Abstract
Defects in nuclear morphology often correlate with the onset of disease, including cancer, progeria, cardiomyopathy, and muscular dystrophy. However, the mechanism by which a cell controls its nuclear shape is unknown. Here, we use adhesive micropatterned surfaces to control the overall shape of fibroblasts and find that the shape of the nucleus is tightly regulated by the underlying cell adhesion geometry. We found that this regulation occurs through a dome-like actin cap that covers the top of the nucleus. This cap is composed of contractile actin filament bundles containing phosphorylated myosin, which form a highly organized, dynamic, and oriented structure in a wide variety of cells. The perinuclear actin cap is specifically disorganized or eliminated by inhibition of actomyosin contractility and rupture of the LINC complexes, which connect the nucleus to the actin cap. The organization of this actin cap and its nuclear shape-determining function are disrupted in cells from mouse models of accelerated aging (progeria) and muscular dystrophy with distorted nuclei caused by alterations of A-type lamins. These results highlight the interplay between cell shape, nuclear shape, and cell adhesion mediated by the perinuclear actin cap.
Keywords: LINC complexes, nucleus
In 1921, Champy and Carleton suggested an apparent correlation between the shape of various types of animal cells and the shape of their respective nuclei (1). Moreover, defects in nuclear shape are routinely used in the lab and in clinical settings as markers of disease and differentiation in human cells and tissues (2). However, remarkably little is known about the factors that determine nuclear morphology in living cells. In particular, the molecular mechanisms that govern the shape of the interphase nucleus are unknown. Here we show that an actin filament structure that forms a cap or dome located above the apical surface of the nucleus tightly controls nuclear shape and identify key associated cytoskeletal regulators of its organization and nuclear shape-determining function. The organization and function of the perinuclear actin cap are deregulated in diseased cells with distorted nuclei.
Results and Discussion
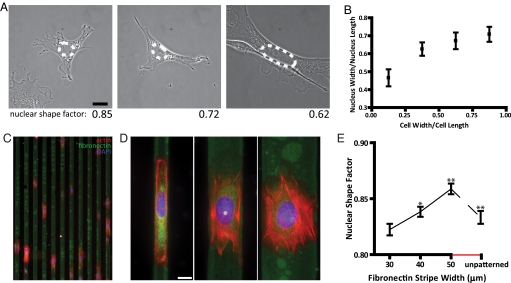
To test the hypothesis of a correlation between the shape of the nucleus and the overall cell shape, mouse embryonic fibroblasts were dispersed on fibronectin (FN)-coated glass substrates. Using morphometric analysis, we found that nuclear shape and cellular shape correlated (Fig. 1 A and B). Shape factor, defined as 4πA/P2 (where A and P are the nuclear area and perimeter), approaches 1 for a rounded nucleus and approaches 0 for an elongated nucleus. Elongated cells typically showed an elongated nucleus of low shape factor; rounded cells showed a rounded nucleus of high shape factor (Fig. 1A). To control cell shape and, therefore, be able to quantify nuclear shape as a function of cell shape, we developed adhesive FN-coated micropatterned stripes of width ranging between 10 and 50 μm, which alternated with stripes covered with nonadhesive polyethylene glycol (PEG) (Fig. 1C) (3–6). Mouse embryonic fibroblasts were allowed to spread, and within 30 min, the majority of the cells had moved to, and aligned in, the direction of the FN stripes (Fig. 1C). Cells were restricted to the width of the underlying FN micropatterns (Fig. 1 C and D). Cells stretched on narrow (e.g., 10 μm) FN stripes had nuclei that were highly elongated along the cell's long axis (Fig. 1D, left) with a low shape factor (Fig. 1E), while cells spread over wider stripes (e.g., 30 μm) displayed nuclei that were significantly more rounded (Fig. 1D, middle). Cells that were “overspread” on 50-μm wide FN stripes, which are wider than the natural size of the cell, had nuclei with shape factors closer to unity than did cells plated on unpatterned, uniformly FN-coated surfaces (Fig. 1 D and E, right). This regulation of nuclear shape by cellular shape was preserved over a wide variety of cells, including Swiss 3T3 mouse fibroblasts.
Fig. 1.
