Abstract
Insulin-like growth factor (IGF) binding proteins provide a layer of complexity to the insulin/IGF signaling system in mammals, but only now, in a recent study in Journal of Biology, has one such protein been functionally characterized in Drosophila.
The first insect protein with the capacity to bind mammalian insulin and insulin-like peptides had a serendipitous discovery eight years ago. A 27 kDa protein from the fall armyworm Spodoptera frugiperda was uncovered as an insulin-binding activity in insect-cell-conditioned media during attempts to purify fragments of the insulin receptor from Sf9 cells [1]. The protein was purified and identified, allowing subsequent identification of its single Drosophila homolog, Imp-L2 [1]. In a recent study in Journal of Biology, Honegger and coworkers [2] present the first, and long-awaited, in vivo functional characterization of this insect insulin/insulin-like growth factor (IGF) binding protein.
Insulin/IGF signaling and IGF binding proteins
The insulin/IGF signaling (IIS) pathway is an evolutionarily conserved neuroendocrine signaling pathway that regulates a plethora of metazoan functions and traits, both during development and in the adult. In model animals ranging from the nematode worm and the fruit fly to the mouse, IIS affects growth and development, metabolic/energy homeostasis, stress resistance, reproduction and lifespan [3-5]. The cellular IIS cascade is initiated by the extracellular binding of an insulin/IGF-like ligand to an insulin-type receptor, resulting in the activation of its intracellular tyrosine kinase domain and the subsequent sequential activation of phosphoinositide 3-kinase (PI 3-kinase) and protein kinase B (Akt) and inactivation of the forkhead box-O transcription factors [3,5]. The active receptor also activates the extra-cellular signal-regulated kinase (Erk), and the Akt branch of the pathway interacts with the target of rapamycin (TOR) pathway [5].
Although there are numerous variants of the intracellular IIS components in mammals, in invertebrates these are mainly encoded by single genes. On the other hand, mammals have only three ligands, insulin, IGF-I and IGF-II [5], whereas there are 38 in the Caenorhabditis elegans genome [6] and seven in Drosophila [7]. Dissecting the functions of all these paralogs may give insights into how this pathway regulates such diverse aspects of animal physiology.
Drosophila and other model organisms have provided valuable insights into the mechanisms and effects of IIS. However, an important aspect of the extracellular regulation of the pathway has not been dissected in Drosophila: the binding of ligands by extracellular binding proteins. In mammals, IGF-I and IGF-II are bound in vivo by IGF binding proteins (IGFBPs) [8]. The effects of IGFBPs on IIS are complex. IGFBPs act as regulators of the activity of IGFs, by prolonging their half-life, altering their local and systemic availability and, through high-affinity binding, sequestering them from the receptor [8,9]. Furthermore, at least some IGFBPs appear to have IGF-independent functions [8].
Mammals have six IGFBPs that can bind IGFs with high affinity, as well as several IGFBP-related proteins (IGFBP-rP) with somewhat lower affinity for IGFs [9]. IGFBPs and IGFBP-rPs belong to a protein superfamily sharing sequence homology predominantly in their amino-terminal portion, which is thought to be involved in IGF binding [9]. The complexity of the IGF-IGFBP system and how it affects IIS has not been examined in invertebrates because no orthologs of IGFBP have been identified – that is, until recently.
Imp-L2: the Drosophila IGFBP
IIS is an important regulator of growth, and overexpression of the Drosophila insulin receptor in the eye during development results in hyperplasia (overgrowth) of the eye. Honegger and coworkers [2] used this phenotype, which had previously been shown to be sensitive to the availability of Drosophila insulin-like peptides (Dilps) [7], to screen for negative regulators of IIS. The authors identified Imaginal morphogenesis protein-Late 2 (Imp-L2) [10] as a strong negative regulator of IIS.
