Abstract
The ubiquitous cellular influence of nitric oxide (NO) is exerted substantially through protein S-nitrosylation. Whereas NO is highly promiscuous, physiological S-nitrosylation is typically restricted to one or very few Cys residue(s) in target proteins. The molecular basis for this specificity may derive from properties of the target protein, the S-nitrosylating species, or both. Here, we describe a protein microarray-based approach to investigate determinants of S-nitrosylation by biologically relevant low-mass S-nitrosothiols (SNOs). We identify large sets of yeast and human target proteins, among which those with active-site Cys thiols residing at N termini of α-helices or within catalytic loops were particularly prominent. However, S-nitrosylation varied substantially even within these families of proteins (e.g., papain-related Cys-dependent hydrolases and rhodanese/Cdc25 phosphatases), suggesting that neither secondary structure nor intrinsic nucleophilicity of Cys thiols was sufficient to explain specificity. Further analyses revealed a substantial influence of NO-donor stereochemistry and structure on efficiency of S-nitrosylation as well as an unanticipated and important role for allosteric effectors. Thus, high-throughput screening and unbiased proteome coverage reveal multifactorial determinants of S-nitrosylation (which may be overlooked in alternative proteomic analyses), and support the idea that target specificity can be achieved through rational design of S-nitrosothiols.
Keywords: cysteine, nitric oxide, S-nitrosothiol, thiol
Protein S-nitrosylation underlies much of the physiological signaling by both nitric oxide (NO) and endogenous S-nitrosothiols, and both hypo- and hyper-S-nitrosylation have been causally implicated in disease (1, 2). Formally, S-nitrosylation occurs either via an oxidative reaction of NO and Cys thiol, in the presence of an electron acceptor (e.g., transition metal or O2), or by the transfer of NO+ (transnitros(yl)ation) from donor S-nitrosothiol (SNO) to acceptor Cys thiol (1). These reactions may be enzyme-catalyzed or otherwise facilitated. For example, hemoglobin and ceruloplasmin support metal-dependent S-nitrosylation (3–5) whereas S-nitroso-hemoglobin (SNO-hemoglobin) and SNO-thioredoxin can transnitrosylate proteins with which they interact directly (anion exchanger 1 and caspase-3, respectively) (6, 7). Transnitrosylation is also implicated in protein S-nitrosylation coupled to NO synthase activity (8–10) and involves the intermediacy of the endogenous low-mass SNO, S-nitrosoglutathione (GSNO), as evidenced by elevated levels of SNO-proteins in mice lacking the GSNO metabolizing enzyme, GSNO reductase (GSNOR) (8–11). GSNOR deletion also increases S(NO)-mediated protein S-nitrosylation and cytostasis in yeast (11, 12), and decreases virulence of several pathogens (13, 14), which suggests that transnitrosylation is an important mediator of nitrosative stress. In addition, metabolism of GSNO to S-nitrosocyteinylglycine and S-nitrosocysteine (CysNO) and CysNO uptake via l-amino acid transporters appear to be required for many biological effects of extracellular (endogenous and exogenous) SNOs (15, 16). S-nitrosylation typically occurs at only one or a few Cys residues in target proteins (2, 17), although the factors underlying this specificity are incompletely understood. Target thiol pKa is often invoked as a critical determinant of transnitrosylation (18), whereas local hydrophobicity may be an important determinant for oxidative S-nitrosylation (by NO itself) (19, 20)–presumably underlying the reported acid-base and hydrophobic motifs (2, 19).
The biotin switch technique (BST), which converts an S-nitrosothiol to an S-biotinylated Cys (21, 22), has greatly facilitated the identification of SNO proteins and specific SNO sites (20, 23) and aided in the discovery of many new (patho)physiological roles for S-nitrosylation. In principle, the characterization of SNO proteomes by this method should reveal classes of (S)NO-reactive proteins and/or common sequences or structural features that would allow for a more complete understanding of S-nitrosylation determinants. However, the assay is biased toward the identification of abundant proteins, and has therefore been limited as a proteomic tool. Functional protein microarrays (24), which have recently been used to measure a wide array of protein functions, including interactions of proteins with other proteins, DNA/RNA, small molecules and phospholipids (24), and also to assay posttranslational modifications [glycosylation and phosphorylation (25, 26)], may address such limitations. Importantly, this methodology provides an unbiased, proteome-wide coverage of potential substrates, obviates the often laborious requirement for target identification by mass spectrometry and enables relative quantification. Arrayed proteins also appear to retain their native structures and activities. We therefore sought to investigate whether protein microarrays could be adapted to studies of S-nitrosylation.