Cell shape controls nuclear shape through underlying adhesion geometry. (A) MEFs on FN-coated glass and their respective nuclear shape factor. Nuclei in elongated cells are typically elongated; nuclei in round cells are typically more rounded. (B) Nucleus shape as a function of cell shape measured by the ratio of nuclear width over nuclear height to cell width over cell length. Cells (180) were examined. (C) MEFs on 10-μm wide FN micropatterns (green). (Scale bar, 20 μm.) (D) Typical organization of actin filaments (red) and shape of DAPI-stained nuclei (blue) of cells plated on 10-μm (Left), 30-μm (Center), and 50-μm wide (Right) FN micropatterns. Nuclei in cells stretched on thin patterns are elongated; nuclei in cells on micropatterns that are wider than their natural size are more rounded than in unpatterned cells. (Scale bar, 10 μm.) (E) Average shape factor of nuclei in MEFs on FN patterns of controlled width and in unpatterned cells (last value). The nuclear shape factor, which is σ = 4πA/P2 where A is the apparent surface area of the nucleus and P its perimeter, is 1 for a perfectly round nucleus and approaches 0 for an elongated nucleus. At least 100 cells were examined for each micropattern width. Significance in the difference in nuclear shape factor between two subsequent micropattern widths is denoted as following: ***, P < 0.001; **, P < 0.01; *, P < 0.05; no star, P > 0.05.
Confocal microscopy showed that DAPI-stained nuclei of cells elongated on micropatterns or spread on uniform surfaces both featured a thin disk-like morphology, with a mean thickness ranging between 2.8 μm for cells on unpatterned surfaces to 3.5 μm for cells on 10-μm stripes; values were confirmed in live cells (not shown).
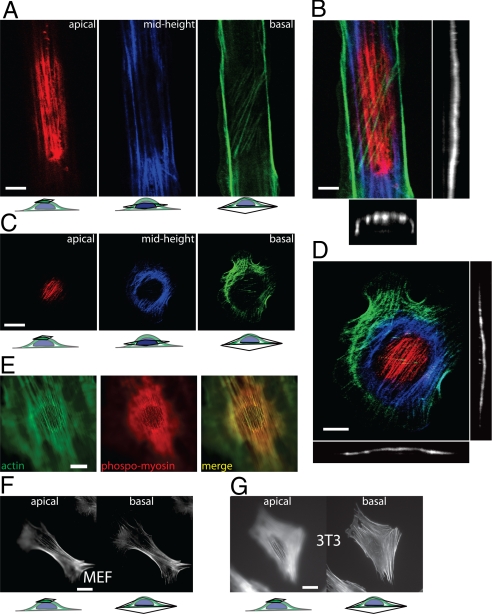
Three-dimensional (3-D) confocal sectional images of actin staining in patterned mouse embryonic fibroblasts (MEFs) revealed thick actin filament bundles organized into a curved shell or cap above the nucleus (Fig. 2A). The 3-D structure of this perinuclear actin cap resembled a dome (Fig. 2 A and B). This cap of actin filaments covers the top of the nucleus with a curvature equal to that of the nucleus' apical outer surface, as readily seen in z-confocal sections through the location of the nucleus (Fig. 2B, right and lower) and in animated 3-D reconstructions of the actin cap (Movies S1 and S2) in patterned cells. Actin filament bundles in the cap above the nucleus were aligned with the overall cell orientation induced by the patterns (Fig. 2A, red bundles in left). Underneath the nucleus, cells displayed either no organized actin or fewer and thinner actin filament bundles (Fig. 2A, diagonal green bundles, right).
Fig. 2.
The perinuclear actin cap. (A) Confocal microscopy sections of the actin filament network at the apical surface (red), mid-height (blue), and basal surface (green) of a MEF on a 10-μm wide FN pattern; see also Movie S1. (B) Full confocal reconstruction (Left) color-coded according to cell height, as well as width cross-section (Bottom) and length cross-section (Right) through the nucleus of the 3-D actin filament organization displayed showing the actin cap in the cell of A; see also Movie S2. (C and D) Confocal sections, reconstruction, side view, and profile view of the perinuclear actin cap at the apical surface, mid-height, and basal surface of a cell on a uniformly FN-coated (unpatterned) surface; see also Movies S3–S6. (E) Colocalization of filamentous actin (Left) and phosphorylated myosin II (Center) in the perinuclear actin cap. (F and G) Apical (Left) and basal (Right) organization of the actin filament network in MEFs and Swiss 3T3 fibroblasts, which both typically display a prominent actin cap.