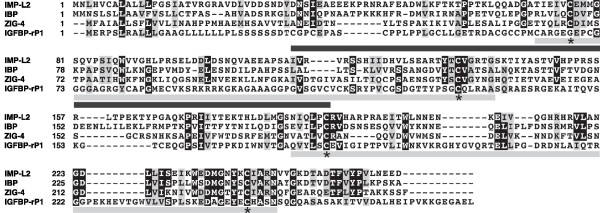
The amino acid sequence of Imp-L2 indicates that it is a secreted protein of the immunoglobulin superfamily [11], with homologs in other invertebrates [1,2]. The carboxy-terminal portion of Imp-L2 is similar to that of the human IGFBP-rP1 (also known as IGFBP7 [9]; Figure 1), leading to the exciting possibility that the screen might have identified a fly IGFBP. Indeed, Imp-L2 had previously been shown to bind human IGF-I, IGF-II and insulin in vitro with high affinity [1], but its binding to Dilps and its potential role in fly IIS had not been examined. Honegger and coworkers [2] therefore set out to determine whether Imp-L2 is functionally equivalent to IGFBPs.
Figure 1.
Sequence comparison of Imp-L2, its invertebrate homologs, and IGFBP-rP1. The sequences of Imp-L2 (Drosophila), Insulin-related peptide binding protein (IBP; S. frugiperda), ZIG-4 (C. elegans) and IGFBP-rP1 (human) were aligned using ClustalW2 [18]. Residues identical or similar in at least three sequences are highlighted in black and gray, respectively. Asterisks below the sequence show the cysteines thought to form two disulfide bridges. The two immunoglobulin-like domains are indicated by a gray bar and the region in IGFBP-rP1 that has the most similarity to IGFBPs by a black bar below the sequences. The annotation was adapted from [2,9].
If Imp-L2 is a functional equivalent of IGFBP, it should negatively regulate growth, and this effect should not be restricted to the cells producing it but should be cell non-autonomous. Indeed, Honegger and coworkers [2] found that weak, ubiquitous overexpression of Imp-L2 yielded smaller flies. When clones of cells in the Drosophila eye were made to overexpress Imp-L2 in an otherwise wild-type fly, their cell specification and patterning were not affected, but the clones were small in size and this reduction also seemed to affect the neighboring cells. Furthermore, overexpression of Imp-L2 in the eye resulted not only in smaller eyes but also in reduction in the size of the whole fly and a developmental delay. Similarly, overexpression in the larval fat body reduced the size of the whole organism. The latter observation may, however, be confounded by the possibility that fat-body-restricted downregulation of IIS could affect energy homeostasis and thus organism growth. Honegger and coworkers [2] also looked at the in vivo levels of phosphatidylinositol (3,4,5)trisphosphate, the secondary messenger produced by PI 3-kinase [5], and demonstrated that, as would be expected of an IGFBP, Imp-L2 overexpression can alter signaling downstream of the insulin receptor.
To further confirm Imp-L2 as a bona fide IGFBP equivalent, Honegger et al. examined its interaction with Dilp2, the most potent growth regulator of all the Dilps [12]. As expected, Dilp2 and Imp-L2 were found to antagonize each other genetically. Weak ubiquitous overexpression of Dilp2 during development caused a body and organ size increase that was exacerbated in flies with only one copy of the Imp-L2 gene. Strong overexpression of either Dilp2 or Imp-L2 alone resulted in lethality, but strong simultaneous overexpres-sion of both allowed wild-type-sized flies to develop. Furthermore, the authors showed that the Imp-L2 protein can bind its native partner, Dilp2, in vitro.
Functions of an IGFBP in flies
The data presented by Honneger and coworkers [2] argue strongly that Imp-L2 is functionally equivalent to mammalian IGFBPs, opening the way to analysis of the functions of this class of IIS regulators in flies. Indeed, the authors reveal a role for Imp-L2 during fly development. Examination of loss-of-function alleles showed that Imp-L2 is required for body size determination during normal growth. Furthermore, Imp-L2 may be important under adverse nutritional conditions. Imp-L2 was induced in the fat body when larvae were starved and loss of Imp-L2 function resulted in a failure to decrease IIS and caused starvation-sensitivity.
A detailed examination of the role of Imp-L2 in adult physiology has yet to be made, but some hints exist as to the function of this protein in the adult. When the germline is ablated late in development, fly lifespan is extended [13]. Concomitantly, the Imp-L2 transcript is upregulated [13], indicating that Imp-L2 may be part of a gonad-derived signaling that modulates whole-body IIS.