Results
Analysis of S-Nitrosylation on Protein Microarrays.
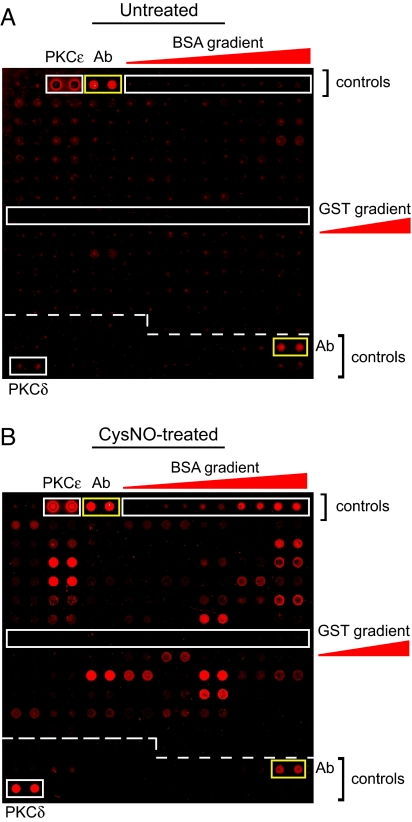
We developed a modified BST assay for detecting SNO-proteins on microarrays that uses an anti-biotin antibody and fluorescently labeled secondary antibody for detection. We then screened a yeast protein microarray (Invitrogen Protoarray) containing ≈4,000 glutathione S-transferase (GST)-tagged ORFs after treatment with 50 μM S-nitrosocysteine (CysNO), a highly reactive SNO (Fig. 1 and supporting information (SI) Fig. S1). Several hundred yeast SNO proteins were identified versus buffer-treated arrays (Fig. 1, Fig. S1, and Table S1). CysNO did not S-nitrosylate GST, which is fused to each ORF (Fig. 1).
Fig. 1.
Detection of S-nitrosylation on protein microarrays. A modified BST (see Materials and Methods; without DTT pretreatment) was applied to yeast Protoarrays that were exposed for 30 min to either buffer alone (A) or 50 μM CysNO (B). A representative block (#12 of 48) of proteins spotted in duplicate is shown for each condition. S-nitrosylation is identified by an increase in fluorescence intensity after CysNO treatment. The positions of positive antibody (Ab) controls (anti-biotin Ab and biotinylated Ab gradient) are indicated by yellow rectangles. Other protein controls (GST, PKCδ, PKCε, and BSA) are indicated by white rectangles. Images are representative of at least three independent experiments.
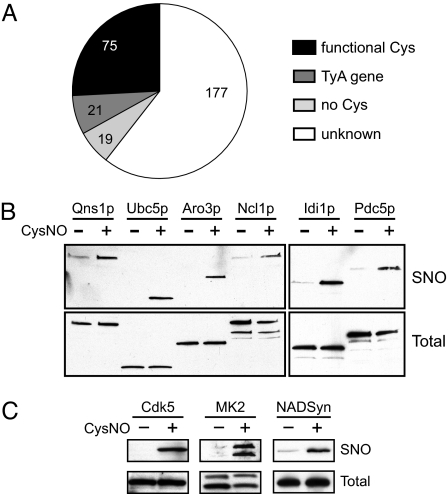
We analyzed the 300 proteins with the highest relative fluorescence intensity from a representative CysNO-treated microarray (Table S1). Approximately two-thirds of these “hits” were detected in two or more independent experiments; variability between experiments could be rationalized by differences in protein content or relative protein concentration among microarray “lots.” Proteins (and predicted gene products) were classified by using the Saccharomyces Genome Database (SGD) (27). Approximately 5% of proteins (19 of 300) lacked a Cys residue and were scored as false positives (Fig. 2A and Table S1). More than half of the false positives were subunits of hetero-oligomeric complexes (Table S1), suggesting that these signals most likely arose from S-nitrosylation of proteins that copurified with the GST-tagged ORFs (28). Known catalytic, redox-active and metal-binding Cys residues were identified in ≈25% of hits; 31 of 300 S-nitrosylated proteins had only a single Cys residue (Table S1), identifying the putative site of S-nitrosylation. Seven ORFs were duplicated twice on the array (resulting in 14 hits), and 21 additional hits corresponded to 12 homologous TyA gene products that are analogous to the retroviral Gag protein (27). In addition, three enzymes of the maltase family (Mal12p, Mal32p, and Yil172cp), several highly homologous homocitrate synthase isozymes (Lys20p and Lys21p), and two ADP-ribosylation factors (Arf1p and Arf2p) were also S-nitrosylated (Table S1). A similar pattern of S-nitrosylation across duplicate and homologous ORFs increased confidence in the microarray analysis.