Unpatterned cells plated either on surfaces uniformly coated with FN or bare glass also displayed an actin cap (Fig. 2 C and D, Fig. S1, and Movies S3–S6) and few thin basal actin bundles underneath the nucleus (Fig. 2 C and D, green bundles in right), which were often not oriented in the same direction as the actin bundles in the actin cap. More than 60% of WT MEFs displayed an actin cap (Fig. 3B). That <10% of WT MEFs showed no actin cap and approximately 30% showed a disrupted cap may be due to the fact the cells were at different points in their cell cycle when imaged. Indeed, postmitotic cells do not display an actin cap for several hours. The presence of a perinuclear actin cap was confirmed in live cells (Movie S7 and Fig. S2). Movies of cells transfected with GFP-lifeact (7) focusing on top of the nucleus show that the filamentous actin cap is a dynamic structure that undergoes large extensions and retractions (Movie S7). The actin cap in live cells orients preferably along the direction of the main axis of the cell (Fig. S2). The actin filament bundles in the actin cap contained serine 19 (S19) phosphorylated myosin II (Fig. 2E) and were terminated by vinculin-containing focal adhesions positioned both at the periphery of the nucleus and at the edge of the cell at the cell's basal surface (Fig. S3). Finally, a similar perinuclear actin cap was observed in Swiss 3T3 fibroblasts (Fig. 2 F and G and Fig. S4) and C2C12 mouse myoblasts, as well as human endothelial cells (HUVECs) and human ovarian epithelial cells, for which the physiological environment is essentially 2-D.
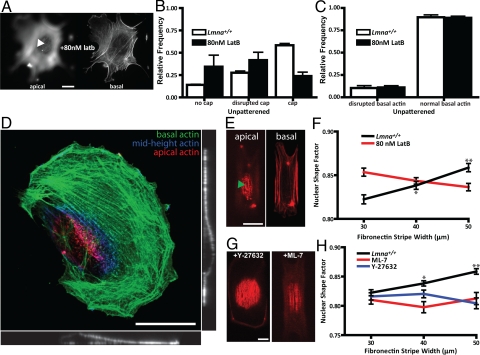
Fig. 3.
Regulation of nuclear shape and the perinuclear actin cap. (A) Representative apical and basal F-actin organization in latrunculin B-treated unpatterned cells. Arrowhead indicates the absence of the actin cap. (Scale bar, 10 μm.) (B) Fraction of control (white bars) and latrunculin B-treated cells (back bars) showing either an organized, a disorganized, or no actin cap. (C) Fraction of control (white bars) and latrunculin B-treated cells (back bars) showing disorganized and organized basal stress fiber architecture. (D) Typical confocal reconstruction, side view, and profile view of the F-actin organization at the apical surface (red), mid-height (blue), and basal surface (green) of a cell treated with a low concentration (80 nM) of latrunculin B showing no organized actin cap, while basal actin is organized. (E) Representative apical and basal F-actin organization in latrunculin B-treated cells on 10-μm wide micropatterns. (F) Averaged shape factor of nuclei in control and latrunculin B-treated MEFs plated on FN micropatterns of controlled width. There was no significant change in nuclear shape factor in latrunculin B-treated cells. (G) Representative top confocal cross-sections showing the organization of the actin cap in ML-7-treated and Y-27632-treated cells on 30-μm micropatterns. (H) Averaged shape factor of nuclei in control patterned MEFs and cells treated with either MLCK inhibitor ML-7 or ROCK inhibitor Y-27632. At least 100 cells were examined in E and G for each micropattern width. Statistical significance in E and G is denoted with stars as in Fig. 1.
To determine whether the actin cap is involved in nuclear shape control and to identify regulators of the actin cap architecture and function, we perturbed the expression and assembly of key cytoskeleton proteins, components of the lamina and the nuclear envelope (NE), and examined the actin cap organization and nuclear shape response to cell shape changes.
