Abstract
Autotransporters are a superfamily of virulence factors produced by Gram-negative bacteria consisting of a large N-terminal extracellular domain (“passenger domain”) and a C-terminal β barrel domain (“β domain”). The mechanism by which the passenger domain is translocated across the outer membrane (OM) is unknown. Here we show that the insertion of a small linker into the passenger domain of the Escherichia coli O157:H7 autotransporter EspP effectively creates a translocation intermediate by transiently stalling translocation near the site of the insertion. Using a site-specific photocrosslinking approach, we found that residues adjacent to the stall point interact with BamA, a component of a heterooligomeric complex (Bam complex) that catalyzes OM protein assembly, and that residues closer to the EspP N terminus interact with the periplasmic chaperones SurA and Skp. The EspP–BamA interaction was short-lived and could be detected only when passenger domain translocation was stalled. These results support a model in which molecular chaperones prevent misfolding of the passenger domain before its secretion and the Bam complex catalyzes both the integration of the β domain into the OM and the translocation of the passenger domain across the OM in a C- to N-terminal direction.
Keywords: autotransporter, Bam complex, outer membrane, protein secretion, virulence factors
Autotransporters are a large superfamily of proteins produced by Gram-negative bacterial pathogens (1). They consist of an N-terminal extracellular domain (“passenger domain”) that often encodes a virulence function and a C-terminal domain (“β domain”) that resides in the outer membrane (OM). Almost all passenger domains are predicted to form a β-helical structure despite the fact that they vary considerably in sequence and range in size from ≈20 to 400 kDa (2). Some passenger domains remain anchored to the cell surface and act as adhesions, whereas others are cleaved following their translocation across the OM and encode degradative enzymes or cytotoxins. While β domains also are highly variable in sequence, they are more consistent in size (≈30 kDa) and form 12-stranded β barrels (3, 4). Based on the observation that the deletion of the β domain leads to retention of the passenger domain in the periplasm, it was originally proposed that the β domain forms a pore through which the passenger domain is secreted (5). Indeed, the term “autotransporter” was coined to embody the idea that a single protein contains all of the functional elements necessary for its own secretion. At least at first glance, crystal structures that show a peptide derived from the passenger domain C terminus embedded inside the β domain seem to support the notion that the β domain functions as a protein-conducting channel (3, 4).
Despite its attractiveness, the autotransporter hypothesis has been challenged by various experimental studies. Crystallographic analysis has shown that the β domain pore is only ≈10 Å in diameter, wide enough to transport only a single polypeptide in an α-helical conformation or two fully extended strands that form a hairpin (3, 4). Molecular dynamics simulations also indicate that the β barrel is unlikely to expand spontaneously, and thus corroborate the conclusion that polypeptides would need to traverse the β barrel pore in an unfolded conformation (6). Paradoxically, however, several studies have provided strong evidence that passenger domains acquire at least a limited amount of tertiary structure in the periplasm or can be fused to folded domains that are secreted efficiently (7–9). A recent study strongly suggested that the passenger domain is extruded across the OM in a C- to N-terminal direction and implied that translocation is initiated by the formation of a hairpin at the C terminus of the passenger domain (10). Neither the interior surface of the β domain nor the C terminus of the passenger domain is conserved, however, and so far no mutation in either of these regions has been clearly shown to produce a translocation defect (11, 12; N. Dautin, unpublished results). Finally, a recent study showed that the C terminus of a passenger domain is incorporated into the β barrel pore before the integration of the β domain into the OM and long before the completion of passenger domain translocation (12). The observation that the embedded polypeptide is required for the insertion of the β domain into the OM and for β barrel stability suggests that it has an important structural role and casts further doubt on the idea that the β domain first folds into a transport channel and then acts on a substrate (4, 12).
To explain the experimental evidence, an alternative hypothesis has been proposed in which as yet unidentified accessory factors play a major role in passenger domain secretion (3, 8, 13). The most logical candidate is the Bam complex (also known as the YaeT or Omp85 complex), a heterooligomer consisting of an integral OM protein (BamA) and several lipoproteins (BamB–E) (14, 15). The Bam complex is required for the correct folding and integration of β barrel proteins (including autotransporter β domains) into the OM of Escherichia coli and Neisseria meningitidis, although its mechanism of action is unknown (14–18). Furthermore, BamA belongs to a superfamily of bacterial, mitochondrial, and chloroplast proteins, some of which promote protein translocation reactions instead of membrane protein integration reactions (19). Based on the available evidence, it has been proposed that the Bam complex might catalyze the integration of the β domain into the OM and the translocation of the passenger domain across the OM in a concerted reaction (13). In this model, the requirement for the β domain in passenger domain secretion would arise from its role in targeting the passenger domain to the factor that mediates the translocation reaction.
