCMV-luciferase [17] was digested with HindIII for the use in a DNA end-joining assay. MCF7 EGFR wildtype or mutant cells were cotransfected with TK-Renilla luciferase (Promega, Madison, WI) and either uncut or HindIII-digested pCMV-luciferase plasmids. Twenty-four hours after transfection, cells were lysed and luciferase activities were determined by Dual-Luciferase Assay System (Promega).
Results
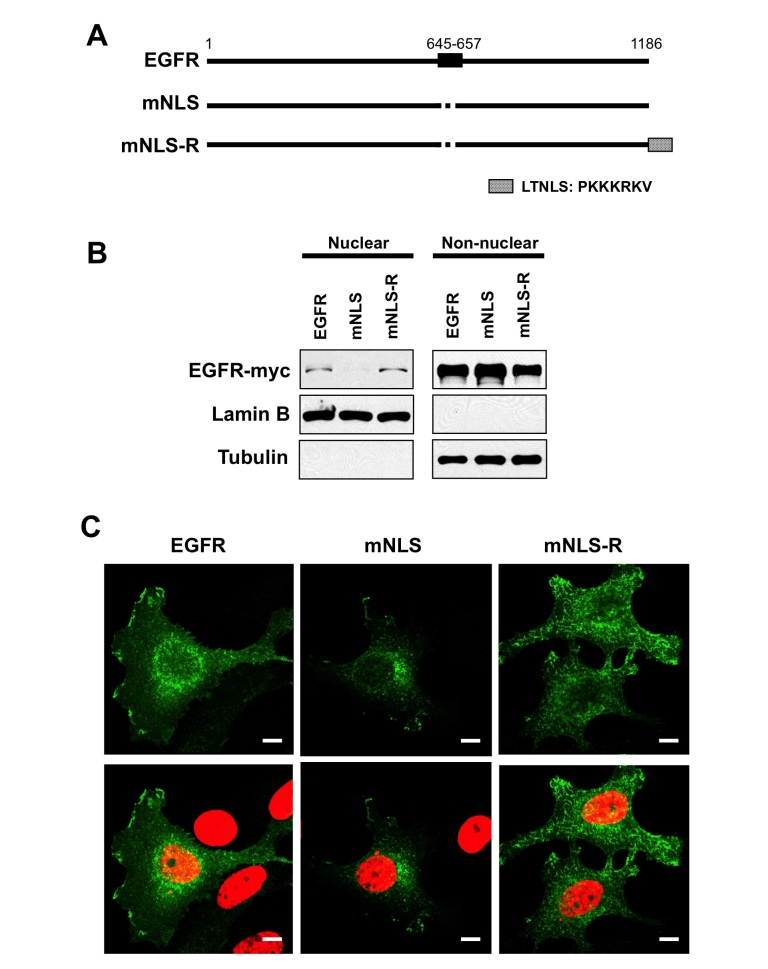
In order to study the function of nuclear EGFR we used an EGFR mutant construct (mNLS), deficient in nuclear localization [14] and then restored its ability to localize to the nucleus. For the NLS reconstituted mutant (mNLS-R), we placed the NLS from the Large T antigen of the SV40 virus at the C-terminus of the mNLS construct to restore the nuclear localization of EGFR (Figure 1A). To test whether these mutants function as intended, we compared the nuclear localization ability of these two constructs with wildtype EGFR. Through transient transfection in HeLa cells and cellular fractionation, consistent to the previous report, the NLS deficient mutant was unable to be detected in the nucleus, and interestingly, re-expression of an NLS to this mutant was able to restore expression to the nucleus (Figure 1B). Consistent with these results, confocal imaging of MCF7 cells, which has a low expression level of EGFR, transiently transfected with these constructs, showed that mNLS was not detected in the nucleus, while the wildtype, and mNLS-R mutant were both able to translocate to the nucleus.
Figure 1.
