Abstract
Midbrain dopamine neurons (mDA) are important regulators of diverse physiological functions, including movement, attention, and reward behaviors. Accordingly, aberrant function of dopamine neurons underlies a wide spectrum of disorders, such as Parkinson's disease (PD), dystonia, and schizophrenia. The distinct functions of the dopamine system are carried out by neuroanatomically discrete subgroups of dopamine neurons, which differ in gene expression, axonal projections, and susceptibility in PD. The developmental underpinnings of this heterogeneity are undefined. We have recently shown that in the embryonic CNS, mDA originate from the midbrain floor plate, a ventral midline structure that is operationally defined by the expression of the molecule Shh. Here, we develop these findings to reveal that in the embryonic midbrain, the spatiotemporally dynamic Shh domain defines multiple progenitor pools. We deduce 3 distinct progenitor pools, medial, intermediate, and lateral, which contribute to different mDA clusters. The earliest progenitors to express Shh, here referred to as the medial pool, contributes neurons to the rostral linear nucleus and mDA of the ventral tegmental area/interfascicular regions, but remarkably, little to the substantia nigra pars compacta. The intermediate Shh+ progenitors give rise to neurons of all dopaminergic nuclei, including the SNpc. The last and lateral pool of Shh+ progenitors generates a cohort that populates the red nucleus, Edinger Westphal nucleus, and supraoculomotor nucleus and cap. Subsequently, these lateral Shh+ progenitors produce mDA. This refined ontogenetic definition will expand understanding of dopamine neuron biology and selective susceptibility, and will impact stem cell-derived therapies and models for PD.
Keywords: dopamine, Sonic hedgehog, lineage, substantia nigra
The midbrain dopaminergic neurons (mDA) are involved in diverse physiological pathways, including motor control, cognition, and reward behaviors. These neurons are clustered in 3 anatomically defined areas: the substantia nigra pars compacta (SNpc), the ventral tegmental area (VTA), and the retrorubral fields (RRF) (1, 2). The SNpc cluster neurons project predominantly to the dorsolateral striatum and are involved in motor function. The VTA neurons project to the prefrontal cortex, amygdala, and nucleus accumbens, circuits involved in emotional and cognitive functions (1, 2). In addition to the anatomical and functional differences, transcriptome as well as marker analyses of neurons in mDA clusters have also revealed some molecular differences (2–5). Finally, neurons in the SNpc are selectively lost in Parkinson's disease (PD) patients (6, 7), as well as in various genetic mouse models (8–11), further highlighting the physiological diversity of mDA. However, little is known about whether mDA diversity is encoded at the progenitor cell level.
Recent lineage studies have revealed a surprising embryonic origin of mDA at the ventral midline of the midbrain (10, 12–15). Using a Shh::Cre driver, we have definitively shown that most mDA are derived from the floor plate (FP), operationally defined by the expression of Shh (10, 15). However, dynamic Shh expression spans a broad domain, and thus it remains to be determined whether this domain is parceled into microdomains, each giving rise to unique cell types. Here, we ask (i) whether different mDA clusters are generated from distinct progenitors and (ii) whether other types of neuron clusters are derived from distinct Shh-expressing progenitors. We extend our previous analyses and provide a comprehensive study describing Shh descendents in the midbrain. Further, using dynamic spatial changes in the Shh expression domain, in conjunction with a genetic inducible fate mapping (GIFM) technique (16), we generate an embryonic progenitor map of the ventral midbrain. By spatially and temporally mapping diverse progenitor domains in the embryonic midbrain, we uncover distinct progenitor pools with differential proclivity to generate mDA clusters.
Results
Shh Expressing Progenitors Give Rise to Multiple Neuron Types in the Midbrain.
Our initial fate-mapping experiments using a Shh::Cre driver revealed that, in addition to mDA, other neuron types also originated from Shh+ progenitors (15). To define the repertoire of neurons derived from the Shh domain, we examined postnatal Shh::Cre,R26R midbrain sections (Fig. 1). Labeled cells were observed in the mDA clusters RRF, SNpc, and VTA/interfascicular nucleus (IF), in the rostral linear nucleus (RLi), medial accessory nucleus (MA3), red nucleus (RN), Edinger Westphal nucleus (EW), and supraoculomotor nucleus and cap (Su3). Some nuclei, such as the nucleus of Darkschewitsch (Dk) and the interstitial nucleus of Cajal (InC), were located just outside the large swath of Xgal-stained cells. Remarkably, some nuclei located even within or more ventral to the general Xgal-stained territory were conspicuously unlabeled. For instance, only sparse Xgal+ cells were visible in the substantia nigra pars reticulata (SNpr) and oculomotor nucleus (3N). No Xgal+ cells are observed in the interpeduncular nucleus (IP; Fig. 1 A–C). Therefore, neurons in these nuclei must originate outside the Shh+ domain.
