Abstract
IL-10-related T cell-derived inducible factor (IL-TIF or IL-21) is a new cytokine structurally related to IL-10 and originally identified in the mouse as a gene induced by IL-9 in T cells and mast cells. Here, we report the cloning of the human IL-TIF cDNA, which shares 79% amino acid identity with mouse IL-TIF and 25% identity with human IL-10. Recombinant human IL-TIF was found to activate signal transducer and activator of transcription factors-1 and -3 in several hepatoma cell lines. IL-TIF stimulation of HepG2 human hepatoma cells up-regulated the production of acute phase reactants such as serum amyloid A, α1-antichymotrypsin, and haptoglobin. Although IL-10 and IL-TIF have distinct activities, antibodies directed against the β chain of the IL-10 receptor blocked the induction of acute phase reactants by IL-TIF, indicating that this chain is a common component of the IL-10 and IL-TIF receptors. Similar acute phase reactant induction was observed in mouse liver upon IL-TIF injection, and IL-TIF expression was found to be rapidly increased after lipopolysaccharide (LPS) injection, suggesting that this cytokine contributes to the inflammatory response in vivo.
IL-10-related T cell-derived inducible factor (IL-TIF) is a new cytokine that was originally identified as a gene induced by IL-9 in murine T lymphocytes and showed a weak but significant amino acid identity with IL-10 (1). Mouse IL-TIF consists of 179 amino acids, including four cysteins and shows 22% sequence identity with IL-10. However, IL-10 and IL-TIF do not share the same receptor complex, as the latter failed to activate signal transducer and activator of transcription-3 (STAT-3) in mouse macrophages or to induce the proliferation of IL-10-responsive MC9 cells. This cytokine is expressed by ConA-activated spleen cells and by T helper cells and mast cells, upon activation by IL-9. A weak constitutive expression also has been detected in thymus and brain (1). The biological activities of IL-TIF remain elusive. In rodents, two IL-TIF-responsive cell lines have been identified. IL-TIF activates STAT factors in PC12, a rat pheochromocytoma cell line often used as a model for neuronal cells, pointing to a putative role for this new cytokine in the nervous system. A similar effect has been observed on Mes13, a kidney mesangial cell line, but not on a series of lymphoid and macrophage cell lines (1), suggesting that IL-TIF acts mainly outside the immune system.
As a further step in our analysis of this new cytokine, we attempted to identify its human counterpart. Here, we describe the cloning of the cDNA encoding the human homolog of mouse IL-TIF. By using recombinant IL-TIF, we showed that this cytokine induces acute phase reactant expression by hepatocytes, suggesting a role for this factor in inflammatory processes.
Materials and Methods
Cell Cultures, Transfections, and Cytokines.
BW5147 murine lymphoma cells were grown in Iscove-Dulbecco's medium supplemented with 10% fetal calf serum (FCS), 50 μM 2-ME, 0.55 mM l-arginine, 0.24 mM l-asparagine, and 1.25 mM l-glutamine. Human embryonic kidney 293 cells-Epstein–Barr virus nuclear antigen (HEK293-EBNA) were grown in DMEM medium supplemented with 10% FCS. HepG2 human hepatoma cells were grown in DMEM medium supplemented with 10% FCS, 0.55 mM l-arginine, 0.24 mM l-asparagine, and 1.25 mM l-glutamine.
Recombinant mouse IL-9 was produced in the baculovirus system and purified as previously described (2). For transient expression, the IL-TIF cDNA was cloned into pCEP-4 plasmid (Invitrogen, Groningen, The Netherlands) under the control of the cytomegalovirus promoter. HEK293-EBNA cells were seeded in 6-well plates (Nunc, Roskilde, Denmark) at three 105 cells/well 1 day before transfection. Transfections were carried out by using the Lipofectamine method (Life Technologies, Gent, Belgium), according to the manufacturer's recommendations with 2 μg of plasmid DNA. After transfection, cells were incubated in 1.5 ml of normal medium for 3 days for maximal production of recombinant human IL-TIF. Human IL-6 (five 108 units/mg) produced in Escherichia coli was kindly provided by W. Sebald (Institut für Physiologische Chemie der Universität Würzburg, Germany). Anti-gp130 and anti-hIL-10Rβ antibodies were purchased from R & D Systems.
hIL-TIF cDNA Cloning.