Establishment of EGFR nuclear targeting deficiency and reconstituted mutant. A, schematic representation of EGFR mutants. The EGFR NLS (NLS; amino acids 645–657) is shown at the top. The NLS mutant (mNLS) is the alanine mutant of the first and second cluster basic amino acids of EGFR tpNLS [14]. The EGFR NLS-reconstituted mutant (mNLS-R) was constructed by placing the LTNLS (NLS from large T antigen of SV40 virus) at the C-terminus. B. HeLa cells were transiently transfected with DNA encoding the indicated plasmids and subjected to biochemical fractionation to separate nuclei from non-nuclear material. The nuclear (30 μg) and non-nuclear (15 μg) extracts were subjected to SDS-PAGE and western blotting. The exogenously expressed EGFR was probed with myc-tag antibody and α-tubulin (non-nuclear marker) and lamin B (nuclear marker) severed as fraction marker and loading control. C, confocal analysis of cellular localization of EGFR mutants. MCF7 cells, low EGFR expressing cells, were transiently transfected EGFR plasmids. Cells were immunostained with an anti-EGFR antibody and FITC-conjugated second antibody then counterstained with DAPI. The cells were visualized by confocal microscopy. Top row shows EGFR signal, bottom row shows merged images of EGFR (green) and DNA (red). Note, bar 10 μm.
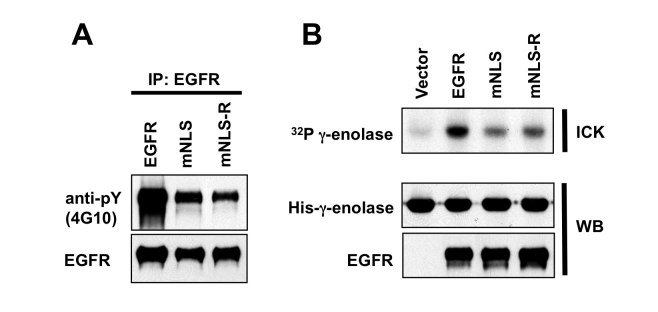
To examine the effects of mutation of the EGFR NLS, and since EGFR is a tyrosine kinase, we next examined whether the kinase activity of the mutants was still intact. We found that the tyrosine phosphorylation of EGFR (Figure 2A) and the tyrosine kinase activity of EGFR, shown by an immunocomplex kinase assay (Figure 2B), was reduced in both mNLS and mNLS-R transfected cells as compared to the wildtype EGFR, suggesting that the NLS of EGFR may be required for maximum tyrosine kinase activity and re-expression of an NLS to the NLS deficient mutant was not enough to restore its tyrosine kinase activity.
Figure 2.
The kinase activities of EGFR NLS mutants are decreased. A. CHO cells were transfected with indicated EGFR-expressing plasmids. EGFR were immunoprecipated and subjected to SDS-PAGE and western blotting with anti-tyrosine antibody (4G10) or anti-EGFR antibody. B. HEK293 cells were transfected with DNA encoding indicated EGFR-expressing plasmids. After 40 hours of transfection, EGFR was immunoprecipated with anti-EGFR antibody and subjected to immuno-complex kinase assay (ICK) and γ-enolase served as substrate. After reaction, proteins were subjected SDS-PAGE and transfer to nitrocellulose paper. After autoradiography, the nitrocellulose paper was subjected to with specific antibodies for γ-enolase (anti-His antibody) or EGFR.
