Abstract
Background:
Essential hypertension is a complex genetic disorder influenced by diverse environmental factors. Of the various physiological pathways affecting the homeostasis of blood pressure, the renin-angiotensin system (RAS) is known to play a critical role. Angiotensin-I converting enzyme (ACE) is a significant component of RAS and an insertion/deletion (I/D) polymorphism in its gene has been implicated in predisposition to hypertension.
Objective:
The present study is aimed to determine the association, if any, of ACE I/D polymorphism with essential hypertension in a rural population of Haryana, India.
Materials and Methods:
The blood samples were collected from the patients visiting M. M. Institute of Medical Sciences, Mullana, Haryana. DNA from the patients (106) and control (110) specimens were isolated, amplified by PCR and analyzed employing agarose gel electrophoresis.
Results:
There was no significant difference in the distribution of DD, II and I/D genotypes of ACE polymorphism in essential hypertensive patients (28.8, 25.5, and 46.2%) and their ethnically matched normal control (24.5, 30, and 45.5), respectively. The two groups also presented with very similar allelic frequencies and were also found to be in Hardy-Weinberg equilibrium.
Conclusions:
The present study demonstrates that ACE I/D polymorphism is not a risk factor for essential hypertension in the hitherto unstudied rural population of Haryana.
Keywords: Angiotensin-I converting enzyme, insertion/deletion polymorphism, essential hypertension, North Indian population
INTRODUCTION
Cardiovascular diseases are becoming a major health burden in developing countries. About 2.6 million Indian people are estimated to die due to coronary heart disease (CHD) alone by the year 2020.[1] Hypertension is one of the important risk factor for the development of CHD. It is a multifactorial and polygenic disorder in which the interaction between several candidate genes and environmental factors play a role. The renin angiotensin system (RAS) is an important regulatory mechanism for maintaining normal blood pressure, fluid and electrolyte balance and its encoding components have been proposed as independent factors for hypertension and other cardiovascular diseases.[2,3]
Angiotensin-I-converting enzyme (ACE) is a zinc metallopeptidase widely distributed on the surface of endothelial and epithelial cells and participates in producing arteriolar constriction and a rise in systolic and diastolic blood pressure. The ACE is encoded by a 21 kb gene that consists of 26 exons and is located on chromosome 17 and contains a polymorphism in the form of either insertion (I) or deletion (D) of a 287 base pair Alu repetitive sequence in intron 16.[4] This polymorphism is shown to be associated with the interpersonal variability and individuals carrying the deletion allele are associated with increased plasma ACE levels. Earlier studies have shown association between this polymorphism and several cardiovascular diseases like myocardial infarction,[5] left ventricular hypertrophy,[6] cardiomyopathy,[2] and hypertension.[7,8] Studies have been carried out on the association between the ACE I/D polymorphism and hypertension in various populations and both positive and negative association have been reported.[9–12] The present study is the first report investigating the role of this important polymorphism in a rural population of Haryana, North India.
MATERIALS AND METHODS
Study population
In the present investigation, the blood samples of 106 essential hypertensive patients and 110 samples of age and sex matched normal, healthy individuals as control group were collected with informed consent from M. M. Institute of Medical, Sciences, Mullana, Haryana. It is prudent to mention that it is the only mutlifacility hospital in Mullana and caters to a rural region within a radius of around 30 km. Patients were initially not on any medication and subsequently, consented for regular check up and treatment. Their follow up is up to date. The blood samples were collected in tubes containing EDTA as an anticoagulant. The samples were transported on ice to the laboratory and were processed on the same day. The isolated DNA samples were stored at −20°C till further analysis.
Various parameters like age, sex, BMI, blood pressure, and dietary patterns were recorded in a questionnaire. Blood pressure (supine) was measured after the subject had rested at least 15 minutes with the help of mercury sphygmomanometer and stethoscope by ausculatory method.[13] The recordings were done at least three times on different days. The hypertension status of the study sample was assessed using standard criteria formulated by Joint National Committee VII.[14] Unrelated subjects living in the same rural background and without any history of hypertension, diabetes and other immunosuppressive conditions were enrolled as control subjects.