In this study we present evidence that the Bam complex facilitates secretion of the passenger domain of EspP, a model autotransporter produced by E. coli O157:H7. The passenger domain of EspP is released from the cell surface by an autoproteolytic mechanism following its translocation across the OM (11). Our experiments were based on the serendipitous discovery that the insertion of 11 aa into a predicted loop in the middle of the EspP passenger domain transiently stalls translocation near the insertion site, thereby creating a translocation intermediate. By incorporating photocrosslinkers at specific locations in the EspP mutant, we were able to probe the environment of the protein during passenger domain translocation and to obtain a snapshot of the secretion reaction. We found that both the segment of the passenger domain near the stall point and the β domain are in contact with components of the Bam complex, while a more N-terminal segment of the passenger domain is in contact with periplasmic chaperones. Along with providing insight into the mechanism of passenger domain secretion, our results also suggest that the folding of the passenger domain exerts a strong influence on the translocation reaction.
Results
Generation of an EspP Passenger Domain Translocation Intermediate.
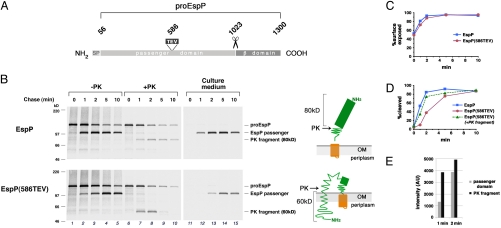
We previously found that the insertion of tobacco etch virus (TEV) protease sites into a segment of EspP that straddles the passenger domain–β domain junction provides a useful tool for monitoring its incorporation into the β barrel pore (11, 12). To monitor the folding of the passenger domain, TEV sites were inserted into several predicted short loops. We reasoned that the TEV protease would cleave EspP only when the segment containing a given TEV site was unfolded. Initially, we wished to examine the effect of the insertions on EspP biogenesis. To this end, AD202 was transformed with a plasmid encoding WT EspP or one of the mutants under the control of a trc promoter, expression of the plasmid-borne gene was induced by the addition of isopropylthiogalactoside (IPTG), and cells were subjected to pulse-chase labeling. Radiolabeled cells were separated from the culture medium by centrifugation, and the cell pellet was divided into two halves. One half was untreated, and the other half was treated with proteinase K (PK). EspP-containing polypeptides were then immunoprecipitated from the culture medium as well as the treated and untreated cells using an antiserum raised against an EspP N-terminal peptide. The exposure of part or all of the passenger domain on the cell surface was evaluated by quantitating the fraction of radiolabeled proEspP (the precleavage form of the protein that contains covalently linked passenger and β domains; Fig. 1A) that was sensitive to PK digestion. The cleavage of proEspP into discrete passenger and β domain fragments was assessed by measuring the fraction of the total signal produced by the cleaved passenger domain. As reported previously (8), cleavage of the WT passenger domain lagged behind its exposure on the cell surface and appeared to occur only after completion of the translocation reaction (Fig. 1B Top; Fig. 1 C and D, squares).
Fig. 1.
A linker insertion transiently stalls EspP passenger domain translocation. (A) Illustration of the primary structure of the EspP precursor showing the signal peptide (residues 1–55), the passenger domain (residues 56–1023), the site of the TEV linker (residue 586), and the β domain (residues 1024–1300). ProEspP is the form of the protein that contains covalently linked passenger and β domains. (B) AD202 transformed with pRLS5 (Ptrc-espP) or pJH97 [Ptrc-espP(586TEV)] were subjected to pulse-chase labeling after the addition of 100 μM IPTG. Half of the cells were treated with PK, and EspP-containing polypeptides were immunoprecipitated from cell and culture medium fractions using an N-terminal anti-EspP antiserum. The cartoons illustrate the source of each PK fragment. (C) The percentage of the WT EspP (squares) or EspP(TEV586) (circles) passenger domain that was surface-exposed. (D) The percentage of the WT EspP (squares) or EspP(TEV586) (circles) passenger domain that was cleaved from proEspP. (E) The radioactive signal of the cleaved passenger domain and PK fragment [in arbitrary units (AUs)] at the 1 min and 2 min time points in the lower panel of (B).