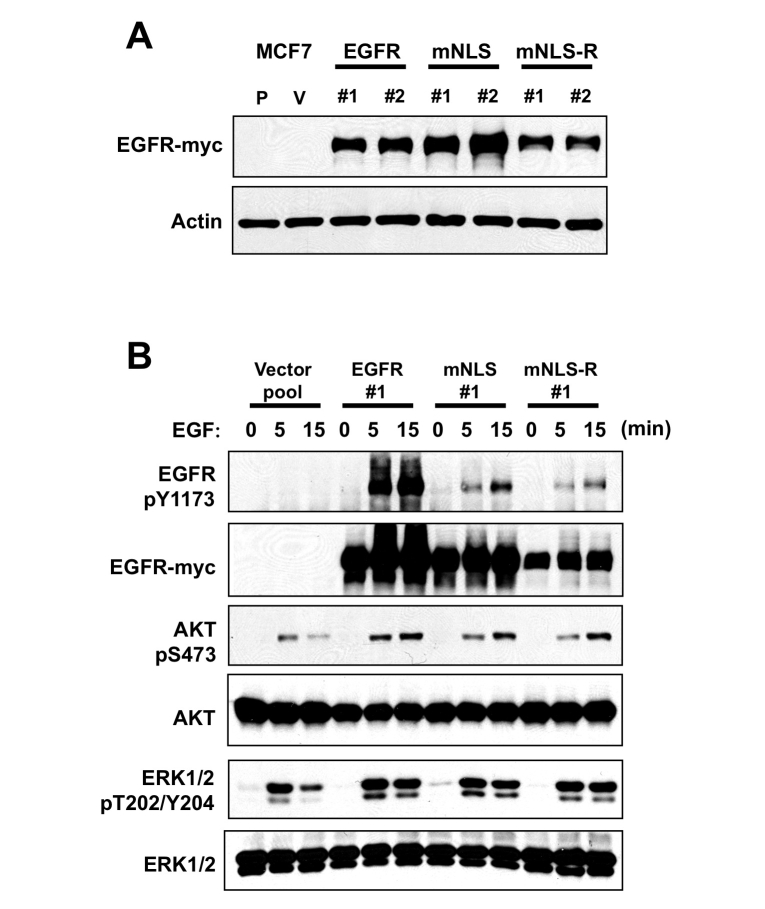
In order to further study the functional difference of nuclear EGFR, we established stable transfectants of wildtype EGFR and the two mutants, mNLS and mNLS-R, in MCF7 cells (Figure 3A). Although EGFR mNLS has a lower kinase activity than wildtype EGFR it has a comparable activity with mNLS-R. Akt and Erk1/2 are common downstream signaling targets of EGFR; therefore, we examined the effects of the NLS deficient mutant and NLS re-expression on their activation (phosphorylation) with EGF. Although EGFR kinase activity and EGF stimulated phosphorylation of Y1173 of EGFR was reduced (Figure 3B), both the NLS deficient mutant and re-expression mutant were still able to induce the phosphorylation of Akt and Erk1/2, indicating that there is still a response to EGF stimulation with the EGFR NLS mutants (Figure 3B).
Figure 3.
EGFR mutants respond to EGF stimulation. A. MCF7-EGFR NLS wild-type, mutant and reconstituted stable expressing cells were established. Western blot with myc-tag antibody indicated EGFR expression. Note, P and V mean parental and vector pool control cells. Two independent clones showed similar EGFR expression though the expression level of mNLS-R was slightly less. B. MCF7-EGFR expressing cells were serum starvation and incubated with 100 ng/ml EGF indicated time. Proteins were harvested and subject to SDS-PAGE and Western bolt with indicated antibodies. EGFR Y1173 is one of EGFR autophosphorylation site; AKT and MAPK are downstream kinases of EGFR. The phosphorylation of EGFR, AKT and MAPK indicated their kinase activities.
To elucidate the physiological relevance of the EGFR NLS deficient mutant we first examined the cell growth of the NLS mutants, mNLS and mNLS-R stable transfectants compared with the wildtype EGFR. While, the EGFR overexpressing cells were able to enhance cell growth as compared to the vector control, both the NLS deficient and the NLS re-expression mutants were not able to induce cell proliferation (Figure 4A). It has been shown that there is a correlation between cisplatin treatment and EGFR nuclear localization [7]; for that reason we next examined the sensitivity of our stable tranfectants to cisplatin. While, EGFR overexpressing cells were found to be resistant to cisplatin compared to the control, the NLS deficient mutant was able to significantly reduce the EGFR resistance to cisplatin. Furthermore, reintroduction of the NLS restored the cells ability to resist cisplatin treatment. Thus, these results indicate that EGFR overexpression renders cells resistant to cisplatin treatment and that nuclear EGFR is required for EGFR-mediated resistance to cisplatin (Figure 4B).
Figure 4.
Nuclear EGFR expression contributes to cisplatin resistance. A. cell proliferation of MCF7-EGFR expressing cells. MCF7-EGFR expressing cells were seeded in the 96 well plates and cell proliferation was measured with MTT assay. Two independent cell lines from different constructs show similar growth rate although only one is shown in the graph in the plot. B. cell survival of MCF7-EGFR expressing cells in response to cisplatin (CDDP) treatment. MCF-EGFR expressing cells were seeded in 96 well plates over night and treated with 25 uM cisplatin for an additional 48 hours. Cell survival was measured by MTT assay.