Human peripheral blood mononuclear cells were cultured for 24 h either with or without anti-CD3 mAb (OKT3, ascites fluid 0.2%). Total RNA was isolated by using guanidinium isothiocyanate lysis and CsCl gradient centrifugation (3). Reverse transcription (RT) was performed on 5 μg of total RNA with an oligo(dT) primer. cDNA corresponding to 50 ng of total RNA was amplified for 25 cycles by PCR with specific primers for murine IL-TIF as follows: sense 5′-AGCTGCTCAACTTCACCCTG-3′ and antisense 5′-CAAGTCTACCTCTGGTCTCAT-3′, with an annealing temperature of 45°C. The sequence of the PCR product was obtained with an automated fluorescence-based system (Applied Biosystems 310) using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (PE Biosystems, Foster City, CA).
The 5′-end of the hIL-TIF message was isolated by using the 5′-RACE kit (Life Technologies) according to the manufacturer's recommendations. In brief, first strand cDNA was prepared by using a primer specific to the hIL-TIF sequence: 5′-TGGCCAGGAAGGGCACCACCT-3′. The cDNA was tailed using terminal transferase and dCTP. cDNA was amplified for 35 cycles by using an internal oligonucleotide specific for hIL-TIF 5′-TGGCCAGGAAGGGCACCACCT-3′ and the 5′-RACE-abridged anchor primer from the kit. Seminested amplification was performed on 5 μl of PCR product (diluted 1/100) using the same hIL-TIF primer and the abridged universal amplification primer from the 5′-RACE kit. Amplification was performed for 30 cycles, with an annealing temperature of 56°C, and using Taq polymerase from Takara (Takara Shuzo, Shiga, Japan). The resulting PCR product was cloned and sequenced. Amplification, cloning, and sequencing were repeated to avoid PCR-induced errors in the sequence.
STAT Activation and Luciferase Assays.
Nuclear extracts were prepared, and analysis of DNA binding activity was performed as described previously (4) by using 32P-labeled oligonucleotide probes corresponding to the γ response region (GRR) of the Fc γ RI gene: upper strand, 5′-ATGTATTTCCCAGAAA-3′ and lower strand, 5′-CCTTTTCTGGGAAATAC-3′. Supershifts were performed by adding antibodies to the incubating mixture of nuclear extracts and labeled DNA probe. A 0.75 μg of anti-STAT-1 antibody (catalog no. G16920, Transduction Laboratories, Lexington, KY), 1 μg of anti-STAT-3 antibody (clone ST3–5G7, Zymed, San Francisco, CA), or 1 μg of anti-STAT-5b antibody (835-X, Santa Cruz) were used per lane.
The reporter pGRR5 was kindly provided by P. Brennan (Imperial Cancer Research Fund, London, UK). This construct contains five copies of the GRR sequence inserted upstream a luciferase gene controlled by the thymidine kinase (TK) promoter. As an internal control, we used the pRL-TK vector (Promega) containing the renilla luciferase gene under the control of the TK promoter. The 106 HepG2 cells were transfected with 15 μg of pGRR5 and with 1 μg pRL-TK (250 V, 74 Ω, 1,200 μ F). The pool of transfected cells was divided in 24-well plates (42,000 cells/well) and one part of the cells was preincubated with anti-IL-10Rβ antibodies (20 μg/ml) whereas the other part was left unstimulated. After 1 h, cells were stimulated either with hIL-TIF (1% HEK293 cell supernatant), with hIL-6 (300 units/ml) or control medium (1% supernatant from mock-transfected HEK293 cells or medium alone). Two hours later, cells were pelleted and lysed. Luciferase activity was monitored with the Dual-Luciferase Reporter Assay System kit (Promega).
RT-PCR Analysis of Acute Phase Protein.
Five 106 HepG2 cells were stimulated for 2, 13, or 24 h with 1% of supernatants from transiently transfected HEK293-EBNA cells. Protein synthesis inhibitor cycloheximide (Sigma) was used at 10 μg/ml. Total RNA was isolated using the TRIzol reagent, according to the manufacturer's recommendations (Life Technologies). RT was performed on 10 μg of total RNA with an oligo(dT) primer. cDNA corresponding to 100 ng of total RNA was amplified for 18 cycles with primers specific for human serum amyloid A (SAA) as follows: sense 5′-AGCTCAGCTACAGCACAGAT-3′ and antisense 5′-CCTGCCCCATTTATTGGCAG-3′ [melting temperature (Tm): 54°C]; for human α1-antichymotrypsin, sense 5′-TGTCCTCTGCCACCCTAACA-3′ and antisense 5′-TAATTCACCAGGACCATCAT-3′ (Tm: 52°C); for human haptoglobin, sense 5′-GTGGACTCAGGCAATGATGT-3′ and antisense 5′-ACATAGAGTGTTAAAGTGGG-3′ (Tm: 52°C); and for human β-actin, sense 5′-GCTGGAAGGTGGACAGCGAG-3′ and antisense 5′-TGGCATCGTGATGGACTCCG-3′ (Tm: 56°C). The postPCR products were analyzed in ethidium bromide-stained agarose gel.