Fig. 2.
Analysis and validation of microarray data. (A) The 300 highest-intensity signals from a CysNO-treated yeast microarray were analyzed (see also Table S1) to identify the proportion of proteins with functional Cys residues, those lacking a Cys residue (false positives), and TyA gene products (which were the most highly represented protein family). (B) For validation of yeast microarray screening, lysates from six yeast strains overexpressing HA-tagged ORFs were treated with and without 50 μM CysNO and analyzed by the BST, followed by Western blotting with an anti-HA antibody. Proteins are in the order of their natural cellular abundance (from left to right). (C) S-nitrosylation of proteins identified by screening of human microarrays was validated in mammalian cells. Rat PC-12, mouse RAW264.7, and human HEK-293 cells were treated with and without 100 μM CysNO for 5 min, with and without 250 μM CysNO for 5 min, and with and without 10 μM CysNO for 15 min, respectively. Lysates were analyzed by the BST, followed by Western blotting for cyclin-dependent kinase 5 (Cdk5; PC-12 cells), MAP kinase-activated protein kinase 2 (MK2; RAW246.7 cells) or 3x-Flag-tagged human NAD+ synthetase (NADSyn, transiently expressed in HEK-293 cells). Rat Cdk5 and mouse MK2 each share at least 98% identity with their human homologues. Images are representative of at least two independent experiments.
We selected six enzymes (Qns1p, Ubc5p, Aro3p, Ncl1p, Idi1p, and Pdc5p) containing catalytically essential Cys thiols (Table S1) for initial validation. Lysates from yeast expressing C-terminally-tagged ORFs [the tag lacks a Cys residue (25)] were treated with CysNO, and S-nitrosylation of each protein was confirmed by the BST (Fig. 2B). We also quantified the S-nitrosylation of two GTPases (Arl1p and Arf1p) in vitro. CysNO readily S-nitrosylated MgGDP-bound Arl1p, but Arf1p S-nitrosylation was only observed with EDTA pretreatment (Fig. S2A), suggesting that removal of the proximal Mg-GDP (Fig. S2B) was required. More generally, the protein microarrays are well suited for relative quantification of protein S-nitrosylation (Table S1). We also assayed human Protoarrays (treated with buffer or GSNO) for S-nitrosylation. Interestingly, we identified several human homologues of yeast SNO-proteins (Table S2A), among which, NAD+ synthetase (NADSyn, a homologue of yeast Qns1p) was previously identified (29). We verified S-nitrosylation of two newly identified SNO-proteins, Cdk5 and MK2, as well as NADSyn, in mammalian cells (Fig. 2C). Thus, the microarray approach may be used for identifying novel SNO targets.
Importantly, we noted that SNO reactivity varied substantially within several classes of yeast proteins possessing catalytic (and presumably highly nucleophilic) Cys thiols, including those of the rhodanese/Cdc25 phosphatase and papain-related Cys-dependent hydrolase families (see below). We sought to further investigate the basis of this differential reactivity.
S-Nitrosylation of the Rhodanese/Cdc25 Phosphatase Family.