First, cells were treated with a low concentration (80 nM) of the F-actin depolymerizing drug latrunculin B (Fig. 3 A–E), which disrupted a significant fraction of the actin caps (Fig. 3B) while leaving the basal stress fibers largely intact (Fig. 3 A and C–E and Movie S8). As a result, the nucleus bulged to an increased thickness, from 2.8–5.8 μm for cells on 10-μm stripes (not shown). We verified that the proportion of cells displaying an actin cap and treated with DMSO was similar to untreated cells (Fig. S5). Latrunculin B treatment led to an almost complete abrogation of the regulation of nuclear shape by cell shape (Fig. 3F). Together, these results suggest that the integrity of the perinuclear actin cap depends on F-actin assembly and that the actin cap plays a critical role in nuclear shape regulation by cell shape.
Since F-actin in the actin cap plays a role in the control of nuclear shape and phosphoryated myosin and actin co-localize in the actin cap (Fig. 2E), we asked whether it was the extent of actin assembly or myosin-mediated contractility that played the dominant role. The treatment of cells with drugs inhibiting actomyosin contractility [Rho kinase inhibitor Y-27632, myosin light chain kinase (MLCK) inhibitor ML-7] disorganized the actin cap and/or diminished the number of actin fibers in the cap (Fig. 3G and Movie S9) compared to control cells. Accordingly, these drugs significantly abrogated nuclear shape regulation (Fig. 3H). These defects were typically accompanied by significant bulging of the nucleus, but to a lesser extent than in cells treated with latrunculin B. Together these results show that the architecture of the actin cap and its control of nuclear shape depend critically on actin assembly and, to a lesser extent, on actomyosin contractility.
The existence of the perinuclear actin cap and its regulation by actin filament assembly and contractility suggest a mechanism by which the shape of the nucleus can be controlled by the underlying cell adhesion geometry. Contractile actin fibers in the perinuclear cap pull the nucleus toward the bottom of the cell, preventing the nucleus from bulging against the cell membrane, and squeeze the nucleus laterally through anchoring by the terminal focal adhesions, which are themselves confined laterally to the micropatterns. The confinement of the nucleus by apical actin fibers could explain the observed thin disk-like shape of the nucleus and the nucleus' inability to move in and out of the plane of focus during nuclear rotation (8).
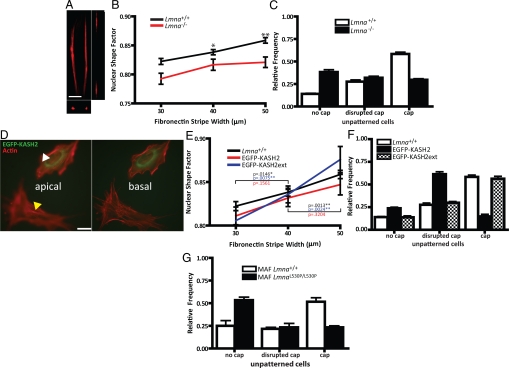
Recent results suggest that the actin cytoskeleton is intimately connected to the outer nuclear membrane through specific nucleocytoskeletal molecular linkers, which are located on the NE and form the linker of nucleoskeleton and cytoskeleton (LINC) complex (9–13). Traditional fluorescence microscopy focusing on the bottom of the cell suggests that the displacement of these NE proteins caused by lamin depletion does not affect actin filament organization (14). Because the NE distribution of LINC complex proteins is altered in embryonic fibroblasts derived from mice lacking A-type lamins, a model of dilated cardiomyopathy and muscular dystrophy (15, 16), we hypothesized that the depletion of lamin A/C from the nucleus would disrupt the organization of the actin cap and the regulation of nuclear shape by cell shape. While the organization of actin fibers away from the nucleus in Lmna−/− cells (imaged by focusing on the bottom of the cell) remained largely unchanged indeed (17, 18), the actin cap disappeared in a majority of Lmna−/− cells (Fig. 4 A and C, Fig. S6, and Movie S10), an effect that was enhanced by patterning Lmna−/− and Lmna+/+ cells (Fig. S7). The thickness of the nucleus increased from 2.8 μm for unpatterned Lmna+/+ cells to 6 μm for unpatterned Lmna−/− cells. Moreover, nuclear morphometric analysis showed that lamin A/C deficiency greatly reduced the control of nuclear shape by cell shape (Fig. 4B). Therefore, the loss of nuclear lamin A/C causes a loss of the actin cap, which attenuates the ability of the nucleus of Lmna−/− cells to respond to overall changes in their cell shape.