One of the linker insertions, which introduces a TEV site at residue 586 and is designated EspP(586TEV), had a striking and completely unexpected effect on EspP biogenesis. Although the C terminus of the mutant passenger domain was exposed on the cell surface as rapidly and efficiently as that of the WT passenger domain, the mutant passenger domain was cleaved much more slowly than its WT counterpart (Fig. 1B Fig. 1 C and D, squares and circles). Furthermore, PK treatment of cells that synthesized EspP(586TEV) yielded different products than those that synthesized WT EspP. Treatment of cells that synthesized the WT protein yielded modest amounts of an ≈80-kDa N-terminal fragment that accumulated at about the same rate as the cleaved passenger domain (Fig. 1B Top, lanes 1–10). The relatively late appearance of the ≈80-kDa fragment suggested that it resulted from the folding of some of the passenger domain into a protease-resistant conformation around the time of passenger domain cleavage (Fig. 1B Top, cartoon). This explanation is supported by the observation that the treatment of soluble passenger domain recovered from the medium at 1 min and 2 min time points yielded roughly the same relative amount of the ≈80-kDa fragment as the treatment of intact cells [supporting information (SI) Fig. S1, lanes 1–4]. In contrast, treatment of cells that synthesized EspP(586TEV) did not yield an ≈80-kDa fragment, but instead yielded larger amounts of a ≈60-kDa fragment that accumulated before the peak of passenger domain cleavage and then declined sharply (Fig. 1B Bottom, lanes 6–10). The absence of the ≈80-kDa fragment suggests that the TEV insertion perturbs protein folding. The fact that the amount of the ≈60-kDa fragment observed at both the 1 min and 2 min time points exceeded the amount of the cleaved passenger domain (Fig. 1E) indicates that it must have been derived largely or exclusively from proEspP. Consistent with this interpretation, PK treatment of the soluble passenger domain did not yield any of the ≈60-kDa fragment (Fig. S1, lanes 5–8).
Several additional observations indicated that the ≈60-kDa fragment resulted from the transient stalling of passenger domain translocation near the site of the TEV insertion (Fig. 1B, cartoon). First, the PK-resistant fragment migrated close to the molecular weight of the EspP passenger domain segment that encompasses residue 56 (the first residue beyond the signal peptide) to residue 586 (57.7 kDa). The PK-resistant fragment was slightly larger than the N-terminal fragment produced by the treatment of EspP(586TEV) with TEV protease (Fig. S2). Thus, in light of evidence that passenger domains are transported across the OM in a C- to N-terminal direction (10) and the observation that the C terminus of the EspP(586TEV) passenger domain is rapidly exposed on the cell surface (Fig. S3), a ≈60-kDa PK-resistant fragment would be expected if translocation stalled at the TEV insertion. The finding that the ≈60-kDa fragment was degraded upon permeabilization of the OM confirms that it was trapped in the periplasmic space (Fig. S4). Second, the accumulation of the ≈60-kDa fragment at early time points, followed by its subsequent disappearance, is consistent with the notion that it is derived from an assembly intermediate. In this regard, it is striking that if the radioactive signals produced by the ≈60-kDa fragment and the cleaved passenger domain are added together, a curve resembling the time course of cleavage of the WT EspP passenger domain can be generated (Fig. 1D, triangles). This observation suggests that the ≈60-kDa fragment is derived from the passenger domain of proEspP molecules that have partially emerged on the cell surface but have not been cleaved because translocation is incomplete. Finally, to further explore the significance of the disappearance of the ≈60-kDa fragment, we harvested cells after a 0 min or 2 min chase, treated them with PK on ice, washed away the protease, and then resumed incubation at 37 °C. Protein biogenesis proceeded normally in the cells that were protease-treated immediately after the pulse-chase labeling, presumably because no protease-sensitive OM proteins are required for passenger domain translocation (Fig. S5, lanes 1–4). Interestingly, while ongoing secretion also could be observed in cells treated after 2 min, the ≈60-kDa fragment produced by the protease persisted for 10 min (Fig. S5, lanes 5–8). It is likely that the fragment persisted because digestion of proEspP prevented the resumption of passenger domain translocation. In any case, this finding rules out the possibility that the ≈60-kDa fragment was derived from a transiently folded form of the secreted passenger domain. Taken together, these results indicate that the insertion at residue 586 creates a bona fide translocation intermediate.
EspP(586TEV) Interacts With the Bam Complex and Chaperones During Transport of the Passenger Domain Across the OM.