Genotyping angiotensin-I converting enzyme insertion/deletion polymorphism
DNA samples were isolated from peripheral blood lymphocytes by the standard modified inorganic method as described by Miller et al.[15] and quantified following standard spectrophotometric analysis. The ACE I/D polymorphism were detected by the polymerase chain reaction using the primers flanking a 287 bp insertion sequence.[4] The optimized reaction conditions consisted of 40 ng of genomic DNA in a reaction volume of 30 μl contains 0.16 μM of each primer, 30 μM of each dNTP, 10 mM Tris-HCL (pH-9.0), 1.5 mM MgCl2, 50 mM KCl, 0.01% gelatin, and 0.3 U of Taq DNA polymerase (Bangalore Genei, Bangalore). Amplification was carried out for 35 cycles, each cycle consisting of denaturation at 94°C for 30 s, annealing at 58°C for 20 s, extension at 72°C for 20 s and finally a 3 m extension at 72°C. The PCR products were resolved in 2% agarose gel and visualized following ethidium bromide staining. All samples, identified as DD after initial amplification, were reconfirmed with an insertion-specific primer pair:
Forward primer: 5’-GCCACTACGCCCGGCTAAT-3’;
Reverse primer: 5’-GATGTGGCCATCACATTCGTCAGAT-3’).
The reaction conditions and amplification parameters for this confirmatory reaction were the same as stated above. Known controls of each genotype were amplified with each set of samples for the ACE I/D polymorphism.
Statistical analysis
Data analysis was done with the help of an SPSS version 7.5. Continous variables are expressed as means ± SD. Intergroup comparisons are made using students † test. Allele frequencies were calculated from genotype frequencies and were compared using chi-squared (χ2) statistics. P value < 0.05 was considered statistically significant.
RESULTS
The clinical details of the hypertensive and control subjects are presented in Table 1. The mean systolic blood pressure (SBP) and diastolic blood pressure (DBP) were significantly higher in the hypertensive subjects than in the control subjects. Interestingly, the family history of hypertension and BMI among the patients was also statistically significant as compared to the normal controls. The incidence of individuals with history of smoking and alcohol consumption was higher among the hypertensive patients as compared to the controls, however the differences were statistically nonsignificant (P >0.05).
Table 1.
Demographic characteristics of subjects
| Patients (%) | Control (%) | P value | |
|---|---|---|---|
| Number | 106 | 110 | |
| Sex (Male:Female) | 63:43 | 67:43 | |
| Age | 53.90 ± 13.90 | 51.96 ± 16.78 | 0.3568 |
| Body mass index (Kg/m3) | 27.47 ± 3.422 | 23.38 ± 3.782 | 0.001 |
| Systolic blood pressure (mm Hg) | 142.95 ± 5.87 | 117.69 ± 3.620 | < 0.0001 |
| Diastolic blood pressure (mm Hg) | 92.44 ± 4.600 | 75.63 ± 4.492 | <0.0001 |
| Alcoholic (regular use) | 14 (13.2) | 8 (7.3) | 0.1798 |
| Smoking | 7 (6.6) | 4 (3.6) | 0.3677 |
| Family history | 10 (9.4) | 3 (2.7) | 0.0383 |
DATA EXPRESSED AS MEAN+SD
Table 2 shows the data pertaining to all the genotypes and the allele distribution in hypertensive patients and normal healthy controls. Both the groups were in the Hardy-Weinberg equilibrium. The frequency of I/D heterozygote as compared to homozygote was higher in both the patients and control group. It was observed that the DD genotype was slightly higher than II genotype in patients as compared to control. The I/D genotypes was 46.2% in hypertensive patients while it was 45.5% in controls. However, the differences were statistically not significant. The frequency of the D allele was only marginally higher in essential hypertensive patients as compared to the normal controls.
Table 2.
Distribution of the genotype and allele frequencies of ACE I/D polymorphism
| Population (n) | Genotype frequencies (%) | Allele frequencies | |||
|---|---|---|---|---|---|
| DD | ID | II | D allele | I allele | |
| Hypertensive (106) | 30 (28.3) | 49 (46.2) | 27 (25.5) | 0.514 | 0.486 |
| Normal controls (110) | 27 (24.5) | 50 (45.5) | 33 (30.0) | 0.473 | 0.527 |
χ2 BASED ON ALLELE FREQUENCY [(DF)=1] (HYPERTENSIVE VS CONTROLS) = 0.694; P = 0.706
Figure 1 depicts a representative agarose gel of various genotypes of the ACE I/D polymorphism in the studied samples. Known DNA samples from II and DD subjects were amplified as controls and yielded expected products of 490 and 190bp, respectively [Figure 1, lanes 1,2]. Sample showing PCR amplified product for both the alleles were labeled as ID genotype [Figure 1, lanes 3, 6, 11, 14].