Fig. 4.
Role of the nuclear lamina and LINC complexes on the presence, organization, and nuclear shaping function of the perinuclear actin cap (A and B). (A) Representative top confocal cross-section showing the organization of the actin cap in a Lmna−/− MEF plated on a 10-μm wide FN micropattern. (B) Averaged nuclear shape factor of MEFs lacking lamin A/C (Lmna−/−) and WT cells as a function of FN micropattern width. Statistical significance is denoted with stars as in Fig. 1. (C) Proportion of WT MEFs and Lmna−/− MEFs showing an organized, disorganized, or no actin cap on unpatterned FN-coated surfaces. (D) Representative basal and apical organization of the actin network in an MEF transfected with EGFP-KASH2 and placed on an unpatterned FN-coated surface. The yellow and white arrowheads indicate the absence of an actin cap in an EGFP-KASH-transfected cell and presence of on actin cap in a control untransfected cell. (Scale bar, 10 μm.) (E) Averaged nuclear shape factor of WT MEFS (black) and MEFs transfected with either EGFP-KASH2 (red) or EGFP-KASH2ext (blue) as a function of micropattern width. (F) Proportion of WT MEFS and MEFs transfected with either EGFP-KASH2 or EGFP-KASH2ext showing an organized, disorganized, or no actin cap on unpatterned FN-coated surfaces. (G) Proportion of WT mouse adult fibroblasts (MAFs) and LmnaL530P/L530P MAFs on unpatterned FN surfaces showing either an organized, a disorganized, or no actin cap on unpatterned FN-coated surfaces.
Since the absence of lamin A/C can have secondary effects on gene expression, we characterized the effects of wild-type (WT) cells expressing EGFP-KASH2 on the organization of the actin cap and on nuclear shape regulation. This construct has recently been shown to displace actin-binding protein Nesprin-2 giant from the NE (14), which may connect the actin cytoskeleton to the nucleus (9, 19–21). We found that the fraction of EGFP-KASH2-transfected cells displaying an organized actin cap was dramatically decreased compared to WT cells (Fig. 4 D and F). Moreover, cells transfected with the control EGFP-KASH2ext, which localizes to the NE but does not displace the LINC complex, did not affect actin cap organization (Fig. 4 D and F). The nuclear shape response of EGFP-KASH2-transfected cells was significantly diminished compared to control and WT cells (Fig. 4E). We also found that lamin A/C distribution is unaffected in cells expressing EGFP-KASH2 (Fig. S8). Hence the disruption of the LINC complexes recapitulates the lamin KO phenotype, including a disrupted actin cap and a reduced ability of the cells to regulate nuclear shape. Together these results indicate that the LINC complexes regulate the organization of the actin cap and its nuclear shaping function.
Similarly, we investigated the organization of the actin filament network in primary fibroblasts harvested from an LmnaL530P/L530P mouse, a model of accelerated aging (progeria) (17, 22, 23). We have recently shown that LINC complex proteins are displaced from the nucleus to the ER and cytoplasm in a majority of LmnaL530P/L530P cells (17). Similarly to Lmna−/− cells, LmnaL530P/L530P cells show no overt disorganization of their basal stress fibers compared to control WT cells (17). However, the actin cap was either disorganized or completely eliminated (Fig. 4G).
Together these results suggest that basal stress fibers and actin filament bundles in the perinuclear actin cap are regulated by distinct pathways; unlike basal stress fibers, the actin cap is physically connected to the nuclear lamina. Actin cap-nucleus connections are altered in cells from mouse models of muscular dystrophy and progeria (17, 24, 25), which disorganizes the actin cap and may explain the deformed shape of the nucleus in these diseased cells.
The organization of lamin A/C and the LINC complexes remained intact in latrunculin-B-treated cells, even with doses up to 500 nM (Fig. S9). Moreover, latrunculin B treatment at low dose alone does not affect the organization of basal stress fibers. Therefore, the elimination of the actin cap seems to be sufficient to eliminate nuclear shape response of WT cells to forced changes in overall cellular shape (Fig. 3 C and D) in a manner similar to that observed in Lmna−/− and LmnaL530P/L530P cells. Therefore, together with the results obtained with EGFP-KASH2, the diminished response of nuclear shape to changes in the shape of Lmna−/− and LmnaL530P/L530P cells is primarily caused by the disorganization of the actin cap itself, which is induced by the displacement of the LINC complexes in these cells.