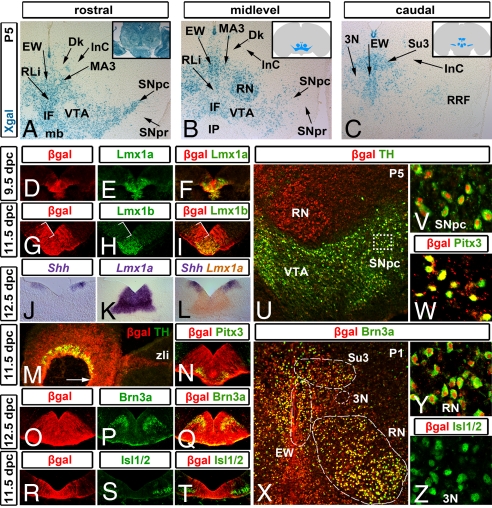
Fig. 1.
Shh+ progenitors give rise to several distinct midbrain neuron populations. (A–C) In Shh::Cre,R26R P5 coronal sections at rostral (A), mid (B), and caudal (C) midbrain levels, Xgal-labeled cells were detected in the retrorubral fields (RRF), the substantia nigra pars compacta (SNpc), the ventral tegmental area (VTA)/interfascicular nucleus (IF) regions, the rostral linear nucleus (RLi), the Edinger Westphal nucleus (EW), the supraoculomotor nucleus and cap (Su3), the red nucleus (RN), and the medial accessory oculomotor nucleus (MA3). Conversely, the oculomotor nucleus (3N), the substantia nigra pars reticulata (SNpr), the nucleus of Darkschewitsch (Dk), the interstitial nucleus of Cajal (InC), and the interpeduncular nucleus (IP) were predominantly devoid of Xgal-labeled cells. In control β-actin::Cre,R26R brains, all midbrain nuclei were labeled (A Inset). (D–I) Embryonic midbrain coronal sections labeled with antibodies that recognize β-gal (Shh fate-mapped cells; red) and Lmx1a (E and F) or Lmx1b (H and I) (green). Early Lmx1a+ progenitors largely colocalize with β-gal+ cells (D–F). During dopaminergic neurogenesis, all Lmx1a/Lmx1b+ progenitors are situated within the β-gal+ territory. Immediately lateral to the Lmx1a/Lmx1b+ progenitor pool, an Lmx1a/Lmx1b–, β-gal+ domain (brackets) predominantly produces other neuron types at these stages (G–I). (J–L) Complementary pattern of Shh and Lmx1a mRNA at 12.5 dpc. Double in situ hybridization for Shh (purple)/Lmx1a (orange) (L) depicts that the lateral Shh domain is largely Lmx1a–, with some sections showing 2- to 3-cell diameter overlap. (M) In 11.5-dpc sagittal sections, the first TH+ dopamine neurons are observed rostrally (arrow) roughly until the zona limitans intrathalamica (zli). In coronal 12.5-dpc sections, the β-gal domain is wider than but inclusive of the Pitx3+ neurons (N), whereas Brn3a+ neurons are primarily embedded within the lateral aspects of the β-gal domain (O–Q). Isl1/2+ oculomotor neurons (3N) reside largely outside of the β-gal domain (R–T) at 11.5 dpc. (U–Z) Postnatal coronal midbrain sections were labeled with β-gal (U–Z; red) and TH (U and V), Pitx3 (W), Brn3a (X and Y), or Isl1/2 (Z) (green). Note that most TH+ and Pitx3+ cells of the SNpc and VTA are β-gal+ (SNpc, 97.76% SEM ±0.36; VTA, 98.34% ±0.46; IF, 97.80% ±0.65; RRF, 96.12% ±1.16; RLi, 97.79% ±0.49 determined as percentage of the β-gal+/Pitx3+ cells). Similarly, the majority of the Brn3a+ cohort that populates the RN, EW, and Su3 is also β-gal+ (RN, 95.81% ±0.38; EW, 97.23% ±0.53; Su3, 90.95% ±2.35). Shown in (X) is a relatively rostral section, in which only the beginning of the Su3 is present. Only a few Isl1/2+ cells of 3N were β-gal+ (Z; 10.75% ±1.72). (V) High-power image of the area indicated by the white box in (U). (Y and Z) High-power images from the regions outlined in (X). Anatomical definitions were in accordance with the mouse brain atlas of Paxinos (see Fig. S8). mb, mammillary body.
The Shh Domain Encompasses Several Spatiotemporally Distinct Progenitor Domains.