Research avenues opened up by Imp-L2
It will be important to establish the similarities and differences between the mammalian IGF-IGFBP system and the Drosophila Dilp-Imp-L2 system. Characterization of the Dilps at the protein level, and of whether and how they form complexes with Imp-L2, will be important. It is interesting in this respect that the homology between IGFBP-rP1 and Imp-L2 does not extend into the amino-terminal, IGFBP-like portion of IGFBP-rP1 (see Figure 1), thought to be required for IGF and insulin binding [9,14]. It will be important to determine functional similarities between Imp-L2 and IGFBP-rP1, especially now that the importance of IGFBP-rP1 as a tumor suppressor has been highlighted [15,16]. Furthermore, it may be interesting to determine whether Imp-L2, like some IGFBPs, has functions independent of Dilp binding, opening up the possibility of using Drosophila to understand how these ligand-independent functions are effected. It will also be interesting to examine whether Imp-L2, like mammalian IGFBPs [8,9], can act both locally and systemically and whether its activity is regulated by proteolysis. A similarity to the mammalian system, in which most IGF-I or IGF-II circulates as part of ternary complexes of IGF, IGFBP3 and the acid-labile subunit (ALS) [8], was uncovered by the recent characterization of the Drosophila ALS [17], which appears to form a trimeric complex with Dilp2 and Imp-L2.
The number of questions that remain only demonstrates how important the work by Honneger and coworkers [2] has been in opening up the field of study of IGFBP in Drosophila. The study of Imp-L2 in such a genetically amenable system will surely yield results relevant to the understanding of mammalian IGFBPs.
Acknowledgments
Acknowledgements
We acknowledge funding by the Wellcome Trust (LP) and a Marie Curie Fellowship (NA). We thank Iain Robinson for critically reading the manuscript.
References
- Sloth Andersen A, Hertz Hansen P, Schaffer L, Kristensen C. A new secreted insect protein belonging to the immunoglobulin superfamily binds insulin and related peptides and inhibits their activities. J Biol Chem. 2000;275:16948–16953. doi: 10.1074/jbc.M001578200. [DOI] [PubMed] [Google Scholar]
- Honegger B, Galic M, Kohler K, Wittwer F, Brogiolo W, Hafen E, Stocker H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J Biol. 2008;7:10. doi: 10.1186/jbiol72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MD, Selman C, McElwee JJ, Partridge L. Separating cause from effect: how does insulin/IGF signalling control lifespan in worms, flies and mice? J Intern Med. 2008;263:179–191. doi: 10.1111/j.1365-2796.2007.01906.x. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- White MF. Regulating insulin signaling and beta-cell function through IRS proteins. Can J Physiol Pharmacol. 2006;84:725–737. doi: 10.1139/Y06-008. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/S0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Mohan S, Baylink DJ. IGF-binding proteins are multifunctional and act via IGF-dependent and -independent mechanisms. J Endocrinol. 2002;175:19–31. doi: 10.1677/joe.0.1750019. [DOI] [PubMed] [Google Scholar]
- Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999;20:761–787. doi: 10.1210/er.20.6.761. [DOI] [PubMed] [Google Scholar]
- Osterbur DL, Fristrom DK, Natzle JE, Tojo SJ, Fristrom JW. Genes expressed during imaginal discs morphogenesis: IMP-L2, a gene expressed during imaginal disc and imaginal histoblast morphogenesis. Dev Biol. 1988;129:439–448. doi: 10.1016/0012-1606(88)90391-0. [DOI] [PubMed] [Google Scholar]
- Garbe JC, Yang E, Fristrom JW. IMP-L2: an essential secreted immunoglobulin family member implicated in neural and ectodermal development in Drosophila. Development. 1993;119:1237–1250. doi: 10.1242/dev.119.4.1237. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/S0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci USA. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Wilson EM, Rosenfeld RG, Oh Y. Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J Biol Chem. 1997;272:30729–30734. doi: 10.1074/jbc.272.49.30729. [DOI] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger AM, Leyland-Jones B, Banerjee K, Spyropoulos DD, Seth AK. Essential roles of IGFBP-3 and IGFBP-rP1 in breast cancer. Eur J Cancer. 2005;41:1515–1527. doi: 10.1016/j.ejca.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Arquier N, Geminard C, Bourouis M, Jarretou G, Honegger B, Paix A, Leopold P. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 2008;7:333–338. doi: 10.1016/j.cmet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]