Figure 1.
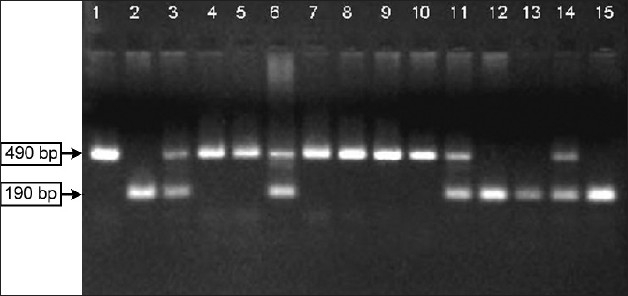
Agarose gel electrophoresis of PCR products showing the amplification for ACE I/D polymorphism. The II genotype was identified by the presence of a single 490 bp product, DD genotype was identified by the presence of a single 190 bp product and ID genotype amplified both 490 and 190bp products. The amplified products in lanes 1 and 2 are DNA sequenced PCR products of known controls of II and DD genotype, respectively
DISCUSSION
Incidence of hypertension is increasing alarmingly in various populations of India and other developing nations.[16–18] It is universally accepted that systemic hypertension is a distinct risk factor for various cardiovascular emergencies, particularly left ventricular failure, myocardial infarction, and stroke. The present study is the first report investigating the association of ACE I/D polymorphism with hypertension in a rural population of Haryana. The strength of present study lies in extensive door-to-door preliminary investigation to identify essential hypertensive patients in villages surrounding the MM University, Ambala, Haryana, over a three-year (2003-2006) period. Preliminary survey identified a total of 2,295 hypertensive subjects, of which 930 were essential hypertensive and were not on any medication. These essential hypertensive patients were persuaded to visit Medical College at Mullana, Haryana for further investigations and 106 (∼11%) of them consented to be part of this study. The consent rate might appear low but one has to keep in mind that these subjects belong to rural area, where rate of illiteracy is very high, and despite three blood pressure measurements on different days indicating elevated levels a few of them refused to accept that they have hypertension.
Pooling of epidemiology studies show that hypertension is present in 25% urban and 10% rural subjects in India.[19] Clearly suggesting that urban conditions somehow increase the prevalence of this disease. Therefore, another forte of the study lies in the fact that patients living in the rural area were studied in the same conditions thus minimizing the influence of urban environment on the disease condition. Family history and body mass index in the hypertensive patients shows statistically significant difference from the control population [Table 1] and it does suggest that genetic factors and body mass index do influence the ability to develop this disease in the studied rural population of Haryana. These observations are in line with earlier report providing evidence that heritable factors in combination with a number of recognized environmental risk factors are important determinants of the pathogenesis and natural history of essential hypertension.[20]
It is important to ascertain gene(s) that are involved in hypertension. This would help in identifying individuals at an increased risk of developing this disease and to initiate appropriate actions in them to avoid development or delay the onset of disease. Genome wide scan and candidate gene approach are two strategies used in dissecting complex genetic diseases.[21] The former, links specific chromosomal region with inheritance of the disease, is technically cumbersome and requires sophisticated infrastructure. The candidate gene approach targets selected gene with defined polymorphism(s) for their association with the disease. The polymorphism could exist as single nucleotide change, insertion/deletion of nucleotide sequence or repetitive DNA elements. A gene and its selected polymorphism preferably should have the following features to make them a candidate target:
The gene product must be functionally relevant to hypertension
Polymorphism within the gene must alter its function
Hypertension needs to link to the chromosomal region harboring the candidate gene.
Available studies demonstrate that the ACE I/D polymorphism fulfills above mentioned criterions in the context of hypertension[7,22–24] and was therefore investigated in the present study.