To determine how different Shh+ progenitors were allocated to assume disparate mDA and other neuronal fates, we took advantage of the fact that the Shh expression domain in the midbrain is dynamic. Shh is expressed initially at the ventral midline at the 6- to 8-somite stage in the mesodiencephalic region. Shortly thereafter, expression appears to spread rostrally to the anterior brain, and caudally into the hindbrain and spinal cord regions (17). In Shh::Cre,R26R embryos 9.0 days postcoitus (dpc), Xgal labeling is observed at the midline of the midbrain (supporting information (SI) Fig. S1). By 9.5 dpc, and through 10.5 dpc, Shh expression and consequent Xgal labeling expands laterally; this lateral expansion is first observed in rostral midbrain sections. Between 10.5 and 11.5 dpc, Shh continues expanding laterally but is downregulated at the midline in the Lmx1a+ progenitors (15, 17, 18). Other FP markers, such as Netrin-1, F-spondin, and Foxa2, are also expressed in a similarly broad domain (Fig. S1). Thus, the cumulative Shh domain encompasses a swath of cells that expresses several traditional FP markers, and accounts for 25%–30% of the dorsoventral axis of the neural tube.
We reasoned that the cumulative Shh domain encompassed progenitors for multiple distinct neuron types. The transcription factors Lmx1a and Lmx1b are coexpressed at the ventral midline of the midbrain and are critical for the normal mDA phenotype (19, 20). In coronal sections of Shh::Cre,R26R 9.5-dpc embryos, β-gal+ cells are colocalized with Lmx1a and Lmx1b (Fig. 1 D–F) (15). As the Shh domain expands laterally, β-gal immunoreactivity is observed in more lateral domains. Thus, by 11.5 dpc, the β-gal domain is wider than, but inclusive of, the Lmx1a/Lmx1b domain (Fig. 1 G–I). At 11.5 and 12.5 dpc, all TH+ as well as Pitx3+ neurons appear to emanate directly from Lmx1a/Lmx1b+ ventricular zone; none are observed emanating from lateral β-gal+, Lmx1a– ventricular zone (along the rostrocaudal axis, TH+ cells extend from the isthmus to the zona limitans). Hence, until 12.5 dpc, the lateral β-gal+, Lmx1a/Lmx1b– domain corresponds largely to the lateral Shh expression domain (Fig. 1 J–N) (15), whose descendents are predominantly not TH+ neurons. Because the RN was prominent in our fate map (Fig. 1B), we tested whether these neurons may originate in the lateral aspects of the Shh domain, by double labeling with Brn3a, a marker for these neurons (21–23). Indeed, a β-gal+/Brn3a+ cohort was observed emanating from lateral β-gal+ progenitors (Fig. 1 O–Q). This cohort of β-gal+/Brn3a+ neurons populates not only the RN, but also EW and Su3 (Fig. 1 X and Y; elaborated further in Fig. 3).
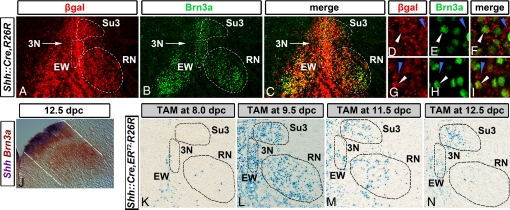
Fig. 3.
Lateral Shh+ progenitors give rise to Brn3a+ neurons. (A–I) Labeling of P1 Shh::Cre,R26R coronal sections with antibodies that recognize β-gal (red) and Brn3a (green) shows many colocalized neurons in the EW, Su3, and the RN (outlined by dotted white lines in A–C). (D–F) and (G–I) show high-power confocal images from the Su3 and EW nuclei, respectively. Though most cells in all 3 nuclei are β-gal+/Brn3a+ (white arrowheads), some are β-gal+/Brn3a– (blue arrowheads), and a few are β-gal–/Brn3a+. (J) Double in situ hybridization for Shh (purple)/Brn3a (orange) showing that Brn3a cohort appears to emanate from the lateral Shh domain, in accordance with previous models (23, 25). (K–N) Shh::CreERT2,R26R embryos were TAM injected at 8.0 (K), 9.5 (L), 11.5 (M), and 12.5 dpc (N) and harvested at 18.5 dpc. In 8.0-dpc injections, few to no Xgal-labeled cells are detected in the EW, Su3, and RN (outlined with dotted lines). In contrast, in 9.5- and 11.5-dpc injections, robust Xgal labeling is observed. In 12.5-dpc injections, very few cells were detected in the RN, and only caudal EW and Su3 regions contained labeled cells. 3N is largely unlabeled in all TAM-induced embryos.
The oculomotor nucleus and the GABAergic nuclei were largely unlabeled in our fate maps (Fig. 1 A–C). Indeed, Isl1/2+ 3N neurons, first emerging at 9.5 dpc, are observed immediately adjacent to the β-gal+ cohort, and few Isl1/2+ cells are colabeled with β-gal in 18.5-dpc sections (Fig. 1 R–T and Z). Additionally, a GATA3+/Lim1/2+ GABAergic cohort, first detected at 10.5 dpc, appears to at least in part originate from Nkx2.2+ progenitors. These progenitors and this postmitotic GABAergic cohort are observed outside the β-gal+ domain in Shh::Cre,R26R embryos (Fig. S2). Consequently, GABAergic nuclei are unlabeled in postnatal fate maps (Fig. 1 A–C).
Shh Domain Encompasses Distinct mDA Progenitor Pools with Different Proclivities.