Expression in E. coli of Mouse IL (mIL)-TIFα.
The mIL-TIFα sequence (corresponding to amino acids Q29 -V179) was amplified by PCR from the cDNA clone using primers TIFN (5′-GCCCTGTGGGCCCATATGCAGGAGGCAAATGCG-3′) and TIFb (5′-TCTTCTCGCTCAGGATCCTTAGACGCAAGCATTTCTC-3′). The PCR product was digested with NdeI and BamHI and cloned into the pET3A plasmid (Stratagene, La Jolla, CA). E. coli strain BL21-codon plus-(DE3)-RIL (Stratagene) was used as the expression host. The cells were grown in Luria–Bertani medium supplemented with ampicillin (100 μg/ml), chloramphenicol (34 μg/ml), and glucose 2% (wt/vol). Expression of IL-TIF was induced with 1 mM isopropyl-β-d-thiogalactoside at a cell density (600 nm) of ≈1.3. Cells were collected by centrifugation 4 h after induction. The cell pellet was disrupted with a high-pressure cell homogenizer, and the IL-TIF inclusion bodies were collected by centrifugation. Inclusion bodies were washed extensively first with Tris⋅HCl 50 mM, NaCl 100 mM, EDTA 1 mM, DTT 1 mM, and sodium deoxycholate 0.5% (wt/vol), pH 8, and finally with the same buffer without detergent.
Inclusion bodies were solubilized overnight at 4°C in 8 M urea, Mes 50 mM, EDTA 10 mM, and DTT 0.1 mM, pH 6.5. The solution was centrifuged for 1 h at 100,000 × g and the supernatant stored at −80°C until use. The purity of the IL-TIF was estimated at ≈90% based on SDS-PAGE and Coomassie blue staining analysis. The concentration of protein was estimated by UV absorbance in urea solution by using a calculated molar absorption coefficient ɛ280 = 3,840 liter/mol × cm.
The IL-TIF protein was refolded by direct dilution of the solubilized inclusion bodies in the following folding mixture: IL-TIF 100 μg/ml, Tris⋅HCl 100 mM, EDTA 2 mM, l-arginine 0.5 M, reduced glutathion 1 mM, and oxidized glutathion 0.1 mM, pH 8. The solution was incubated for 20 h at 4°C. The folding mixture was then concentrated by ultrafiltration in an Amicon chamber with a YM3 membrane before purification on a Superdex75 (Amersham Pharmacia Biotech) gel filtration column. The protein was eluted with Tris⋅HCl 20 mM and NaCl 50 mM, pH 7.
In Vivo Induction of Murine SAA and IL-TIF Expression.
Various amounts of recombinant mIL-TIF (50, 12.5, 3.2, 0.8, and 0.2 μg) were injected i.p. into endotoxin-resistant C3H/HeJ female mice (10–12 weeks old). Mice were killed 6 h later. Mice receiving 50 μg of recombinant mIL-TIF were killed after 1, 3, 6, 12, or 24 h. The liver was directly frozen in liquid nitrogen. Total RNA was isolated by using the TRIzol reagent, according to the manufacturer's recommendations (Life Technologies). Total RNA (10 μg) was fractionated in a 1.3% agarose gel containing 2.2 mol/liter formaldehyde before transfer onto a Hybond-C Extra nitrocellulose membrane (Amersham Pharmacia Biotech). The murine SAA probe was labeled by using the Rediprime DNA labeling kit from Amersham. Hybridization and washes were performed as described (5). The murine SAA probe was obtained by PCR with specific oligonucleotides as follows: sense 5′-TCTGCTCCCTGCTCCTGGGA-3′ and antisense 5′-TCCAGGAGGTCTGTAGTAAT-3′. The PCR was performed on cDNA synthesized from liver RNA isolated as described above and the PCR product was cloned.
For IL-TIF induction, 2 μg of E. coli LPS (Sigma, Bornem, Belgium) were injected i.p. into BALB/c female mice (12 weeks old). Mice were killed 2 h later and their organs were frozen in liquid nitrogen. Total RNA was isolated by using the TRIzol reagent (Life Technologies). RT was performed on 10 μg of total RNA with an oligo(dT) primer. cDNA corresponding to 100 ng of total RNA was amplified for 27–35 cycles with primers specific for mIL-TIF as follows: sense 5′-CTGCCTGCTTCTCATTGCCCT-3′ and antisense 5′-CAAGTCTACCTCTGGTCTCAT-3′ (Tm: 55°C). The PCR products were analyzed by agarose gel electrophoresis.
Results
Characterization of the Human IL-TIF cDNA.