The frequencies of different genotypes were found to be similar in patient and the control population [Table 2]. The frequencies of both the alleles (I/D) are quite high in the control and cases, thus obviating the possibility that the frequency of the rare allele is a cause for concern in the studied sample. Lack of association between ACE I/D polymorphism and essential hypertension have been reported by investigators in Indian and other populations of the world.[25–28,40] Ethnic background is known to influence the ACE I/D polymorphism globally.[29,30] A significant association of the ACE high producing D allele with hypertension in African, Americans, Chinese, and Japanese populations have already been reported.[8,22–24] However, two studies from Australia[31] and Pakistan[32] recorded the association of I allele with hypertension. The association of I allele with hypertension in Pakistan population was attributed to limited number of individuals studied[32] and to the presence of high levels of inbreeding.[25]
The frequency of D allele of ACE I/D polymorphism in different hypertensive populations of India varied within 0.522 to 0.409 [Table 3]. The highest frequency was reported in a Sikh group from Punjab that also showed an association between the D allele and the hypertension. Similar observations have also been made on populations from other states of India.[30] The frequency of D allele in the studied patient and control populations were well within the reported range for the North Indian populations [Table 3]. Contrary to the earlier findings, no association between D allele and essential hypertension was observed in the rural population of Haryana. We believe the number of patients studied in other Indian populations showing positive associations with D allele [Table 3] were very small to allow any meaningful conclusion.
Table 3.
The genotypic distribution and allele frequencies of the ACE I/D polymorphism in essential hypertension in different population of the world
| Population studied (n) | Genotype distribution | Frequencies | Reference | ||||
|---|---|---|---|---|---|---|---|
| DD | ID | II | D allele | I allele | |||
| North Indian (106) | 30 | 49 | 27 | 0.514 | 0.496 | Present study | |
| European | |||||||
| Italian (86) | 31 | 41 | 14 | 0.60 | 0.40 | Teresa et al.[35] | |
| Kyrgyz (180) | 20 | 91 | 69 | 0.36 | 0.64 | Polupanov et al.[36] | |
| Dutch (257) | 80 | 138 | 39 | 0.58 | 0.42 | Schut et al.[37] | |
| Turkish (109) | 49 | 59 | 01 | 0.73 | 0.27 | Agachan et al.[38] | |
| Solvenian (413) | 132 | 199 | 82 | 0.57 | 0.43 | Galvnik et al.[39] | |
| German (621) | 167 | 309 | 145 | 0.517 | 0.483 | Mondry et al.[40] | |
| Asians | |||||||
| Japanese (87) | 17 | 26 | 44 | 0.35 | 0.65 | Ishigami et al.[41] | |
| Chinese (189) | 18 | 77 | 94 | 0.70 | 0.30 | Lee et al.[42] | |
| Indians | |||||||
| Punjabi, Punjab (100) | 17 | 58 | 25 | 0.46 | 0.54 | Randhawa et al.[23] | |
| Sikh, Punjab (45) | 12 | 21 | 11 | 0.522 | 0.477 | Pasha et al.[28] | |
| Jat, Haryana (30) | 03 | 21 | 06 | 0.45 | 0.55 | Pasha et al.[28] | |
| Dogra, HP (52) | 12 | 21 | 19 | 0.432 | 0.567 | Pasha et al.[28] | |
| Kumaonese (Uttranchal) (33) | 07 | 13 | 13 | 0.409 | 0.590 | Pasha et al.[28] | |
| Assamese (52) (Assam) | 08 | 28 | 16 | 0.424 | 0.576 | Pasha et al.[28] | |
Identifying association between a gene and a complex genetic disease is difficult. One possible reason for this is the involvement of a large number of genes in the etiology of essential hypertension. Furthermore, these genes may interact with each other in different combinations to give rise to a similar disease phenotype. The magnitude of this problem makes the frequency of any polymorphism contributing to a disease phenotype marginally higher in disease group compared with unaffected controls.[33] Linkage analysis has limited power to detect such small effects[34] and case control studies with matched controls from the same population had greater probability of detecting such minute effects.[21] The inability to find association between ACE I/D polymorphism and hypertension in the present study strongly point out that ACE gene is not playing a predominant role in the pathophysiology of this disease in our population and is not a good predictor of susceptibility to hypertension. Similar observations have also been made in a Meta analysis studying the role of genetic polymorphisms in hypertension.[40] Since hypertension is a complex genetic disorder, it is assumed that there could be other genetic and environmental factors that interact and influence the development of this disease.