Because the cumulative Shh domain is broad, we next sought to refine our fate maps by labeling distinct subsets of Shh+ progenitors. We therefore took advantage of the dynamic Shh expression domain; using an inducible Shh::CreERT2 strain (24), we were able to label spatiotemporally distinct subsets of Shh+ progenitors by administering tamoxifen (TAM) at different gestational times. Following TAM administration, for a period of up to 36 h (16), the cytoplasm-sequestered CreERT2 protein can translocate to the nucleus, and effectively recombine loxP sites in the R26R locus.
TAM administration at 7.5 dpc labeled the most medial progenitors at the ventral midline. In 7.5 (injection) → 10.5-dpc (harvest) Shh::CreERT2,R26R embryos, Xgal cells appeared to occupy only the most medial region of the Lmx1a domain (hereafter called the medial domain; Fig. 2A); adjacent Lmx1a+ progenitors seemed largely Xgal– (hereafter called the intermediate domain). This suggests that functional TAM activity in this context was extinguished before the expansion of the Shh domain observed at 9.5 dpc (Fig. S1). Further, these data also show that an expansion of the initial Shh domain is likely due to inductive mechanisms rather than the growth of the initial Shh+ pool.
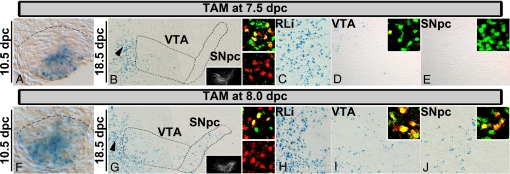
Fig. 2.
Medial and intermediate Shh domains give rise to distinct mDA neurons. (A–J) In Shh::CreERT2,R26R embryos, 7.5 (injection) → 10.5 dpc (harvest), Xgal+ cells are detected in the most medial region of the Lmx1a+ domain (delineated by the dotted line according to Lmx1a staining on an adjacent section) (A). In 7.5 (injection) → 18.5 dpc (harvest), Xgal-labeled cells are observed predominantly in the rostral linear nucleus (RLi; arrowhead in B; C). These cells (β-gal in red) are positive for Pitx3 (green; C Upper Inset) but largely negative for TH (green; C Lower Inset). Some labeling is detected in the VTA/IF regions (B and D). Labeled cells are β-gal+ (red)/Pitx3+ (green) (D Inset, a single β-gal+/Pitx3+ cell). Very few labeled cells are observed in the SNpc (B and E). (E Inset) No Pitx3 (green)+/β-gal (red)+ cells. (C–E) High-magnification images of the section in (B). (F–J) In 8.0 (injection) → 10.5 dpc (harvest; F), rostrally, Xgal+ cells span, and occasionally extend, the entire Lmx1a+ domain. A 8.0 (injection) → 18.5 dpc (harvest) results in labeling of cells in the RLi (arrowhead in G; H), VTA/IF (G and I), and SNpc (G and J). Double labeling for β-gal (red) and Pitx3 (green) (Upper Insets) or TH (green; H Lower Inset) in the corresponding regions. (B and G Insets) TH on adjacent sections relative to the Xgal labeling.
To determine the fate of cells derived from the medial domain, we examined later-stage embryos (Fig. 2 B–E, and Figs. S3 and S4). In 7.5 (injection) → 18.5 dpc (harvest; Fig. 2 B–E and Fig. S4), labeled cells were detected predominantly in the RLi (n = 8/8) (Fig. 2 B and C), a nucleus characterized by cells expressing Pitx3, Lmx1b, and Nurr1, but little to no TH (insets in Fig. 2C, and Fig. S5). Some labeling was observed in the VTA/IF regions (n = 8/8), and these labeled cells were β-gal+/Pitx3+ and β-gal+/TH+ mDA (Fig. 2 B and Inset in D). Remarkably, little labeling was observed in the SNpc proper (Fig. 2 B and Inset in E; n = 6/8; in 2 embryos moderate numbers of cells in the SNpc were observed, presumably because these embryos were slightly more developmentally advanced at the time of TAM administration). In more caudal sections, only scattered labeled cells were observed in medial mDA regions (Fig. S4), perhaps because the caudal medial domain is narrower (Fig. S3), and possibly has less neurogenic ability. These findings suggest that the medial-most Lmx1a+ progenitors contribute little to the SNpc but instead produce mostly TH– RLi neurons, and some VTA/IF mDA (Table S1).