We next used a site-specific in vivo photocrosslinking method to identify accessory factors that interact with EspP(586TEV) while passenger domain translocation is stalled. This technique uses the coexpression of an amber suppressor tRNA and an amino acyl-tRNA synthetase from Methanococcus jannaschii that are orthogonal to the E. coli translation machinery to incorporate an “unnatural” amino acid at an amber codon engineered into a protein of interest (20). An amber codon was introduced at one of eight positions in the passenger domain or at one of two positions in the β domain of EspP and EspP(568TEV) (Fig. 2C). AD202 was transformed with two plasmids that encode an amber mutant and the amber-suppression system (pDULEBpa) (21), IPTG was added to induce expression of the EspP derivative, and cells were subjected to pulse-chase labeling. Amber suppression led to the incorporation of the photoactivable amino acid analog p-benzoyl-L-phenylalanine (Bpa) at the site of the amber codon. Bpa can be crosslinked only to proteins that lie within ≈4 Â. After showing that amber suppression was relatively efficient (≈20%–30%) and confirming that the translocation of the EspP(586TEV) passenger domain stalled under amber suppression conditions (Fig. S6), we established a crosslinking protocol in which two equal samples were removed from radiolabeled cultures after a 1 min chase. One sample was exposed to UV light, and EspP-containing polypeptides were immunoprecipitated using the N-terminal antiserum. Interestingly, while no crosslinked products were observed after UV-irradiation of samples that contained a derivative of WT EspP, UV-irradiation of five of the EspP(586TEV) derivatives yielded one or two major high–molecular weight adducts (Fig. S7). Due to slight differences in suppression efficiency and larger differences in protein stability, the level of each amber codon–terminated EspP fragment varied considerably (Fig. S7, asterisks).
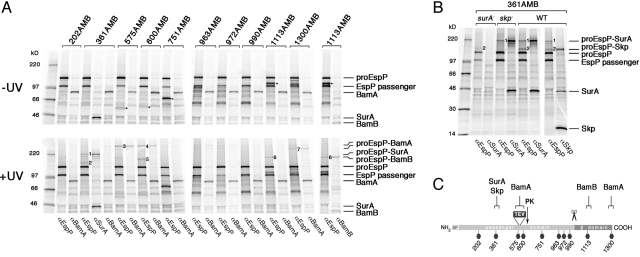
Fig. 2.
Crosslinking of an EspP secretion intermediate to the Bam complex, SurA and Skp. (A) AD202 was transformed with pDULEBpa and a derivative of pRI23 [Plac-espP(586TEV)] harboring an amber codon at the indicated position. Cells were pulse-labeled and subjected to a 1 min chase after the addition of 200 μM IPTG. Half of each sample was UV-irradiated, and equal portions were used for immunoprecipitation with antisera specific for an EspP N-terminal peptide, BamA, BamB, or SurA. Major crosslinked adducts are numbered 1–7. Truncated forms of the protein that resulted from translation termination at the amber codon are denoted by an asterisk. (B) The experiment in (A) was repeated using AD202 (WT), HDB130 (AD202 surA-), and HDB131 (AD202 skp-). Only the UV-irradiated samples are shown. (C) Summary of the data in (A) and (B) showing the proteins that were crosslinked at each position and the position at which PK cleaves the translocation intermediate.
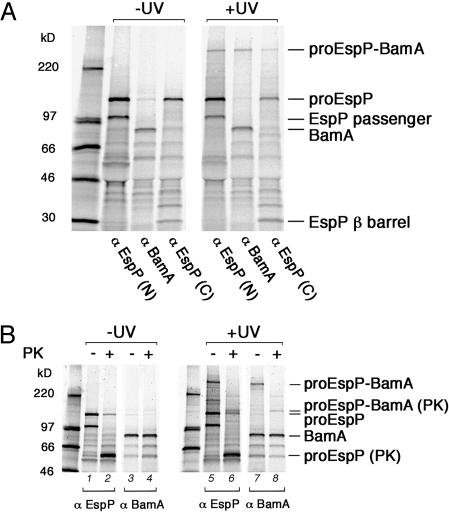
Based on their mobility on SDS/PAGE (>220 kDa), we surmised that the largest adducts, which were observed when the crosslinker was placed at residue 575, 600, or 1300 (adducts 3, 4, and 7, respectively), contained proEspP crosslinked to BamA. To test our hypothesis, we subjected identical aliquots from all of the samples generated in the experiment described above to immunoprecipitation using either the N-terminal anti-EspP antiserum or an anti-BamA antiserum. As we predicted, all 3 adducts were immunoprecipitated by both antisera (Fig. 2A, Bottom Panel). The observation that adduct 3 also was immunoprecipitated by an antiserum generated against an EspP C-terminal peptide led to the conclusion that it contained proEspP and not the cleaved passenger domain (Fig. 3A). Likewise, adduct 7 must have contained proEspP, because the crosslinker was located in the β domain. Curiously, adducts 3 and 4 migrated more slowly than adduct 7, and all 3 adducts had an apparent molecular weight greater than the sum of proEspP (≈135 kDa) and BamA (≈85 kDa). Although in theory each adduct might contain more than one proEspP molecule linked to a single copy of BamA, they appear to migrate too rapidly to contain multiple copies of proEspP. Furthermore, if each adduct contained multiple copies of proEspP (and thus arose through multiple independent crosslinking events), then one or more smaller adducts that migrate closer to the combined molecular weight of proEspP and BamA should have been observed, due to the inherent inefficiency of crosslinking. Thus, the most likely explanation of the results is that each adduct contains a single copy of proEspP but migrates aberrantly on SDS/PAGE. Adduct 7 contains the N and C termini of BamA (Fig. S8) and thus presumably migrates faster than adducts 3 and 4 because the crosslinking of BamA to each domain of proEspP affects mobility differently.