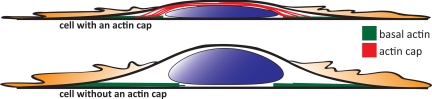
In conclusion, our results suggest that the shape of the nucleus is controlled by cell shape through a previously unreported actin filament structure, the perinuclear actin cap, which is located above and around the interphase nucleus. The actin cap consists of thick, dynamic, actomyosin filament bundles, which form a highly organized dome that covers the top of the nucleus, is connected to the NE through the LINC complex, and is mostly aligned with the overall cell orientation (Fig. 5). Actin filament bundles in the actin cap are structurally and functionally distinct from dorsal stress fibers located at the top of the lamella (26) and the contractile actin ring located at the division plane during mitosis (27). The perinuclear organization of the actin cap depends on the integrity of the nuclear lamina and LINC complexes, as well as actin filament assembly and contractility.
Fig. 5.
Schematic of the actin filament architecture at the basal and apical surface of an adherent cell. (Top) Apical actin fibers (red) wrap around the apical outer surface of the nucleus, forming the perinuclear actin cap; (bottom) either the depletion of nuclear lamin A/C, the disruption of LINC complexes, actin filament disassembly, or the inhibition of actomyosin contractility disrupts the organization of the actin cap and its nuclear shaping function.
Experimental Procedures
Cell Culture, Drug Treatments, and Transient Transfection.
Unless stated, reagents were purchased from Sigma. WT and KO MEFs were cultured in DMEM [American Type Culture Collection (ATCC)] supplemented with 10% FBS (ATCC) and 100 U penicillin/100 μg of streptomycin and maintained at 37 °C in a humidified, 5% CO2 environment. ROCK inhibitor Y-27632 (concentration, 30 μM), myosin II inhibitor blebbistatin (2.5 or 25 μM), MLCK inhibitor ML7 (25 μM), and actin filament depolymerizing drug latrunculin B (80 nM) were diluted using the stock medium. Cells were incubated with latrunculin B for 30 min before measurements and 45 min for the other drugs. Transient transfection was carried out as described (14) using Fugene HD transfection reagent (Roche) according to the manufacturer's specifications.
FN Micropatterning.
Custom masks were used to create master micropatterns. Briefly, silicon wafers were cleaned by Piranha clean (3:1 H2SO4:H2O2) and then baked at 200 °C for 15 min. Omni-Coat (MicroChem) was spun onto the treated glass according to manufacturer specifications. SU-8 2010 photoresist was then spun onto the silicon at 3,000 rpm for a thickness of 10 μm. The wafers were exposed to UV light through the mask and developed. Finally, the wafers were hard-baked overnight. PDMS (Corning) was poured over the master patterns and baked for at least 6 h at 60 °C. Stamps were then cut out and washed with ethanol before each use. Custom 35-mm glass slides (MatTek) were functionalized in 5% (in ethanol) mercaptopropyl-trimethoxysilane (Sigma) in the dark for 20 min. The slides were washed in ethanol and dried completely with nitrogen. The PDMS stamps were coated with 200 μL 50 μg/mL FN. After 10 min, the stamps were completely dried with nitrogen and then put on the treated glass slides. After 1 min, the stamps were removed, and the slides were treated in 1 mg/mL polyethylene glycol (PEG) for 2 h in the dark. The PEG solution was aspirated, and cells were seeded on the micropatterns.
Morphometric Measurements and Immunofluorescence Microscopy.