When TAM is administered at 8.0 dpc, both medial and intermediate Lmx1a+ progenitors are labeled, in accordance with the expansion of the Shh domain at this time (Fig. S1). In 8.0 (injection) → 10.5-dpc (harvest) Shh:::CreERT2,R26R embryos, Xgal cells occupied most of the Lmx1a domain, particularly in rostral sections (Fig. 2F). In 8.0 (injection) → 18.5-dpc (harvest) embryos, fate-mapped cells were observed in the RLi (n = 3/3; Fig. 2 G and H, and Fig. S4). More labeled cells were observed in the VTA/IF regions, and large numbers of labeled cells were observed in the SNpc (n = 3/3; Fig. 2 G, I, and J). Cells in these regions were β-gal+/Pitx3+ and β-gal+/TH+ mDA (Fig. 2 I and J Upper Insets). In caudal sections, fewer labeled cells were observed in mDA regions (Fig. S4). In 8.5- and 9.5-dpc TAM-injected embryos (n = 4/4), fate-mapped cells were located in the RLi (Pitx3+) and throughout the rostrocaudal extent of mDA (TH+/Pitx3+; Table S1).
By 11.5 dpc, Shh is largely downregulated in the Lmx1a domain, most clearly in rostral sections (15), and by 12.5 dpc, Shh downregulation in this domain is evident in most sections (Fig. 1 J–L and Fig. S1). At these stages, Shh is maintained in lateral regions, hereafter referred to as the lateral domain. At these stages, the lateral Shh domain largely corresponds to Nkx6.1+ progenitors (Fig. S1). In 13.5- and 14.5-dpc embryos some TH+ mDA could be seen in proximity to these laterally located progenitors. Indeed, some TH+ mDA appear to emanate from the lateral, β-gal+ ventricular zone in Shh::cre,R26R embryos (Fig. S6). Likewise, some postmitotic Lmx1b+ cells were observed in proximity to the lateral, Lmx1b– ventricular zone. The dorsal limit of postmitotic Lmx1b+ cells, like TH, largely coincided with the lateral margin of β-gal+ territory (Figs. S6 and S7). Lmx1b+ cells in these lateral regions displayed low levels of Pitx3 and possibly contribute, at least in part, to the low Pitx3 mDA cohort observed in 14.5- and 18.5-dpc embryos (Figs. S5 and S6). To determine the fate of lateral Shh progenitors, we administered TAM at 12.5 dpc. In accordance with the Shh expression pattern, 12.5 (injection) → 14.5 dpc (harvest) revealed preferential labeling of lateral progenitors. Some β-gal+/TH+ neurons were observed emanating directly from these lateral progenitors (Fig. S6). In 12.5 (injection) → 18.5-dpc (harvest) experiments, fate-mapped cells were located in the RLi and VTA/IF regions. A few if any labeled cells were present in the SNpc (Fig. S6 and Table S1). These experiments point to the existence of a lateral Shh+ mDA progenitor pool. However, the stochastic marking achieved by using these tools, coupled with the confounding labeling of some centrally located progenitors, preclude an accurate estimation of the size of this laterally derived mDA cohort.
A Brn3a+ Cohort Is Derived from the Lateral Shh Domain.
Cumulative fate maps using the Shh::Cre strain suggest that a Brn3a cohort, populating the RN, EW, and Su3, is derived from the lateral Shh domain (Figs. 1 O–Q and 3 A–J, and Figs. S6 and S7). To definitively test this, we examined labeling of these clusters in Shh::CreERT2,R26R embryos receiving TAM at different time points. In 7.5 and 8.0 (injection) → 18.5-dpc embryos, a few to no labeled cells were observed in these clusters (Fig. 3K). In contrast, in 8.5-, 9.5-, and 11.5-dpc-injected embryos (wherein lateral Shh+ progenitors are also recombined), several labeled cells were observed in all 3 clusters (Fig. 3 L and M; Table S1). In 12.5-dpc-injected embryos, only few cells were detected in the RN, in accordance with the end of the RN neurogenetic interval (25); more caudally, several labeled cells were detected in the EW and Su3 regions (Fig. 3N). These data suggest that the Brn3a cohort is largely derived from the lateral, but not medial or intermediate, Shh domains.
Discussion
A key motivation for detailing the developmental underpinnings of mDA diversity is the exciting potential for stem cell-derived models and therapeutics for PD (26). First, iPS-derived mDA from PD patient fibroblasts have been reported and will likely serve as valuable in vitro human cell models for this disease (27). Second, attempts to rescue rodent models of PD with mDA derived from mouse embryonic stem (ES) cells that have been programmed with developmental regulatory genes have been successful (28–30). To further improve both of these techniques, it is critical to refine ES cell manipulations to specifically produce SNpc (A9)-type mDA, the neurons that are prominently lost in PD patients (31, 32). Thus, defining the embryonic progenitor pools that contribute to distinct subsets of mDA in vivo is an important step toward our understanding and treatment of diseases of the dopamine system.