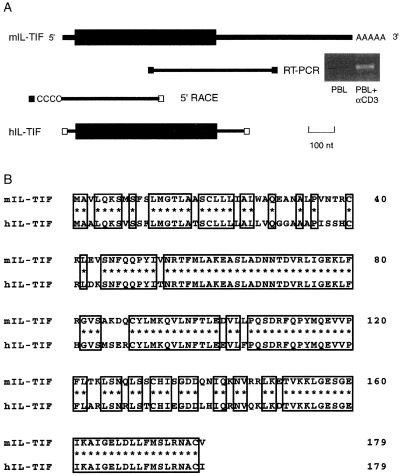
Because the IL-TIF mRNA was strongly expressed by ConA-activated mouse T-cells, we used anti-CD3-stimulated peripheral blood mononuclear cells as a source for its human homolog. A series of oligonucleotide primers were designed from the murine IL-TIF sequence and used in RT-PCR reactions. As shown in Fig. 1A, by using one primer in the middle of the mouse ORF and another primer from the 3′-untranslated region, we were able to amplify a message of 454 nucleotides that was present in anti-CD3-activated, but not in resting peripheral blood mononuclear cells. This fragment was sequenced and the 5′-end of the cDNA was isolated by the 5′-RACE method. Finally, oligonucleotides that flanked the coding sequence allowed for the amplification of a 690-nucleotide fragment, including a 537-bp ORF that encodes a 179-aa protein. Human IL-TIF has the same length and shares 79% amino acid identity with mIL-TIF (Fig. 1B) and 25% identity with human IL-10. Further analysis of the GenBank databases showed that the last exon of IL-TIF is present on a bacterial artificial chromosome clone derived from chromosome 12q15 (accession no. AC007458; 191,111 bp; bacterial artificial chromosome RPCI11–444B24). Based on this bacterial artificial chromosome sequence, the IL-TIF gene is located at ≈90 kb of the IFNγ gene and at <30 kb of the AK155 gene, another IL-10-related cytokine gene that was described recently (6).
Figure 1.
Human IL-TIF cDNA cloning and protein sequence. (A) Total RNA was extracted from human peripheral blood mononuclear cells cultured for 24 h with or without anti-CD3 mAb. RT-PCR was performed with oligonucleotides specific for mIL-TIF (■). (Upper) The mIL-TIF mRNA is represented with the ORF as a black box. The 5′-end was amplified by a 5′-RACE experiment with an oligo specific to human IL-TIF (□) and with a second oligo specific to the dC tail. Finally, the full-length ORF of the hIL-TIF cDNA was amplified using specific primers (□). (B) Alignment of murine IL-TIF and human IL-TIF protein sequences: conserved residues are boxed.
Identification of hIL-TIF-Responsive Cell Lines.
Recombinant human IL-TIF protein was produced by transient transfection of HEK293 cells. Such HEK293 cell supernatants were used to search for new IL-TIF responsive cell based on the hypothesis that hIL-TIF, like mIL-TIF, would induce activation of STAT transcription factors in target cells. Using an electrophoretic mobility shift assay with the FcγRI-derived GRR sequence, which binds to all STAT factors, we screened a number of cell lines for the nuclear translocation of STAT transcription factors after 15 min of hIL-TIF stimulation.
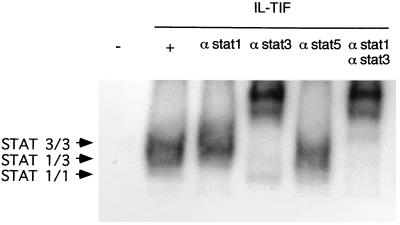
In this assay, an IL-TIF-induced bandshift was observed with the hepatoma cell line, HepG2. Further characterization of STAT factors that are activated in response to IL-TIF in HepG2 cells was achieved by supershift experiments with antibodies specific for STAT-1, -3, and -5. As shown in Fig. 2, anti-STAT-3 antibodies supershifted most of the retardation complexes, and the weak remaining complexes were supershifted by anti-STAT-1, whereas anti-STAT-5 antibodies had no effect. Thus, STAT-3, and to a lesser extent STAT-1, are the major STAT transcription factors activated by IL-TIF in this hepatoma cell line. Similar results were obtained with another human hepatoma cell line, HepG3, and with the rat hepatoma, H4IIE, using both human and mouse IL-TIF. As previously observed with mouse cell lines, IL-TIF failed to activate STAT transcription factors in lymphoid cell lines such as Epstein–Barr virus-transformed B cells. By contrast, four of nine melanoma cell lines showed STAT-3 activation in response to IL-TIF, indicating that the activity of this new cytokine is not restricted to hepatocytes (data not shown).
Figure 2.