Actin filament and focal adhesion architecture were examined by immunofluorescence confocal microscopy. Cell were allowed to spread on patterned or unpatterned FN-coated surfaces overnight, were fixed with 2% paraformaldehyde for 1 h, and stained for nuclear DNA, focal adhesion protein vinculin, extracellular matrix molecule FN, nuclear protein lamin A/C, phosphorylated myosin, and cytoskeletal filamentous actin. For staining, cells were permeabilized with 0.1% Triton X-100 for 10 min. BCS, 10%, in PBS was used to block nonspecific binding for 20 min. The antibodies used were: anti-FN antibody (Chemicon) at 1:100, anti-vinculin antibody (Sigma) at 1:40, anti-phosphomyosin light chain 2 (Ser 19) antibody (Cell Signaling Technology) at 1:50, and anti-lamin A/C antibody (MAb3538; Chemicon). To visualize actin and nuclear DNA, Alexa-Fluor phalloidin 488 and 300 nM DAPI (Invitrogen) were used, respectively. In live cells, F-actin organization was visualized using GFP-lifeact, as described (7, 28).
Fluorescent images were either collected using a Cascade 1K CCD camera (Roper Scientific) mounted on a Nikon TE2000E microscope with a 60× Plan Fluor lens (N.A. 1.4) or using a Zeiss 510 laser-scanning confocal microscope. 3-D images were analyzed and processed using a combination of LSM Image Browser (Zeiss), Metaporph, and ImageJ (National Institutes of Health). Special attention was paid to use small increments between focal sections (<0.15 μm) and to scan the same cell starting at slightly different heights, not to miss actin structures underneath the nucleus.
DAPI-stained nuclei were individually traced by hand, and size, length of minor axis, length of major axis, and shape factor were measured using Metamorph software (Universal Imaging). For cells plated on unpatterned surfaces, the ratios of width to length of cells (see Fig. 1B) were binned between 0.00–0.25, 0.26–0.50, 0.51–0.75, and 0.76–1.00, respectively.
The number of cells examined for each FN strip width, drug treatment, and type of cell is shown in the figures. We verified that addition of DMSO (1:500 volume dilution) alone did not affect the proportion of cells showing an actin cap and the organization of the actin cap (Fig. S5). Mean values, standard error of measurement (SEM), and statistical analysis of shape factors were calculated and plotted using Graphpad Prism (Graphpad Software). Two-tailed unpaired t-tests were conducted to determine significance caused by protein depletions or drug treatments.
Acknowledgments.
This work was partly supported by National Institutes of Health grant nos. R01GM084204 and U54CA143868, and the Muscular Dystrophy Association. We thank Roland Wedlich-Soldner for his generous gift of GFP-lifeact. We also thank Dr. Yiider Tseng and members of the Wirtz and Searson labs for technical advice and reagents.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908686106/DCSupplemental.
References
- 1.Champy C, Carleton HM. Memoirs: Observations on the shape of the nucleus and its determination. J Cell Sci. 1921;s2–65:589–625. [Google Scholar]
- 2.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci USA. 2001;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 5.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 6.Roca-Cusachs P, et al. Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys J. 2008;94:4984–4995. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riedl J, et al. Lifeact: A versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JS, Chang MI, Tseng Y, Wirtz D. Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol Biol Cell. 2005;16:871–880. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crisp M, et al. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart CL, Roux KJ, Burke B. Blurring the boundary: The nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 11.Worman HJ, Gundersen GG. Here come the SUNs: A nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Libotte T, et al. Lamin A/C dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell. 2005;16:3411–3424. doi: 10.1091/mbc.E04-11-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelmsen K, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raharjo WH, Enarson P, Sullivan T, Stewart CL, Burke B. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J Cell Sci. 2001;114:4447–4457. doi: 10.1242/jcs.114.24.4447. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale CM, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JS, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmakumar VC, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- 21.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 22.Mounkes L, Kozlov S, Burke B, Stewart CL. The laminopathies: Nuclear structure meets disease. Curr Opin Genet Dev. 2003;13:223–230. doi: 10.1016/s0959-437x(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 23.Mounkes LC, Kozlov S, Hernandez L, Sullivan T, Stewart CL. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature. 2003;423:298–301. doi: 10.1038/nature01631. [DOI] [PubMed] [Google Scholar]
- 24.Lammerding J, et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lammerding J, et al. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardo M, Nurse P. Equatorial retention of the contractile actin ring by microtubules during cytokinesis. Science. 2003;300:1569–1574. doi: 10.1126/science.1084671. [DOI] [PubMed] [Google Scholar]
- 28.Wildt B, Wirtz D, Searson PC. Programmed subcellular release for studying the dynamics of cell detachment. Nat Methods. 2009;6:211–213. doi: 10.1038/nmeth.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]