Rhodanese enzymes catalyze the transfer of sulfane sulfur from thiosulfate to cyanide (to form thiocyanate) (30). A predominant feature of this enzyme class is an active-site loop that forms a pocket with the catalytically essential Cys thiol at its center (Fig. S3A). Several rhodaneses are inhibited by SNOs (Table S2B), and notably, two of the most highly SNO-reactive proteins, Uba4p and Tum1p, have rhodanese-like domains (Fig. 3A). Two additional rhodanese-like proteins (Rdl1p and Rdl2p) were identified by BLASTP searches (31) against either Tum1p or Uba4p. Of these, only Rdl2p appeared on the array, but its reactivity toward CysNO was low compared with Tum1p and Uba4p (Fig. 3A and Table S1). The rhodanese superfamily also includes the SNO target Cdc25 phosphatase (Table S2B), which shares little primary sequence homology to rhodanese but possesses a rhodanese-like fold and conserved active-site loop (30). BLASTP analysis identified three proteins with structural homology to Cdc25 (Fig. 3A), including the phosphatase Ygr203wp (Ych1p; yeast Cdc25 homologue 1). However, Ych1p appeared resistant to S-nitrosylation (Fig. 3A). The catalytic loops (and adjoining helices) of Ych1p and Rdl1p are superimposable but lack primary sequence homology (Fig. S3A).
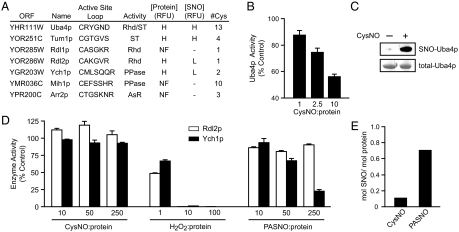
Fig. 3.
S-nitrosylation of rhodanese/cdc25 phosphatase superfamily members. (A) A summary of yeast ORFs belonging to the Rhodanese/Cdc25 phosphatase superfamily, including the sequences of their active-site loops, enzymatic activity (Rhd, rhodanese; ST, sulfane sulfur transferase; PPase, phosphatase; AsR, arsenite reductase), relative abundance on the microarray and degree of S-nitrosylation by CysNO (H, high; L, low; NF, not found). Note that the rhodanese activities of Rdl1p and Rdl2p have not been reported previously, and that (as for Rdl2p, in D), Rdl1p is inhibited by neither CysNO nor PASNO. (B) Rhodanese activity was measured after incubation of Uba4p (100 μM) with the indicated molar ratios of CysNO:protein for 10 min. (C) Uba4p (25 μM) was treated with 500 μM CysNO and analyzed by the BST (Upper). Total-Uba4p was visualized by Coomassie staining (Lower). (D) Ych1p (phosphatase) and Rlp2p (rhodanese) activities were measured after incubation of 10 μM enzyme for 10 min with the indicated molar ratios of CysNO, H2O2 or S-nitroso-4-mercaptophenyl acetic acid (PASNO; Fig. S5), followed by additional steps described in SI Materials and Methods. (D) Ych1p (1 μM) was treated for 10 min with 250 μM CysNO or PASNO and protein S-nitrosylation was quantified by Hg-coupled photolysis-chemiluminescence. Data in B and C are mean ± SEM (n = 3) and data in D are mean of three replicates.
The microarray data suggested that Uba4p and Tum1p (but not Rdl2p or Ych1p) would be S-nitrosylated and inhibited by CysNO, and to validate these results, we purified the recombinant proteins from Escherichia coli. The rhodanese activity of Uba4p (32) was dose-dependently inhibited by CysNO and GSNO (Fig. 3B and Fig. S3B); the enzyme was S-nitrosylated by CysNO and GSNO (Fig. 3C and Fig. S3C) and the catalytic Cys (C397) was identified as a SNO-site (Fig. S3D). We found that Rdl2p (but not Tum1p) also had rhodanese activity, and we confirmed that neither Rdl2p nor the phosphatase Ych1p were inhibited or S-nitrosylated by CysNO (Fig. 3 D–E). On the other hand, H2O2 was a potent inhibitor of both enzymes (Fig. 3D), suggesting differential reactivity of active-site cysteines toward alternative redox-active compounds. We hypothesized that SNO-bearing substrates or substrate mimetics might serve as inhibitors of these enzymes. SNO-thiosulfate is unstable (33), but a previously unidentified phospho-tyrosine mimetic, S-nitroso-4-mercaptophenylacetic acid (PASNO; Fig. S4) inhibited and S-nitrosylated Ych1p (Figs. 3 D–E). Ych1p has two Cys thiols (C90 and C116), and the catalytic Cys (C90) was identified as the locus of these effects, because the inhibition of C116S Ych1p by PASNO was comparable with wild type (Fig. S3E). By comparison, PASNO did not inhibit Rdl2p (Fig. 3D).