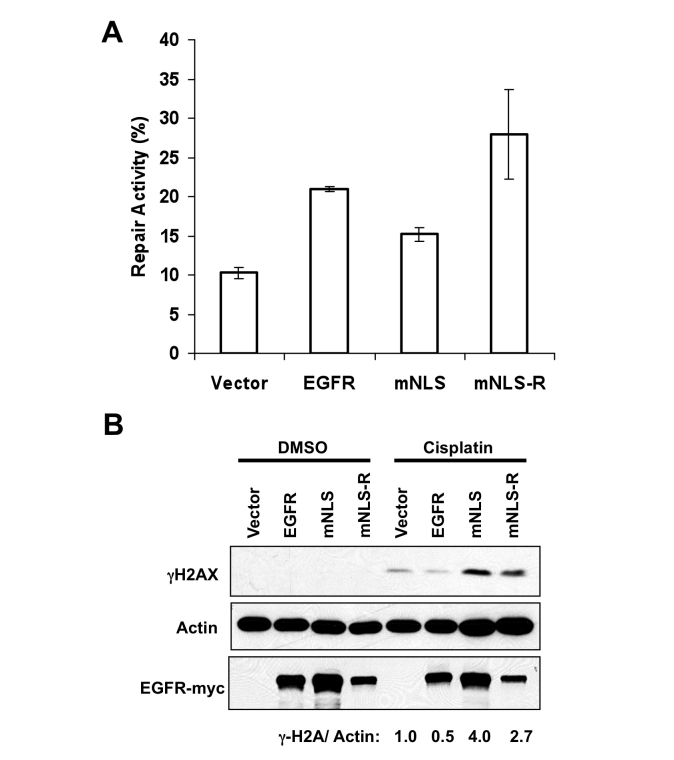
It is also known that nuclear EGFR is involved in DNA repair [5–7] and that cisplatin is able to induce DNA damage [18]; therefore we next examined DNA repair in the stable transfectants. To do this, we used a similar assay as previously described by Das, et. al. to test for repair of double strand breaks [6]. We digested a circular plasmid expressing luciferase with the HindIII restriction enzyme and then transfected the linear DNA into the MCF7 stable transfectants. If DNA repair were intact, we would expect that the transfected DNA would be repaired to its circular plasmid form enabling expression of luciferase. We found that luciferase expression in the mNLS cells was reduced compared to the EGFR wildtype expressing cells. In addition, re-introduction of the NLS in the mNLS-R cells demonstrated increased luciferase activity (Figure 5A), indicating that nuclear localization of EGFR can enhance DNA repair. We saw a similar tendency with transient transfection (data not shown). Since cisplatin induces DNA damage, and our EGFR wildtype expressing cells, but not the mNLS expressing cells, were resistant to cisplatin we next examined the effect of cisplatin on DNA damage in our stable transfectants. Using γH2AX as a DNA damage marker [19], we found that the EGFR NLS deficient mutant expressing cells showed an increase in DNA damage with cisplatin treatment compared to EGFR wildtype expressing cells, and that this increase in DNA damage was reduced in the NLS re-expression mutant (Figure 5B). Thus, these results indicate that nuclear EGFR is required for the increased DNA repair and reduced DNA damage of EGFR wildtype expressing cells.
Figure 5.
Nuclear EGFR expression contributes to double strand break repair. A. circular or HindIII-digested linear CMV-firefly luciferase plasmids were transfected into MCF7-EGFR expressing cells. TK-Renilla luciferase was co-transfected as tranfection efficiency control. Luciferase expression was measured after 24 hours of transfection. Repair activity was calculated by luciferase expression from linear plasmid divided with circular plasmid. B. MCF7-expressing cells were treated with 100 uM cisplatin 12 hours. Proteins wear harvested and subjected to Western blot with indicated antibodies. γH2A stands for S139 phosphorylated H2A.X and it is an indicator of DNA damage. γH2A expression after cisplatin treatment was calculated by densitometric analysis.
Discussion
EGFR overexpression occurs in many cancer types [1] and it has been shown that nuclear EGFR can be a poor prognostic factor [4, 8–11], indicating the importance of understanding nuclear EGFR's role in cancer. Cisplatin, which acts by targeting DNA, is frequently used to treat cancers; Unfortunately, resistance to cisplatin treatment is a common occurrence [20]. Here we provide a basis by which nuclear expression of EGFR in cancers contribute to resistance to cisplatin treatment, or other DNA damaging drugs.