STAT activation by IL-TIF in HepG2 cells. HepG2 cells were stimulated for 15 min with hIL-TIF (1% 293-EBNA cell supernatant) or with a supernatant from mock-transfected cells. Gel shift assays were performed as described in Materials and Methods, with the Fcγ RI-derived GRR probe. STAT-containing complexes were supershifted with anti-STAT-1, -3, or -5 antibodies as indicated.
IL-TIF Induces Acute Phase Reactant Production.
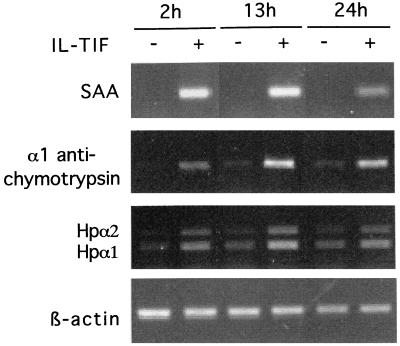
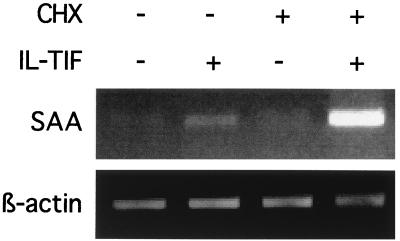
Because activation of STAT-3 by cytokines such as IL-6 is known to result in acute phase protein induction in hepatoma cells (7, 8), we tested the possibility that IL-TIF exerts the same activity. HepG2 cells were stimulated with and without IL-TIF, and RT-PCR was used to assess the expression of SAA, α1-antichymotrypsin, and haptoglobin. As shown in Fig. 3, IL-TIF strongly induced the expression of SAA, α-1 antichymotrypsin, and, to a lesser extent, haptoglobin. By contrast, IL-10 was not active in this assay (data not shown). To determine whether IL-TIF directly up-regulates SAA expression or whether this process requires protein synthesis, HepG2 cells were stimulated with hIL-TIF in the presence of cycloheximide. As shown in Fig. 4, cycloheximide did not block SAA induction, indicating that protein synthesis is not required for hIL-TIF activity. In the presence of cycloheximide, the effect of IL-TIF is even increased. This might result from the inhibition of production of proteins of the SOCS family, which are induced by STAT factors and act as a negative feedback on cytokine activities (9).
Figure 3.
Induction of acute phase protein by IL-TIF in vitro. HepG2 cells were stimulated for the indicated periods of time with hIL-TIF (1% 293-EBNA cell supernatant) or with a supernatant from mock-transfected cells. RT-PCR amplification was performed with oligonucleotides specific for SAA, α1-antichymotrypsin, haptoglobin, and β-actin. PCR products were analyzed by agarose gel electrophoresis. The two bands observed for haptoglobin were sequenced and correspond to previously described allelic variations (23).
Figure 4.
Protein synthesis is not required for SAA up-regulation by IL-TIF. HepG2 cells were stimulated for 2 h with hIL-TIF (1% supernatant from 293-EBNA cells transfected with the IL-TIF cDNA in pCEP4 expression plasmid) or with a supernatant from mock-transfected cells, and with or without 10 μg/ml cycloheximide. RT-PCR amplification was performed with oligonucleotides specific for SAA and β-actin. PCR products were analyzed by agarose gel electrophoresis.