S-Nitrosylation of the Papain-Related Cys-Dependent Hydrolase Family.
An additional SNO-reactive enzyme family was suggested by the apparent S-nitrosylation of numerous glutamine amidotransferases (GATases) (34), including Qns1p (Fig. 2B), Trp3p, and Sno2p (Table S1). The glutaminase active site of these enzymes features a catalytic triad (e.g., Cys-His-Glu) in which the Cys is situated in a “nucleophilic elbow” at the N terminus of an α-helix (Fig. S5A). This motif is conserved in the SNO-sites of the protease papain and the GATase domain of CTP synthetase (Table S2B). Over 40 yeast proteins are members of this Cys-dependent hydrolase family, including GATases (of the nitrilase superfamily), deubiquitinating enzymes (DUBs) (including proteases and C-terminal hydrolases specific to conjugates of ubiquitin and ubiquitin-like proteins), and proteins of the DJ-1 superfamily (Table S3). In our arrays, the SNO-reactivity of the nitrilases was much higher than DUBs or DJ-1-related proteins (Fig. 4A and Table S3); we selected a protein from each of these classes for validation.
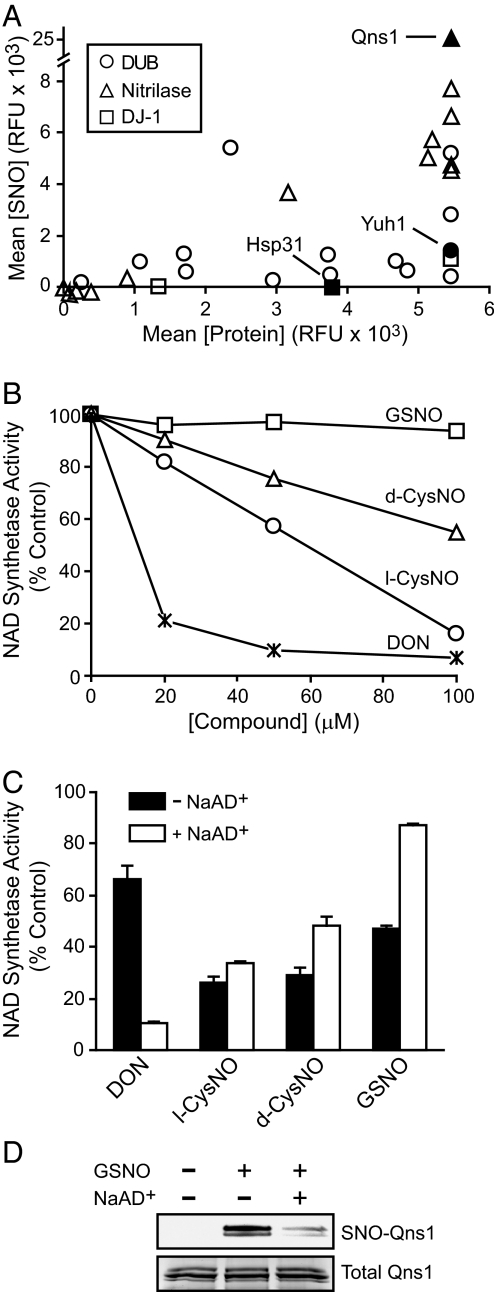
Fig. 4.