We have previously shown that the nuclear localization sequence (NLS) located in the juxtamembrane region of EGFR has a strong nuclear targeting ability [14]. While introduction of the Large T Antigen NLS at the C-terminal (mNLS-R) restored the ability to translocate to the nucleus and enhance DNA repair, it was not able to restore the kinase activity or enhance cell proliferation. Thus, NLS of EGFR may be associated with two distinct functions, tyrosine kinase activity and nuclear localization.
While we show that nuclear EGFR is required for DNA repair it is still not clear exactly how EGFR is involved. It is known that EGFR is able to interact with DNA-dependent Protein Kinase (DNA-PK), both the catalytic subunit and the Ku70/80 regulatory subunits [12]. Furthermore, Dittmann et. al. showed that after ionizing radiation, heat, H2O2 or cisplatin treatment, EGFR, Ku70/80 and phosphatase I all translocate to the nucleus, and at the same time there is an increase in DNA-PK activity [7]. More recently, the same group also showed that the radioprotectors, O-phospho-l-tyrosine (P-Tyr) and Bowman-Birk proteinase inhibitor (BBI) both stimulate nuclear EGFR localization and DNA-PK activity [21, 22]. They and others found that P-Tyr stimulation of EGFR nuclear localization was associated with phosphorylation of EGFR on T654 by PKCɛ [22, 23]. Futhermore, BBI was also able to stimulate the complex formation between EGFR p53 and MDC1 (Mediator of DNA Damage Checkpoint Protein 1). Thus, regulation of DNA repair by nuclear EGFR may be complicated and the question remains as to what other proteins may be involved in nuclear EGFR dependent DNA repair, and whether the tyrosine phosphorylation activity of EGFR plays a role.
At this point, the mechanism of cisplatin induced EGFR nuclear localization is also unclear. It has been shown previously that cisplatin can induce the activation of EGFR, which requires EGFR kinase activity [24]. Both cisplatin and UV radiation have been shown to induce p38MAPK-mediated internalization of EGFR and with cisplatin treatment internalization of EGFR has been shown to be dependent on p38MAPK phosphorylation of T699 on EGFR [25–27]. P38MAPK mediated endocytosis has been shown to involve the GDI (guanyl-nucleotide dissociation inhibitor)/Rab5 complex and phosphorylation of the Rab5 effectors EE1A and Rabenosyn-5, regulating their recruitment to the cell membrane [27–29]. The mechanism of UV radiation mediated internalization of EGFR has been shown to go through Rab5 containing endosomes and to involve p38MAPK phosphoryation of both EE1A and GDI [27]. Thus, it may be interesting to determine if cisplatin induces EGFR internalization in a similar manner and if p38MAPK –mediated internalization of EGFR is required for EGFR induced nuclear localization. There may be other factors that are involved in cisplatin induced EGFR nuclear localization.
It will be critical to determine if nuclear EGFR is also involved in resistance to other therapeutic agents. As mentioned previously, ionizing radiation was shown to induce nuclear localization of EGFR as well as association of EGFR with DNA-PK [7]. It is possible that other chemotherapeutic agents, such as doxorubicin and etoposide may work in the same way.
Lastly, DNA-PK has been suggested to be a therapeutic target and is poor prognostic indicator for B-cell chronic lymphocytic leukemia [30]. Our results here indicate that targeting DNA-PK may also be advantageous in combination with cisplatin or other similar drugs as it may sensitize tumors to DNA damaging agents.
Acknowledgments
This study was supported by an R01 (CA1109311), a NSC stem cell grant (96-3111-B-039), and a MDACC Cancer Center Support Grant (CA16672) to MCH.