The results of the present study has triggered two very valid questions, i) what is the effect of different ACE I/D genotypes on the progression of the disease? and ii) are different drug regimens required for individuals with different ACE I/D polymorphism? Interestingly, our preliminary observations do suggest that hypertensive patients with DD phenotypes require higher doses of ACE inhibitor in their drug regimen as compared to their II counterparts (data not shown). The information will be of immense use in tailoring individualized therapy to hypertensive patients based on the ACE I/D genotypes.
CONCLUSION
Our study suggests that the ACE I/D polymorphism is not a risk factor for the development of essential hypertension in the studied rural population from Haryana.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Nistar S. Prevention of coronary heart disease in South Asia. Lancet. 2002;360:1015–8. doi: 10.1016/S0140-6736(02)11088-9. [DOI] [PubMed] [Google Scholar]
- 2.Raynolds MV, Bristow MR, Bush EW, Abraham WT, Lowes BD, Zisman LS. Angiotensin-converting enzyme DD genotype in patients with ischemic or idiopathic dilated cardiomyopathy. Lancet. 1993;342:1073–5. doi: 10.1016/0140-6736(93)92061-w. [DOI] [PubMed] [Google Scholar]
- 3.Higaki J, Baba S, Katsuya T, Sato N, Ishikawa K, Mannami T. Deletion allele of angiotensin-converting enzyme gene increases risk of essential hypertension in Japanese men. Circulation. 2000;101:2060–5. doi: 10.1161/01.cir.101.17.2060. [DOI] [PubMed] [Google Scholar]
- 4.Rigat B, Hubert C, Corvol P, Soubrier F. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCPI) (dipeptidyl carboxypeptidase) Nucleic Acids Res. 1992;20:1433. doi: 10.1093/nar/20.6.1433-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig E, Corneli PS, Anderson JL, Marshall HW, Lalouel JM, Ward RH. Angiotensin-converting enzyme gene polymorphism is associated with myocardial infarction but not with development of coronary stenosis. Circulation. 1995;91:2120–4. doi: 10.1161/01.cir.91.8.2120. [DOI] [PubMed] [Google Scholar]
- 6.Schunkert H, Hense HW, Holmer SR, Stender M, Perz S, Keil U, Lorell BH. Association between a deletion polymorphism of the angiotensin-converting enzyme gene and left ventricular hypertrophy. N Engl J Med. 1994;330:1634–8. doi: 10.1056/NEJM199406093302302. [DOI] [PubMed] [Google Scholar]
- 7.Duru K, Farrow S, Wang JM, Lockbetteb W, Kurtz T. Frequency of a deletion polymorphism in the gene for angiotensin converting enzyme is increased in African-americans with hypertension. Am J Hypertens. 1994;7:759–62. doi: 10.1093/ajh/7.8.759. [DOI] [PubMed] [Google Scholar]
- 8.Barley J, Blackwood A, Miller M, Markandu ND, Carter ND, Jeffery S. Angiotensin Converting Enzyme gene I/D polymorphism blood pressure and the rennin-angiotensin system in Caucasian and afro-Caribbean peoples. J Hum Hypertens. 1996;10:31–5. [PubMed] [Google Scholar]
- 9.Jeng JR, Harn HJ, Jeng CY, Yueh KC, Shieh SM. Angiotensin I converting enzyme gene polymorphism in Chinese patients with hypertension. Am J Hypertens. 1997;10:558–61. doi: 10.1016/s0895-7061(97)00036-8. [DOI] [PubMed] [Google Scholar]
- 10.Higashimori K, Zhao Y, Higaki J, Kamitani A, Katsuya T. Association analysis of a polymorphism of the angiotensin converting enzyme gene with essential hypertension in the Japanese population. Biochem Biophys Res Commun. 1993;191:399–404. doi: 10.1006/bbrc.1993.1231. [DOI] [PubMed] [Google Scholar]
- 11.Vassilikioti S, Doumas M, Douma S, Petidis K, Karagiannis A. Angiotensin converting enzyme gene polymorphism is not related to essential hypertension in Greek population. Am J Hypertens. 1996;9:700–02. doi: 10.1016/0895-7061(95)00449-1. [DOI] [PubMed] [Google Scholar]