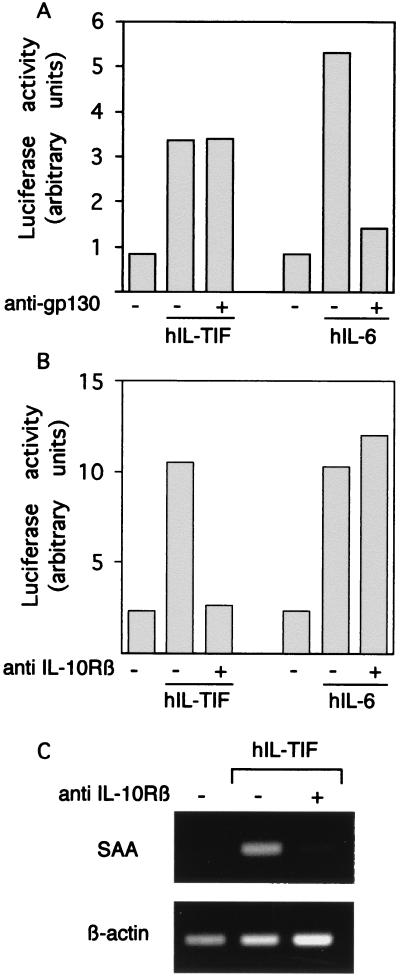
Because IL-TIF and IL-6 seemed to have similar activities on acute phase reactants, we addressed the possibility that IL-TIF activity was mediated through gp130. This transmembrane protein is the main signaling component of the IL-6 receptor complex and is shared by other receptors, such as those of leukemia inhibiting factor, IL-11, ciliary neurotrophic factor, and oncostatin M (7). HepG2 cells were stimulated with IL-TIF or IL-6 in the presence of polyclonal anti-gp130 antibodies that block the activity of gp130-interacting cytokines. To quantitatively assess STAT activation, HepG2 cells were transfected with a luciferase reporter construct that included five STAT-binding sites in addition to the TK promoter. As shown in Fig. 5A, both IL-TIF and IL-6 stimulated STAT-dependent promoter activity in HepG2 cells but anti-gp130 antibodies blocked only IL-6 activity. Another candidate as a component of the IL-TIF receptor is the IL-10Rβ chain, which is known to be involved in the IL-10R (10) but whose expression extends beyond the IL-10-responsive cells. In the presence of anti-IL-10Rβ antibodies, IL-TIF activity was completely blocked whereas that of IL-6 was unaffected, indicating that this receptor chain is indeed involved in IL-TIF signaling (Fig. 5B). The same inhibitory effect was observed at the level of SAA expression by HepG2 in response to IL-TIF (Fig. 5C), confirming that IL-10Rβ can mediate acute phase reactant induction in vitro.
Figure 5.
IL-TIF activity requires IL-10Rβ chain. (A and B) HepG2 cells, transfected with the pGRR5 luciferase construct, were preincubated with 20 μg/ml anti-gp130 antibodies (A) or anti-IL-10Rβ antibodies (B) and then stimulated by IL-6 (300 units/ml), IL-TIF (1% supernatants from IL-TIF-transfected HEK293 cells), or left unstimulated. Luciferase activity was monitored 2 h later. The results are expressed in arbitrary units including standardization by using renilla luciferase internal control. The data correspond to the mean of duplicate cultures and are representative of three independent experiments. Supernatants from mock-transfected HEK293 cells had no effect in the assay (data not shown). (C) HepG2 cells were preincubated with anti-IL-10Rβ antibodies and stimulated with IL-TIF as indicated above. After 2 h, total RNA was extracted and RT-PCR amplification was performed with oligonucleotides specific for SAA and β-actin. PCR products were analyzed by agarose gel electrophoresis.
In Vivo Activity and Production of IL-TIF.
To assess the ability of IL-TIF to regulate acute phase proteins in vivo on normal hepatocytes, recombinant murine IL-TIF was injected i.p. into endotoxin-resistant C3H/HEJ mice. Total RNA was extracted from the liver of these animals, and SAA expression was analyzed by Northern blotting. As shown in Fig. 6A, a high dose of mIL-TIF (50 μg) induced SAA expression as soon as 1 h after i.p. injection of the cytokine. The maximal effect was reached after 6 h, and SAA expression decreased at 24 h after injection. Injection of various doses of IL-TIF showed that 3 μg of IL-TIF still resulted in maximal induction, whereas the SAA message was still detectable with 0.8 μg of IL-TIF (Fig. 6B). An increase in SAA protein also was detected by ELISA in the serum of mice that received seven daily injections of 30 μg of IL-TIF (data not shown).
Figure 6.
In vivo SAA induction by IL-TIF. (A) Kinetics of induction of SAA by IL-TIF in vivo. Mice were injected i.p. with 50 μg of recombinant mIL-TIF. Mice were killed after various periods of time, and total RNA was extracted from the liver. Total RNA (10 μg) was fractionated by agarose gel electrophoresis and transferred to a nitrocellulose filter before hybridization with the SAA cDNA probe. Ethidium bromide-stained 18S and 28S ribosomal RNA show the equal loading of the gel. (B) Dose-dependent induction of SAA by IL-TIF. Mice were injected i.p. with various amounts of recombinant mIL-TIF and were killed 6 h after injection. Total liver RNA (10 μg) was fractionated by agarose gel electrophoresis and transferred to a nitrocellulose filter before hybridization with the SAA cDNA probe. Ethidium bromide-stained 18S and 28S ribosomal RNA show the equal loading of the gel.
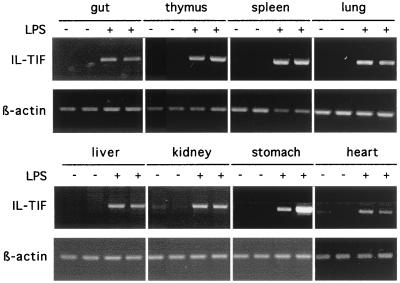
IL-TIF was initially identified as a T cell-derived cytokine whereas most cytokines that regulate the liver acute phase response are mainly produced during inflammation. To determine whether IL-TIF also could be produced upon inflammatory stimuli in vivo, we injected LPS into BALB/c mice and analyzed IL-TIF mRNA expression after 2 h in various organs. As shown in Fig. 7, LPS injection induced IL-TIF expression in all organs examined, indicating that IL-TIF is actually involved in inflammatory responses.