The floor plate (FP) is a key ventral organizer in the CNS, operationally defined by the molecule Shh. Previously, we have shown that the hindbrain and spinal cord FP are neurogenically inert, but surprisingly, the midbrain FP is highly neurogenic and generates mDA. Here, we extend these studies to show that in the embryonic midbrain, the Shh-expressing domain is dynamic, and that at different times the Shh domain defines progenitor populations with distinct proclivities to generate different neuron types (Fig. 4 and Fig. S8). Using conventional fate mapping in conjunction with a GIFM approach, we develop a model in which the medial Shh+ progenitors give rise to mDA located in the VTA/IF regions, as well as RLi neurons. Remarkably, these medial progenitors do not contribute efficiently to the SNpc. In contrast, when medial and intermediate progenitors are labeled, both VTA/IF and SNpc dopamine neurons are generated. Lateral Shh+ progenitors define the primordium of a Brn3a+ cohort that populates the RN, EW, and Su3. Subsequently, some lateral Shh+ progenitors appear to produce mDA. Together, these data reveal that the midbrain FP is a dynamic organizer that makes extensive and temporally distinct cellular contributions to the tissue that it patterns, and in that way resembles another key Shh-expressing organizer—the ZPA of the limb (24).
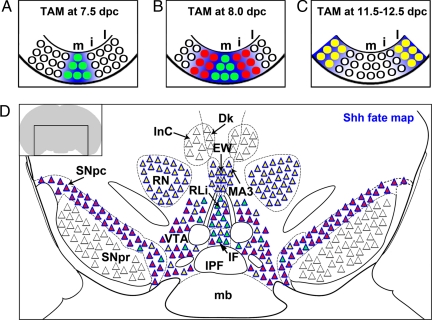
Fig. 4.
A model for the embryonic origin of midbrain nuclei derived from spatiotemporally separable Shh+ progenitors. (A–C) Schematic representation of Shh+ domains during midbrain development. Based upon cumulative and inducible genetic fate-mapping results, we propose that midbrain Shh+ progenitors (green, red, and yellow circles) occupy 3 spatiotemporally separable domains: medial (green; Lmx1a+), intermediate (red; Lmx1a+), and lateral (yellow; largely Lmx1a–). Blue background approximates Xgal-labeled domains after indicated TAM injections. Note that the time of labeling of progenitors does not necessarily correlate to the birthdates of descendent neurons derived from those progenitors. (D) Schematic of the adult organization of major rostral midbrain nuclei with respect to their embryonic origin. The color within each triangle corresponds to the progenitor domain from which these neurons are derived. The medial progenitors (green circles in A and B) predominantly contribute to the RLi (mostly TH–) and sparsely to the VTA/IF regions, but remarkably less to the SNpc. More caudally, these progenitors contribute few neurons to the VTA (see Fig. S8). The intermediate progenitors (red circles in B) contribute to the RLi and mDA in the VTA/IF regions and SNpc. The lateral Shh+ progenitors (yellow circles in C) delineate the primordium of a Brn3a+ cohort that populates the EW, Su3 (see also Fig. S8), and the RN (this nucleus is largely unlabeled in 12.5-dpc TAM injections, but is derived from the lateral Shh domain at 10.5 and 11.5 dpc). The lateral progenitors also generate some mDA. Although in close proximity to Shh descendants (blue triangle contour) in postnatal sections, several populations are largely unlabeled (black triangle contour) in our experiments. Thus, these cells must be derived outside of Shh+ progenitors. These cells belong to the 3N (Isl1/2+), IP, and GABAergic neurons of the SNpr, PRN, Dk, and InC (see also Fig. S8). A schematic of a fate map at a more caudal level is shown in Fig. S8. IPF, interpeduncular fossa.
The dynamic expansion of the midbrain Shh domain could conceivably occur by one of 2 mechanisms. First, it is possible that expansion of the Shh domain results from the growth of the initial Shh-expressing population. Second, it is possible that Shh expansion occurs by inductive mechanisms, such that the initial Shh-expressing population induces neighboring cells to express Shh. Here, by restricting labeling to medial progenitors, we reveal that these labeled progenitors do not expand concomitantly with the Shh domain. Thus, our data favors a model wherein the expansion of the Shh+ FP occurs primarily by inductive mechanisms rather than growth of the initial population. Although further studies will be required to define these inductive mechanisms in mouse, studies in zebrafish spinal cord have suggested that Shh itself, first expressed in the medial FP, may play an inductive role in the expansion of the FP (33).
Our data support the presence of 3 distinct dopaminergic progenitor pools (Fig. 4 and Fig. S8). A medial pool, at the immediate midline, gives rise to rostral linear neurons and some mDA neurons mainly in the VTA/IF regions. Strikingly, only few SNpc mDA are derived from this domain. When medial and intermediate Shh+ progenitors are labeled, SNpc neurons are observed. We deduce that SNpc neurons are largely derived from intermediate progenitors (Fig. 4 and Fig. S8). Because medial and intermediate domains roughly comprise the Lmx1a/b domain, our data suggest that Lmx1a/b progenitors are, at least, parceled into 2 subgroups having distinct proclivities. Consistent with this notion, molecular subdivisions have been observed within the Lmx1a/b territory. For example, high levels of Corin appear to define medial, but not intermediate, Lmx1a+ progenitors (34). In contrast, gene expression patterns of Wnt1 and RALDH1 appear to define mainly intermediate, but not medial, Lmx1a+ progenitors (13, 18). Further, Wnt1-derived fate maps do reveal neurons in the SNpc (13). A third, lateral, Shh+ progenitor domain is situated largely outside the Lmx1a domain. Although clonal analysis will be required for definitive verification, our data suggest that this lateral Shh domain generates a Brn3a+ cohort, and then appears to switch competence and give rise to a cohort of mDA that are restricted mainly to the VTA/IF regions. In this regard, it is interesting to revisit studies involving conditional Otx2 mutants, wherein large numbers of ectopic mDA appear to be generated laterally at the expense of the RN (35). Otx2 may thus play a pivotal role in determining the competence of this lateral progenitor pool. Taken in the context of these studies, our data suggest that medial, intermediate, and lateral progenitors have unique intrinsic properties that result in the production of distinct cohorts of neurons.