- 12.Chiang FT, Lai ZP, Chern TH, Tseng CD, Hsu KL. Lack of association of the angiotensin converting enzyme gene polymorphism with essential hypertension in a Chinese population. Am J Hypertens. 1997;10:197–201. doi: 10.1016/s0895-7061(96)00345-7. [DOI] [PubMed] [Google Scholar]
- 13.Rose GA, Blackburn H. Cardiovascular Survey Methods. WHO Monograph Series. 1968;56:90–5. [PubMed] [Google Scholar]
- 14.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones DW, Materson BJ, Oparil S, Wright JT, Roccella EJ. Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 15.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das SK, Sanyal K, Basu A. Study of urban community survey in India: Growing trend of high prevalence of hypertension in a developing country. Int J Med Sci. 2005;2:70–8. doi: 10.7150/ijms.2.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane D, Beevers DG, Lip GYH. Ethnic differences in blood pressure and the prevalence of hypertension in England. J Hum Hypertens. 2002;16:267–73. doi: 10.1038/sj.jhh.1001371. [DOI] [PubMed] [Google Scholar]
- 18.Wang TJ, Vasan RS. Epidemiology of uncontrolled hypertension in United States. Circulation. 2005;112:1651–2. doi: 10.1161/CIRCULATIONAHA.104.490599. [DOI] [PubMed] [Google Scholar]
- 19.Gupta R. Trends in hypertension epidemiology in India. J Hum Hypertens. 2004;18:73. doi: 10.1038/sj.jhh.1001633. [DOI] [PubMed] [Google Scholar]
- 20.Bhavani BA, Padma T, Shastry BKS, Reddy NKS. Gender specific association on insertion/deletion polymorphism of the human angiotensin converting enzyme gene with essential hypertension. Int J Hum Genet. 2004;4:207–13. [Google Scholar]
- 21.Lander ES, Schork NJ. Genetic dissection of complex traits science. 1994;265:2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 22.Chiang FT, Chern TH, Lai ZP, Tseng CD, Hsu KL. Age and gender dependent association of the angiotensin-converting enzyme gene with essential hypertension in Chinese population. J Hum Hypertens. 1996;10:823–6. [PubMed] [Google Scholar]
- 23.Morrise T, Takeguchi Y, Takeda R. Angiotensin converting enzyme polymorphism and essential hypertension. Lancet. 1994;343:125. doi: 10.1016/s0140-6736(94)90859-1. [DOI] [PubMed] [Google Scholar]
- 24.Nakano Y, Oshima T, Hiranga H, Matsuura H, Kajiyama G, Kambe M. DD genotype of the angiotensin – converting enzyme gene is a risk factor for early onset of essential hypertension in Japanese patients. J Lab Clin Med. 1998;131:502–06. doi: 10.1016/s0022-2143(98)90058-0. [DOI] [PubMed] [Google Scholar]
- 25.Randhawa NK, Kumar A, Matharaoo K Bhanwer AJS. Association studies of angiotensin converting enzyme gene insertion/deletion polymorphism with hypertension in Punjabi population. Int J Hum J Genet. 2006;6:317–21. [Google Scholar]
- 26.Harrap SB, Davison HR, Connor JM, Soubrier F, Fraser R, Foy CJ, Walt GC. The angiotensin converting enzyme gene and predisposition of high blood pressure. Hypertension. 1993;21:455–60. doi: 10.1161/01.hyp.21.4.455. [DOI] [PubMed] [Google Scholar]
- 27.Jeunemaitre X, Lipton PR, Hunt SC, Williams RR, Lalouel JM. Absence of linkage between the angiotensin-converting enzyme gene and human essential hypertension. Nature Genet. 1992;1:72–5. doi: 10.1038/ng0492-72. [DOI] [PubMed] [Google Scholar]
- 28.Kamdar S, Daniel H, Fogarty P, Lawson M, Munroe P, Caulfield M. ACE insertion/deletion (I/D) polymorphism in Vincentian African Caribbean with essential hypertension. J. Hum. Hypertens. 1994;8:611. [PubMed] [Google Scholar]