Fig. 3.
BamA interacts specifically with proEspP. AD202 transformed with pDULEBpa and a derivative of pRI23 harboring an amber codon at residue 575 were subjected to pulse-chase labeling and UV irradiation as described in the legend to Fig. 2. (A) Equal aliquots of both UV-irradiated and untreated samples were subjected to immunoprecipitation using antisera specific for EspP N- and C-terminal peptides and BamA. (B) Cells that were UV-irradiated or untreated were divided into 2 portions, one of which was treated with PK. Samples were then subjected to immunoprecipitation using anti-EspP N-terminal and anti-BamA antisera.
Because the other adducts were considerably smaller than adducts 3, 4, and 7, we conjectured that they contained proEspP crosslinked to a protein other than BamA. Adduct 1, which was observed when the crosslinker was placed at residue 361, migrated roughly halfway between proEspP and adducts 3 and 4. The size of this adduct suggests that the interacting protein was ≈40–50 kDa. Because residue 361 is predicted to reside in the periplasm when passenger domain translocation stalls, we surmised that the interacting protein might be SurA, a ≈46-kDa periplasmic chaperone associated with the Bam complex (22). To test this possibility, we subjected a third aliquot derived from cells that produced EspP(586TEV/361AMB) in the experiment described above to immunoprecipitation using an anti-SurA antiserum. Consistent with our hypothesis, adduct 1 was recognized by anti-EspP and anti-SurA, but not by anti-BamA (Fig. 2A). Adduct 1 also was not observed in a surA- strain (Fig. 2B). Adduct 2, which is relatively small, was immmunoprecipitated by an antiserum generated against the 17-kDa chaperone Skp and was absent in a skp- strain, indicating that it resulted from an interaction between Skp and residue 361 (Fig. 2B). Adduct 6, which was observed when the crosslinker was placed in the β domain at residue 1113, migrated roughly halfway between proEspP and adduct 7. Because the β domain is presumably in or near the OM when passenger domain translocation stalls, we surmised that the interacting protein might be a ≈30- to 50-kDa component of the Bam complex, such as BamB or BamD. Using antisera generated against these proteins, we found that adduct 6 contained BamB (Fig. 2A, last 2 lanes). We have not yet identified the interacting protein in adduct 5.
We next wished to confirm that proEspP(586TEV) and BamA interact when translocation of the passenger domain across the OM is stalled. Toward this end, AD202 transformed with a plasmid-encoding EspP(586TEV) with an amber mutation at position 575 and pDULEBpa were pulse-labeled and collected after a 1 min chase. Half of each sample was UV-irradiated, as described above. Then both the UV-irradiated and untreated samples were divided in half again, and one portion was treated with PK. Finally, all samples were subjected to immunoprecipitation with the N-terminal anti-EspP antiserum and anti-BamA. Consistent with the results shown in Figs. 1B and Fig. S2, proEspP(586TEV) was converted to a ≈60-kDa PK-resistant fragment in the absence of UV irradiation (Fig. 3B, lanes 1 and 2). Conversely, BamA was not digested by the protease (Fig. 3B, lanes 3 and 4). In the UV-irradiated samples, subsequent PK treatment reduced the high–molecular weight adduct to a ≈150-kDa band that was immunoprecipitated by both antisera (Fig. 3B, lanes 6 and 8). The resistance of BamA to PK digestion implies that the protease cleaved the proEspP moiety. Thus, based on its size, the ≈150-kDa band most likely corresponds to BamA crosslinked to the PK-resistant fragment of proEspP(586TEV) that we observed when passenger domain secretion was stalled.
The EspP(586TEV)–BamA Interaction Is Transient.