References
- 1.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 2.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 3.Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94:184–188. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, Hung MC. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 6.Das AK, Chen BP, Story MD, Sato M, Minna JD, Chen DJ, Nirodi CS. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–5274. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 7.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 8.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 9.Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, Rimm D, Burtness BA. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 10.Hoshino M, Fukui H, Ono Y, Sekikawa A, Ichikawa K, Tomita S, Imai Y, Imura J, Hiraishi H, Fujimori T. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 11.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, Huo LF, Miller S, Hung MC. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2008 doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J Biol Chem. 1998;273:1568–1573. doi: 10.1074/jbc.273.3.1568. [DOI] [PubMed] [Google Scholar]
- 13.Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J Cell Biol. 2007;179:183–186. doi: 10.1083/jcb.200705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 15.Yan DH, Spohn B, Hung MC. Delivery of DNA to tumor cells using cationic liposomes. Methods Mol Biol. 2004;245:125–136. doi: 10.1385/1-59259-649-5:125. [DOI] [PubMed] [Google Scholar]
- 16.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 17.Chen JS, Liu JC, Shen L, Rau KM, Kuo HP, Li YM, Shi D, Lee YC, Chang KJ, Hung MC. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther. 2004;11:740–747. doi: 10.1038/sj.cgt.7700752. [DOI] [PubMed] [Google Scholar]
- 18.Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Perez JM. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med Chem. 2007;7:3–18. doi: 10.2174/187152007779314044. [DOI] [PubMed] [Google Scholar]
- 19.Clingen PH, Wu JY, Miller J, Mistry N, Chin F, Wynne P, Prise KM, Hartley JA. Histone H2AX phosphorylation as a molecular pharmacological marker for DNA interstrand crosslink cancer chemotherapy. Biochem Pharmacol. 2008;76:19–27. doi: 10.1016/j.bcp.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 20.Siddik ZH. Cisplatmode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 21.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. The radioprotector Bowman-Birk proteinase inhibitor stimulates DNA repair via epidermal growth factor receptor phosphorylation and nuclear transport. Radiother Oncol. 2008;86:375–382. doi: 10.1016/j.radonc.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Dittmann K, Mayer C, Wanner G, Kehlbach R, Rodemann HP. The radioprotector O-phospho-tyrosine stimulates DNA-repair via epidermal growth factor receptor- and DNA-dependent kinase phosphorylation. Radiother Oncol. 2007;84:328–334. doi: 10.1016/j.radonc.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Wanner G, Mayer C, Kehlbach R, Rodemann HP, Dittmann K. Activation of protein kinase Cepsilon stimulates DNA-repair via epidermal growth factor receptor nuclear accumulation. Radiother Oncol. 2008;86:383–390. doi: 10.1016/j.radonc.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 24.Benhar M, Engelberg D, Levitzki A. Cisplatin-induced activation of the EGF receptor. Oncogene. 2002;21:8723–8731. doi: 10.1038/sj.onc.1205980. [DOI] [PubMed] [Google Scholar]
- 25.Vergarajauregui S, San Miguel A, Puertollano R. Activation of p38 mitogen-activated protein kinase promotes epidermal growth factor receptor internalization. Traffic. 2006;7:686–698. doi: 10.1111/j.1600-0854.2006.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winograd-Katz SE, Levitzki A. Cisplatin induces PKB/Akt activation and p38(MAPK) phosphorylation of the EGF receptor. Oncogene. 2006;25:7381–7390. doi: 10.1038/sj.onc.1209737. [DOI] [PubMed] [Google Scholar]
- 27.Zwang Y, Yarden Y. p38 MAP kinase mediates stress-induced internalization of EGFR: implications for cancer chemotherapy. EMBO J. 2006;25:4195–4206. doi: 10.1038/sj.emboj.7601297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalli V, Vilbois F, Corti M, Marcote MJ, Tamura K, Karin M, Arkinstall S, Gruenberg J. The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex. Mol Cell. 2001;7:421–432. doi: 10.1016/s1097-2765(01)00189-7. [DOI] [PubMed] [Google Scholar]
- 29.Mace G, Miaczynska M, Zerial M, Nebreda AR. Phosphorylation of EEA1 by p38 MAP kinase regulates mu opioid receptor endocytosis. EMBO J. 2005;24:3235–3246. doi: 10.1038/sj.emboj.7600799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willmore E, Elliott SL, Mainou-Fowler T, Summerfield GP, Jackson GH, O'Neill F, Lowe C, Carter A, Harris R, Pettitt AR, Cano-Soumillac C, Griffin RJ, Cowell IG, Austin CA, Durkacz BW. DNA-dependent protein kinase is a therapeutic target and an indicator of poor prognosis in B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14:3984–3992. doi: 10.1158/1078-0432.CCR-07-5158. [DOI] [PubMed] [Google Scholar]