- 29.Sekerli E, Katsanidis D, Papadopoulou V, Makedou A, Vavatsi N, Gatzola M. Angiotensin-I converting enzyme gene and I/D polymorphism distribution in the Greek population and a comparison with other European populations. J Genet. 2008;1:91–3. doi: 10.1007/s12041-008-0013-7. [DOI] [PubMed] [Google Scholar]
- 30.Pasha MA, Khan A, Kumar R, Ram R, Grover S, Shrivatsva K, Selvamurthy W. Variations in angiotensin converting enzyme gene insertion/deletion polymorphism in Indian populations of different ethni corigins. J Bio Sci. 2002;27:67–70. doi: 10.1007/BF02703684. [DOI] [PubMed] [Google Scholar]
- 31.Zee RY, Lou YK, Griffiths LR, Morrise BJ. Association of a polymorphism of angiotensin–I converting enzyme gene with essential hypertension. Biochem Biophys Res Commun. 1992;184:9–15. doi: 10.1016/0006-291x(92)91150-o. [DOI] [PubMed] [Google Scholar]
- 32.Ismail M, Akhter N, Nasir M, Firesat S, Yub Q, Khaliq S. Association between the angiotensin converting enzyme gene insertion/deletion polymorphism and essential hypertension in young Pakistan patients. J Biochem Mol Bio. 2004;35:252–5. doi: 10.5483/bmbrep.2004.37.5.552. [DOI] [PubMed] [Google Scholar]
- 33.Gray IC, Campbell DA, Spurr NK. Single nucleotide polymorphisms as tools in human genetics. Hum Mol Genet. 2000;9:2403–8. doi: 10.1093/hmg/9.16.2403. [DOI] [PubMed] [Google Scholar]
- 34.Risch M, Merikangas A. The future of genetic studies of complex human diseases Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 35.Teresa M, Steela P, Barlassina C, Maumta P, Lanzani C. ACE and α adducin polymorphism as marker of individual response to diuretic therapy. Hypertension. 2003;41:398–403. doi: 10.1161/01.HYP.0000057010.27011.2C. [DOI] [PubMed] [Google Scholar]
- 36.Polupanov A, Halmatov A, Pak O, Romanova T, Kim E, Cheskidova N, Aldashev A. The I/D polymorphism of the angiotensin converting enzyme gene as a risk factor for ischemic stroke in patients with essential hypertension in Kyrgyz population. Areh Turk Soc Cardial. 2007;35:347–53. [Google Scholar]
- 37.Schut A, Gyseles B, Stricker B, Hofman A, Witteman J, Pals H. Angiotensin Converting Enzyme gene insertion/deletion polymorphism and the risk of heart failure in hypertension subjects. Eur Heart J. 2004;25:2143–8. doi: 10.1016/j.ehj.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Agachan B, Isbir T, Yilmaz H, Akoh E. Angiotensin converting enzyme I/D, angiotensinogen T174M-M235T and angiotensin –II type I receptorA1166C gene polymorphism in Turkish hypertensive patients. Exp Mol Med. 2003;6:545–9. doi: 10.1038/emm.2003.71. [DOI] [PubMed] [Google Scholar]
- 39.Glavnik N, Petrovic D. M235T Polymorphism of the angiotensin gene and I/D polymorphism of the angiotensin-I converting enzyme gene in essential arterial hypertension in Caucasians. Folia Biol Prague. 2007;53:69–70. [PubMed] [Google Scholar]
- 40.Mondry A, Loh M, Lihu P, Zuhu A, Nagel M. Polymorphism of the insetion/deletion ACE and M235T AGT genes and hypertension surprising new findings and meta analysis of data. BMC Nephrology. 2005;6:1. doi: 10.1186/1471-2369-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishigami T, Iwamoto T, Tamura K, Yamaguchi S, Iwaswa K, Uchino K, Umemura S, Ishii M. Angiotensin converting enzyme gene polymorphism and essential hypertension in Japan. Ethnic difference in ACE genotype. Am J Hypertens. 1995;1:95–7. doi: 10.1016/0895-7061(94)00184-D. [DOI] [PubMed] [Google Scholar]
- 42.Lee EJ. Population genetics of the angiotensin converting enzyme in Chinese. Br J Clin Pharmacol. 1994;37:212–4. doi: 10.1111/j.1365-2125.1994.tb04264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