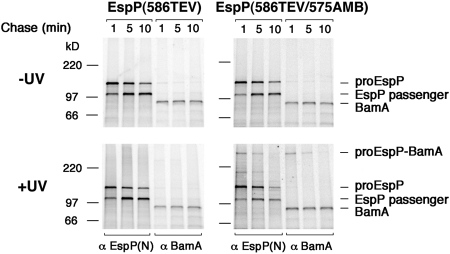
If the proEspP(586TEV) that interacted with BamA were a bona fide translocation intermediate (as opposed to a misfolded or aberrant form of the protein), then the crosslinked products should be short-lived, and crosslinking should depend on the incorporation of Bpa. To test this prediction, cells synthesizing EspP(586TEV) or EspP(586TEV/575AMB) were pulse labeled and collected after a 1, 5, or 10 min chase. Half of the cells were UV irradiated, and all samples were subjected to immunoprecipitation using the N-terminal anti-EspP and anti-BamA antisera. Consistent with our prediction, a high–molecular weight adduct that was recognized by both anti-EspP and anti-BamA antisera was observed only in UV-irradiated cells that produced EspP(586TEV/575AMB) (Fig. 4). Furthermore, the intensity of the high–molecular weight adduct was highest after a 1 min chase and declined over time. Thus, the time-dependence of the crosslinked product was similar to that of the PK-resistant fragment of proEspP(586TEV). Interestingly, the high–molecular weight adduct disappeared faster than proEspP; ≈60% of the proEspP that was observed at 1 min remained after 5 min, but only ≈40% of the adduct remained at the later time point. This observation suggests that residue 575 is in proximity to BamA only before the termination of translocation and subsequent processing of the passenger domain.
Fig. 4.
The EspP–BamA interaction is transient. AD202 was transformed with pDULEBpa and pRI23 or pRI23 harboring an amber codon at residue 575. Cells were pulse-labeled and subjected to a 1, 5, or 10 min chase after the addition of 200 μM IPTG. One of the 2 samples obtained at each time point was UV irradiated, and immunoprecipitation was performed using anti-EspP N-terminal and BamA antisera.
Discussion
In this report we describe evidence that an autotransporter passenger domain interacts with BamA during its translocation across the OM. Our study was promoted by the observation that the insertion of a small linker into a predicted loop in the EspP passenger domain stalls translocation near the insertion site. Because the pause was transient, the stalled form of the protein represents a bona fide translocation intermediate. Several observations that emerged from site-specfic photocrosslinking experiments indicated that the passenger domain of the insertion mutant [EspP(586TEV)] interacted with BamA specifically during the pause in the transport reaction. First, only the pro form of EspP(586TEV) was crosslinked to BamA; no crosslinking to the cleaved passenger domain, which is produced after the completion of translocation, was observed. Second, the EspP–BamA interaction was seen almost exclusively at early time points and coincided temporally with the delay in translocation. Third, only residues close to the pause site were crosslinked to BamA. The crosslinking pattern shows that a specific segment of EspP was bound to BamA. In this regard, it is interesting that no crosslinking between BamA and WT EspP was observed. The data suggest that under normal physiological conditions the translocation reaction is very fast and processive, and that the BamA–EspP passenger domain complex is short-lived. Taken together, our results strongly suggest that the Bam complex plays a direct role in passenger domain translocation.
In addition to revealing an interaction between the EspP(586TEV) passenger domain and BamA, our crosslinking studies also show that the β domain interacts with BamA and BamB during the stalling of passenger domain translocation. Although previous studies have shown that the depletion of Bam complex components leads to defects in the membrane integration of OM proteins (14–18) and that BamA recognizes unfolded OM proteins in vitro (16, 23), our results provide direct evidence of an interaction between the Bam complex and an OM protein in vivo. Interestingly, the crosslinking of the final residue of EspP to BamA is consistent with the finding that BamA recognizes a conserved motif at the C terminus of β barrel proteins (23). The crosslinking of the EspP β domain to the Bam complex upon the stalling of passenger domain translocation suggests that the transport reaction is closely coordinated in space and time with the insertion of the β barrel into the OM. Nevertheless, whether the β domain is fully integrated into the OM before the completion of passenger domain secretion is unclear. The observation that passenger domain cleavage does not occur during stalling suggests that even if the β domain is fully integrated, a conformational change that is required for processing is inhibited until translocation is complete.