Figure 7.
Inducibility of IL-TIF expression upon LPS injection. BALB/c mice were injected either with 200 μl of 10 μg/ml LPS or the same volume of PBS and were killed 2 h later. Total RNA was prepared from different organs from two mice per condition. RT-PCR amplification was performed with oligonucleotides specific for mIL-TIF and for β-actin. PCR products were analyzed by agarose gel electrophoresis. Similar results were found in three independent experiments.
Discussion
In the present paper, we report the cloning of the human IL-TIF cDNA and show that this new cytokine is able to up-regulate acute phase reactant production by liver cells. Production of acute phase proteins is considered as a survival mechanism for the short term, but its maintenance for longer periods contributes to chronic inflammation and may have negative clinical consequences (11, 12). This liver response is mainly due to cytokine release by activated macrophages and other cells, particularly IL-1, tumor necrosis factor, and IL-6 (13–16).
The finding that IL-TIF up-regulates acute phase reactant production extends the number of cytokines that might contribute to this process. In addition, we show that the activity of this factor is mediated by IL-10Rβ. This transmembrane protein is required for IL-10 signaling (10), and IL-10Rβ-deficient mice recapitulate the phenotype of IL-10-deficient mice, namely a high susceptibility to inflammatory bowel disease and splenomegaly (17). Our data indicate that this molecule is not only a component of the IL-10 receptor complex but might be shared between a larger family of cytokine receptors for IL-10-related factors. This observation also suggests that further analysis of IL-10Rβ-deficient mice might unmask unique characteristics as compared to IL-10 deficient animals, that could reflect IL-TIF-dependent processes.
In addition, it is likely that other members of the IL-10 family that could also use IL-10Rβ will be described in the near future. Recently, a new cytokine called AK155 was identified because of its up-regulation during viral infection of T lymphocytes (6). This protein shares 27% amino acid identity with IL-10 and shows a similar homology with IL-TIF. Interestingly, the AK155 gene is located on chromosome 12q15, at 40 kb from the IFNγ gene, whereas the IL-TIF gene is located in the very same region, at 90 kb from the IFNγ gene (L.D., unpublished data). This suggests that chromosome 12q15 bears an IFN/IL-10-related cytokine cluster that might be involved in immune and inflammatory responses. In this regard, it must be stressed that chromosome 12q15 has been linked to inflammatory bowel disease and to asthma (18–21). Further analysis of polymorphisms in the human IL-TIF gene and the production of IL-TIF-deficient or transgenic animals should allow us to determine precisely the role of this new factor in inflammatory processes.
IL-TIF was originally identified as a gene up-regulated by IL-9 in a murine T cell lymphoma. This suggested that IL-TIF might be responsible for some of the in vivo activities of IL-9 such as lymphoma induction, asthma susceptibility, anti-parasite immune response, or B1 lymphocyte expansion (22). Although IL-9 also up-regulated IL-TIF expression in T helper cell clones and mast cell lines in vitro, we failed to detect any up-regulation of this gene in IL-9 transgenic mice, suggesting that it does not play a major role in the in vivo biological activities of IL-9. However, IL-TIF is produced by normal T cells upon ConA activation in mice, or anti-CD3 stimulation in humans, suggesting a role associated with antigen-specific immune responses. In this report, we show that LPS induces IL-TIF mRNA expression in vivo within 2 h in various organs, pointing to other cell types as potential IL-TIF-producers. This finding contrasts with our initial observation that murine spleen cells stimulated for 24 h with LPS in vitro failed to express IL-TIF (1). This discrepancy between in vivo and in vitro IL-TIF induction might reflect an indirect mechanism of gene induction. Further studies will be necessary to elucidate the mechanisms regulating the expression of this new cytokine during inflammatory processes and to determine potential applications of IL-TIF, itself or its antagonists, in modulating inflammation in vivo.
Note Added in Proof.
The human IL-TIF sequence has been recently identified from expressed sequence tag databases by Xie et al. (24). It was proposed to rename the protein IL-21.
Acknowledgments
This work was supported in part by the Belgian Federal Service for Scientific Technical and Cultural Affairs, the Opération Télévie and the Actions de recherche concertées, Communauté Française de Belgique, Direction de la recherche scientifique. J.-C.R. is a research associate with the Fonds National de la Recherche Scientifique, Belgium.