Our fate maps reveal that nuclei with diverse functions, such as the Brn3a+ RN, EW, and Su3, are derived from the lateral Shh domain (Fig. 4 and Fig. S8). In contrast, the Isl1/2+ 3N cohort is relatively unlabeled in our fate maps. We posit a model in which a common progenitor pool first generates Isl1/2+ motorneurons and subsequently generates the Brn3a cohort, akin to the sequential generation of motor and serotonin neurons observed in the hindbrain (36, 37). In our study, Isl1/2+ 3N neurons appear to arise from progenitors immediately adjacent to the Shh domain (at 9.5 dpc). As Shh expression spreads into these adjacent progenitors, Isl1/2+ neurons ceased to be produced. Instead, these progenitors, now included in the Shh::Cre recombinase domain, subsequently generate the Brn3a+ (β-gal+) cohort (10.5 dpc onward). This model is consistent with rodent birthdating studies showing that the neurogenetic interval of the 3N precedes that of the RN (25). Accordingly, the Isl1/2+ and Brn3a+ neurons are in register along the ventricular-pial axis of the 12.5-dpc embryo. As development proceeds, several fate-mapped Brn3a+ cells settle more ventrally than Isl1/2+ cells, forming the RN. Smaller cohorts of fate-mapped Brn3a+ cells populate the EW that lies medial to 3N, as well as the Su3, which remains dorsal to the 3N. Thus, the early emerging 3N is ultimately surrounded by the later emerging Brn3a cohort.
Using a genetic fate-mapping approach, we have defined distinct progenitor pools in the embryonic midbrain (Fig. 4 and Fig. S8). We reveal that different populations of mDA may be derived from distinct progenitors. Additionally, we propose the progenitor domains for several other important midbrain nuclei. Future refinements using other recombinase driver strains as well as intersectional genetic approaches (38, 39) would allow further dissection of the mDA primordium. These should also reveal progenitor distinctions along the rostrocaudal axis. Accurate embryonic midbrain maps will facilitate our understanding of the biology of the dopaminergic system, the interpretation of gain- and loss-of-function phenotypes of various mouse mutants, and the spectrum of human neurological conditions related to the neurotransmitter dopamine.
Materials and Methods
Mice.
Mice bearing a GFPcre or CreERT2 fusion that was knocked in to the Shh locus (hereafter designated Shh::Cre or Shh::CreERT2) (24) were crossed to R26R indicator mice. In inducible fate-mapping experiments, 4–9 mg/40 g TAM (dissolved in corn oil) was delivered by IP injection as previously described (40). Nine to 10 a.m. on the day of the vaginal plug was designated as 0.5 days postcoitum (dpc). For key time points, embryos from at least 2 litters were analyzed. Embryos were harvested and analyzed between 10.5 and 19.5 dpc. Mice were maintained and killed according to the protocols approved by the Northwestern Animal Care and Use Committee. Two embryos that received vehicle (oil only) showed no Xgal labeling in the brain.
Xgal Histochemistry, in Situ Hybridization, and Fluorescent Immunolabeling.
Tissue preparation, detection of Xgal, mRNA in situ hybridization, and immunofluorescence were performed as described (15, 38). Antibodies used include goat β-gal (Biogenesis), guinea pig Lmx1b, rabbit Lmx1a, rat BrdU (Serotec), rabbit and sheep tyrosine hydroxylase (Pel Freeze), rabbit Pitx3 (Zymed), rabbit GABA (Sigma), goat GATA3 (Santa Cruz), goat Foxa2 (Santa Cruz), rabbit Brn3a, mouse Isl1/2 [Developmental Studies Hybridoma Bank (DSHB)], mouse Lim1/2 (DSHB), mouse Nkx6.1 (DSHB), and mouse Nkx2.2 (DSHB). Epifluorescent or confocal images were processed in Adobe Photoshop CS2.
Supplementary Material
Acknowledgments.