Besides supporting the hypothesis that passenger domain translocation is facilitated by the Bam complex, our characterization of a translocation intermediate provides additional insight into the secretion mechanism. The observation that the N terminus of the passenger domain was protected from PK digestion and could be crosslinked to periplasmic proteins (SurA and Skp) during the stalling of the transport reaction is consistent with other evidence indicating that the passenger domain is secreted in a C- to N-terminal direction (10). The finding that during stalling most of the passenger domain C-terminal to the stall point was degraded by PK (Fig. S3) also shows that translocation is initiated at the C terminus. Furthermore, the observation that proteolysis of the stalled form of EspP(586TEV) prevents completion of translocation (Fig. S5) indicates that the passenger domain segment that is exposed on the cell surface plays a key role in the transport reaction. This result suggests that translocation requires the presence of a loop and/or a folded structure that progressively traps the passenger domain in the extracellular space.
An intriguing question that emerges from this study concerns the mechanism by which the insertion of a small peptide in the EspP passenger domain transiently stalls translocation. Although putative protein translocation intermediates have previously been constructed by the engineering of tightly folded globular domains into substrates, the isolation of a translocation intermediate through the insertion of a linker has not been reported. In all probability, the insertion affects the local folding of the passenger domain, thereby impeding translocation. Indeed, the observation that the mutation prevents accumulation of the ≈80-kDa PK-resistant fragment associated with WT EspP (Fig. 1B) indicates that the mutation affects protein folding at least at one level. Furthermore, if passenger domains fold vectorially in a C- to N-terminal fashion, as has been proposed (2), then it is easy to imagine how the mutation might produce a local effect. It is unclear, however, whether the insertion prevents the acquisition of a conformation that is required for translocation or, conversely, promotes the formation of a meta-stable translocation-incompetent conformation. Even if the passenger domain must be in a specific conformation to traverse the OM, the observation of an ≈80-kDa PK-resistant fragment only after translocation is complete suggests that the passenger domain is not fully folded as it emerges into the extracellular milieu.
BamA is homologous to bacterial two partner secretion (TPS) transporters, which have been proposed to function as protein-conducting channels for another family of large β-helical proteins, and it is tempting to speculate that BamA promotes autotransporter passenger domain secretion by a similar mechanism. The crystal structure of FhaC, a Bordetella pertussis TPS transporter, reveals a plugged 16-stranded β barrel monomer (24). Even if the plug were removed, however, it is unclear if this type of channel would be wide enough to accommodate the folded segments secreted by the autotransporter pathway. Moreover, if the passenger domain were transported through an aqueous channel formed by a BamA monomer while the hydrophobic exterior of the β domain was integrated into the OM by another mechanism, then it is unclear how the passenger domain would be released from the channel. In an alternative model, a novel type of translocation channel might be assembled from BamA oligomers or multiple Bam complex subunits. Furthermore, the Bam complex might catalyze the folding of the β helix in the periplasm, thereby creating a structure that drives the translocation reaction. The periplasmically disposed POTRA domains of BamA have been proposed to promote β barrel assembly by β augmentation (25), and they might assemble β helices in a stepwise fashion by a similar mechanism. Thus, the Bam complex might function as a chaperone that stimulates assembly reactions which, in turn, promote the integration of autotransporter β domains into the OM and the transport of passenger domains across the OM. In this scenario, SurA and Skp might maintain N-terminal segments of the passenger domain in a loosely folded conformation until they interact with the Bam complex.
Materials and Methods
Bacterial Strains, Growth Conditions, and Reagents.
The strains used in this study were AD202 (MC4100 ompT::kan) (26), HDB130 (AD202 surA::cat), and HDB131 (AD202 Δskp). All cultures were grown at 37 °C in M9 medium containing 0.2% glycerol and all of the L-amino acids except methionine and cysteine (40 μg/mL). Ampicillin (100 μg/mL) and tetracycline (5 μg/mL) were added as needed. Antisera generated against EspP N- and C-terminal peptides have been described previously (27), and anti-SurA, anti-Skp, and anti-BamB antisera were provided by Rajeev Misra, Natacha Ruiz, and Dan Kahne, respectively. A rabbit polyclonal antiserum was generated against His-tagged BamA purified as described in SI Text.
Plasmid Construction.
Plasmid pRLS5 has been described previously (27). A PstI site was introduced 1,758 bp downstream of the espP start codon using oligonucleotide Pst586(+) and its complement and the Stratagene QuikChange mutagenesis kit; all oligonucleotides are listed in Table S1. PstTev(+) and PstTev(−), which encode a TEV cleavage site, were subsequently cloned into the PstI site to generate plasmid pJH97. Oligonucleotides that encode a 10-histidine tag (10H-link and GC-10H-link) were cloned into the EagI site of pRLS5 and pJH97 to generate pRI20 and pRI21, respectively. The modified espP genes were then excised from pRI20 and pRI21 with EcoR I and placed behind the lac promoter in plasmid RB11 (28) to generate pRI22 and pRI23. Amber mutations were introduced into these two plasmids by site-directed mutagenesis using the AMB oligonucleotides and their complements.