Abbreviations
- IL-TIF
IL-10-related T cell-derived inducible factor
- CHX
cycloheximide
- GRR
γ response region
- SAA
serum amyloid A
- STAT
signal transducer and activator of transcription
- mIL
mouse IL
- LPS
lipopolysaccharide
- RT
reverse transcription
- TK
thymidine kinase
- HEK293-EBNA
human embryonic kidney 293 cells-Epstein–Barr virus nuclear antigen
- Tm
melting temperature
Footnotes
Data deposition: The sequence of the human IL-TIF cDNA reported in this paper has been submitted to the European Molecular Biology Laboratory (EMBL)/GenBank database (accession no. AJ277247).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170291697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170291697
References
- 1.Dumoutier L, Louahed J, Renauld J-C. J Immunol. 2000;164:1814–1819. doi: 10.4049/jimmunol.164.4.1814. [DOI] [PubMed] [Google Scholar]
- 2.Druez C, Coulie P, Uyttenhove C, Van Snick J. J Immunol. 1990;145:2494–2499. [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Jeene Publishing; 1993. [Google Scholar]
- 4.Demoulin J-B, Uyttenhove C, Van Roost E, de Lestré B, Donckers D, Van Snick J, Renauld J-C. Mol Cell Biol. 1996;16:4710–4716. doi: 10.1128/mcb.16.9.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Snick J, Goethals A, Renauld J-C, Van Roost E, Uyttenhove C, Rubira M R, Moritz R L, Simpson R J. J Exp Med. 1989;169:363–368. doi: 10.1084/jem.169.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knappe A, Hor S, Wittmann S, Fickenscher H. J Virol. 2000;74:3881–3887. doi: 10.1128/jvi.74.8.3881-3887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinrich P, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wegenka U M, Lutticken C, Buschmann J, Yuan J, Lottspeich F, Muller-Esterl W, Schindler C, Roeb E, Heinrich P C, Horn F. Mol Cell Biol. 1994;14:3186–3196. doi: 10.1128/mcb.14.5.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gisselbrecht S. Eur Cytokine Netw. 1999;10:463–470. [PubMed] [Google Scholar]
- 10.Kotenko S V, Krause C D, Izotova L S, Pollack B P, Wu W, Pestka S. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumann H, Gauldie J. Immunol Today. 1994;15:74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 12.Uhlar C M, Whitehead A S. Eur J Biochem. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 13.Andus T, Geiger T, Hirano T, Kishimoto T, Heinrich P C. Eur J Immunol. 1988;18:739–746. doi: 10.1002/eji.1830180513. [DOI] [PubMed] [Google Scholar]
- 14.Heinrich P C, Castell J V, Andus T. Biochem J. 1990;265:621–636. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinarello C A. N Engl J Med. 1984;311:1413–1418. doi: 10.1056/NEJM198411293112205. [DOI] [PubMed] [Google Scholar]
- 16.Richards C, Gauldie J, Baumann H. Eur Cytokine Netw. 1991;2:89–98. [PubMed] [Google Scholar]
- 17.Spencer S D, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan A, Sordat B, Gibbs V C. J Exp Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ober C, Cox N J, Abney M, Di Rienzo A, Lander E S, Changyaleket B, Gidley H, Kurtz B, Lee J, Nance M, et al. Hum Mol Genet. 1998;7:1393–1398. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- 19.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger J D, Lathrop G M, Bell J I, Jewell D P. Nat Genet. 1996;14:199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 20.Duerr R H, Barmada M M, Zhang L, Davis S, Preston R A, Chensny L J, Brown J L, Ehrlich G D, Weeks D E, Aston C E. Am J Hum Genet. 1998;63:95–100. doi: 10.1086/301929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes K C, Neely J D, Duffy D L, Freidhoff L R, Breazeale D R, Schou C, Naidu R P, Levett P N, Renault B, Kucherlapati R, et al. Genomics. 1996;37:41–50. doi: 10.1006/geno.1996.0518. [DOI] [PubMed] [Google Scholar]
- 22.Renauld J-C, Van Snick J. In: The Cytokine Handbook. Thompson A, editor. London: Academic; 1998. pp. 313–331. [Google Scholar]
- 23.van der Straten A, Herzog A, Cabezon T, Bollen A. FEBS Lett. 1984;168:103–107. doi: 10.1016/0014-5793(84)80215-x. [DOI] [PubMed] [Google Scholar]
- 24.Xie, M. H., Aggarwal, S., Ho, W. H., Foster, J., Zhang, Z., Stinson, J., Wood, W. I., Goddard, A. D. & Gurney, A. L. (2000) J. Biol. Chem., in press. [DOI] [PubMed]