We thank Dr. Cliff Tabin for Shh:CreGFP and Shh:CreERT2 mice and Shh cDNA; Dr. Michael German for anti-Lmx1a; Drs. Thomas Muller and Carmen Birchmeier for anti-Lmx1b; and Dr. Eric Turner for anti-Brn3a antibodies. We thank Dr. Doug Kim for useful comments and Dr. Abdelhak Belmadani and Bor-Shuen Wang for imaging and technical assistance. R.A. was supported by the Whitehall Foundation, Brain Research Foundation, and the American Parkinson's Disease Association. M.J. was supported by the Parkinson's Disease Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904285106/DCSupplemental.
References
- 1.Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- 2.Bjorklund A, Dunnett SB. Dopamine neuron systems in the brain: An update. Trends Neurosci. 2007;30:194–202. doi: 10.1016/j.tins.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Grimm J, Mueller A, Hefti F, Rosenthal A. Molecular basis for catecholaminergic neuron diversity. Proc Natl Acad Sci USA. 2004;101:13891–13896. doi: 10.1073/pnas.0405340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerfen CR, Baimbridge KG, Thibault J. The neostriatal mosaic: III. Biochemical and developmental dissociation of patch-matrix mesostriatal systems. J Neurosci. 1987;7:3935–3944. doi: 10.1523/JNEUROSCI.07-12-03935.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendez I, et al. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson's disease. Brain. 2005;128:1498–1510. doi: 10.1093/brain/awh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fearnley JM, Lees AJ. Ageing and Parkinson's disease: Substantia nigra regional selectivity. Brain. 1991;114:2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 7.Damier P, Hirsch EC, Agid Y, Graybiel AM. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson's disease. Brain. 1999;122:1437–1448. doi: 10.1093/brain/122.8.1437. [DOI] [PubMed] [Google Scholar]
- 8.Smidt MP, et al. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- 9.Hwang DY, Ardayfio P, Kang UJ, Semina EV, Kim KS. Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Brain Res Mol Brain Res. 2003;114:123–131. doi: 10.1016/s0169-328x(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 10.Kittappa R, Chang WW, Awatramani RB, McKay RD. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sgado P, et al. Slow progressive degeneration of nigral dopaminergic neurons in postnatal Engrailed mutant mice. Proc Natl Acad Sci USA. 2006;103:15242–15247. doi: 10.1073/pnas.0602116103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono Y, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: Midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- 13.Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Bonilla S, et al. Identification of midbrain floor plate radial glia-like cells as dopaminergic progenitors. Glia. 2008;56:809–820. doi: 10.1002/glia.20654. [DOI] [PubMed] [Google Scholar]
- 15.Joksimovic M, et al. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci. 2009;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- 16.Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: Establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- 17.Echelard Y, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 18.Wallen A, et al. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp Cell Res. 1999;253:737–746. doi: 10.1006/excr.1999.4691. [DOI] [PubMed] [Google Scholar]
- 19.Andersson E, et al. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 20.Smidt MP, et al. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- 21.Fedtsova NG, Turner EE. Brn-3.0 expression identifies early post-mitotic CNS neurons and sensory neural precursors. Mech Dev. 1995;53:291–304. doi: 10.1016/0925-4773(95)00435-1. [DOI] [PubMed] [Google Scholar]
- 22.Agarwala S, Ragsdale CW. A role for midbrain arcs in nucleogenesis. Development. 2002;129:5779–5788. doi: 10.1242/dev.00179. [DOI] [PubMed] [Google Scholar]
- 23.Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- 24.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Prakash N, et al. Nkx6–1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development. 2009;136:2545–2555. doi: 10.1242/dev.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKay R, Kittappa R. Will stem cell biology generate new therapies for Parkinson's disease? Neuron. 2008;58:659–661. doi: 10.1016/j.neuron.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- 29.Friling S, et al. Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc Natl Acad Sci USA. 2009;106:7613–7618. doi: 10.1073/pnas.0902396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabar V, et al. Therapeutic cloning in individual parkinsonian mice. Nat Med. 2008;14:379–381. doi: 10.1038/nm1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isacson O, Kordower JH. Future of cell and gene therapies for Parkinson's disease. Ann Neurol. 2008;64:S122–S138. doi: 10.1002/ana.21473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- 33.Schauerte HE, et al. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development. 1998;125:2983–2993. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- 34.Jönsson ME, Ono Y, Björklund A, Thompson LH. Identification of transplantable dopamine neuron precursors at different stages of midbrain neurogenesis (Translated from English) Exp Neurol. 2009;219:341–354. doi: 10.1016/j.expneurol.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Puelles E, et al. Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nat Neurosci. 2003;6:453–460. doi: 10.1038/nn1037. [DOI] [PubMed] [Google Scholar]
- 36.Pattyn A, et al. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacob J, et al. Transcriptional repression coordinates the temporal switch from motor to serotonergic neurogenesis. Nat Neurosci. 2007;10:1433–1439. doi: 10.1038/nn1985. [DOI] [PubMed] [Google Scholar]
- 38.Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- 39.Jensen P, et al. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter NL, Awatramani RB, Farley FW, Dymecki SM. Ligand-activated Flpe for temporally regulated gene modifications. Genesis. 2005;41:99–109. doi: 10.1002/gene.20101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.