Pulse-Chase Labeling and Photocrosslinking.
Overnight cultures were washed and diluted into fresh M9 at an OD550 of 0.02. When the cultures reached an OD550 of 0.2, EspP synthesis was induced by the addition of 100 μM or 200 μM IPTG, as noted. After 30 min, cells were pulse labeled for 30 s with 30 μCi/mL Tran35S-label (MP Biomedical). Cold methionine and cysteine (1 mM) were then added. In experiments that did not involve photocrosslinking, cells were pipetted over ice at various time points, pelleted (3,000 × g, 6 min, 4 °C), and resuspended in M9 salts. The supernatants were centrifuged again (3,000 × g, 6 min, 4 °C) to obtain cell-free culture medium fractions. Half of the resuspended cells (or culture medium) were treated with 200 μg/mL of PK on ice, and PK digestion was stopped by the addition of 2 mM PMSF. Proteins in all samples were collected by TCA precipitation. In photocrosslinking experiments, 1 mM Bpa was added along with the inducer, and cells were radiolabeled as described above. At each time point, 8 mL aliquots were pipetted into a 15 mL tube and chilled on ice (untreated sample) or placed in a 6-well plate on ice and UV-irradiated (365 nm; Spectroline UV-Lamp SB-100P) for 7 min at a distance of 3–4 cm. Cells were concentrated by centrifugation (3,500 × g, 10 min, 4 °C) and resuspended in M9 before PK treatment or TCA precipitation. All immunoprecipitations were performed as described previously (12), and proteins were resolved by SDS/PAGE on 8%–16% minigels (Invitrogen). Radioactive proteins were detected using a Fuji BAS-2500 phosphorimager. Cell surface exposure was defined as 1-{proEspP(+PK)/[proEspP(−PK) + passenger(T)]}, where passenger(T) represents the sum of the passenger domain on the cell surface and in the culture medium. Passenger domain cleavage was defined as 1-{passenger(T)/[proEspP(-PK) + passenger(T)]}. In all calculations, the signal from each band was normalized to account for differences in the number of radioactive amino acids.
Supplementary Material
Acknowledgments.
We thank Natacha Ruiz and Tom Silhavy for providing the BamA antisera used in preliminary experiments and Janine Peterson for providing technical assistance. We also thank Nathalie Dautin and Susan Buchanan for their critical reading of the manuscript. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907912106/DCSupplemental.
References
- 1.Henderson IR, et al. Type V secretion pathway: The autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Junker M, et al. Pertactin β-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc Natl Acad Sci USA. 2006;103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oomen CJ, et al. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard TJ, et al. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14:1214–1220. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohlner J, et al. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 6.Khalid S, Sansom MS. Molecular dynamics simulations of a bacterial autotransporter: NalP from Neisseria meningitidis. Mol Memb Biol. 2006;23:499–508. doi: 10.1080/09687860600849531. [DOI] [PubMed] [Google Scholar]
- 7.Brandon LD, Goldberg MB. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J Bacteriol. 2001;183:951–958. doi: 10.1128/JB.183.3.951-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skillman KM, et al. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol Microbiol. 2005;58:945–958. doi: 10.1111/j.1365-2958.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- 9.Jong WS, et al. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol Microbiol. 2007;63:1524–1536. doi: 10.1111/j.1365-2958.2007.05605.x. [DOI] [PubMed] [Google Scholar]
- 10.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71:1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 11.Dautin N, et al. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26:1942–1952. doi: 10.1038/sj.emboj.7601638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the β-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol. 2008;67:188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein HD. Are bacterial “autotransporters” really transporters? Trends Microbiol. 2007;15:441–447. doi: 10.1016/j.tim.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, et al. Identification of a multicomponent complex required for outer membrane protein biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembled outer membrane protein in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voulhoux R, et al. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz N, et al. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Jain S, Goldberg MB. Requirement for YaeT in the outer membrane assembly of autotransporting proteins. J Bacteriol. 2007;189:5393–5398. doi: 10.1128/JB.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentle I, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 21.Farrell IS, et al. Photo-crosslinking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 22.Sklar JG, et al. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert V, et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clantin B, et al. Structure of the membrane protein FhaC: A member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the OmpT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 27.Szabady RL, et al. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newitt JA, Bernstein HD. A mutation in the E. coli secY gene that produces distinct effects on inner membrane protein insertion and protein export. J Biol Chem. 1998;273:12451–12456. doi: 10.1074/jbc.273.20.12451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.