S-nitrosylation and inhibition of cysteine-dependent hydrolases. (A) Fluorescence intensities from a CysNO-treated microarray were plotted against the relative protein abundance for yeast Cys hydrolase motif-containing proteins. (B) Qns1p (0.25 μM) was treated for 30 min with increasing concentrations of 6-diazo-5-oxo-L-norleucine (DON) or various S-nitrosothiols, and NAD+ synthetase activity was measured. (C) Qns1p activity was measured after treatment with 100 μM inhibitors as in B with or without NaAD+. (D) One hundred microliters of Qns1p (0.1 mg/mL) in NAD+ synthethase assay buffer (see Materials and Methods) with or without NaAD+ was incubated with GSNO for 30 min at 30 °C and analyzed by the BST. Data in B are mean (n = 3), and data in C are mean ± SEM. Image in D is representative of two independent experiments.
We first measured the inhibition of the NAD+ synthetase Qns1p by SNOs compared with the activity-based inhibitor 6-diazo-5-oxo-L-norleucine (DON) under pseudocatalytic conditions (including the cosubstrates nicotinic acid adenine dinucleotide (NaAD+) and MgATP). Qns1p was dose-dependently inhibited by DON and CysNO but not by GSNO (Fig. 4B), despite the apparently robust S-nitrosylation of Qns1p by GSNO on the microarray (Table S1). In addition, l-CysNO was a more potent inhibitor than d-CysNO (Fig. 4B). Reaction with the catalytic Cys by both the substrate l-Gln and the inhibitor l-CysNO occurs on a sp2-hybridized electrophilic atom (C and N, respectively) at approximately the same distance from the α-carbon (Fig. S4) (35); accordingly, the stereoselective inhibition by l- versus d-CysNO likely reflects a more favorable recognition of l-CysNO by the Qns1p active site.
Qns1p has two domains, with distinct active sites: The GATase domain catalyzes the hydrolysis of l-Gln to glutamate and ammonia, and the NAD+ synthetase domain utilizes the ammonia for the amidation of NaAD+ (36). In GATase domain-containing proteins, substrate binding to the ammonia-accepting active site typically stimulates glutaminase activity as a result of conformational changes in the glutaminase catalytic pocket (37). Accordingly, hydrolysis of glutamine by Qns1 is stimulated 50-fold (with an increase in kcat) upon NaAD+ binding (36). We reasoned that the low reactivity of GSNO (Fig. 4B) might reflect allosteric effects of NaAD+ on the GATase domain. Without NaAD+, DON was a relatively poor inhibitor, consistent with reduced glutaminase activity. On the other hand, the absence of NaAD+ greatly potentiated Qns1p inhibition by GSNO and nearly abolished the differential inhibition by l- versus d-CysNO (Fig. 4C). S-nitrosylation of Qns1p by GSNO was similarly modulated by NaAD+ (Fig. 4D), suggesting that the active site C175 is the site of inhibitory S-nitrosylation. After NaAD+ binding, GSNO appears unable to access the glutaminase active-site cleft. Furthermore, the features of Qns1p that appear to favor S-nitrosylation by l- versus d-CysNO are also evidently lost in the absence of NaAD+, although the catalytic Cys appears to maintain its thiolate character (i.e., low pKa) irrespective of NaAD+.
In addition, we compared the reactivities of DJ-1 family member Hsp31p [which is induced by and protects against oxidative stress (38)] and the USP Yuh1p [a ubiquitin C-terminal hydrolase (39)], neither of which were S-nitrosylated on the arrays (Fig. 4A and Table S3). These proteins each have a single Cys, and we verified that recombinantly expressed and purified proteins had approximately one free thiol as measured by Ellman's reagent. Purified Hsp31p was inert to CysNO or GSNO treatment, verifying the array results, but Yuh1p was S-nitrosylated and inhibited by CysNO (and to a lesser extent by GSNO) (Fig. S5 B and C). Thus, the microarray analysis produced a potentially false-negative result; nonetheless, the apparently low reactivity of SNOs toward the majority of yeast USPs is consistent with reports that their catalytic cysteines are buried in the absence of substrate (39).
Discussion
Low-mass SNOs mediate cellular signal transduction and nitrosative stress (10), and SNO deficiency has been identified with disease states (1, 40, 41). A principal mechanism through which SNOs elicit cellular effects involves the S-nitrosylation of Cys residues at active or allosteric sites within proteins. The molecular basis of SNO specificity is still poorly understood. Our experimental analysis indicates that low thiol pKa may be necessary but is not sufficient to support protein transnitrosylation by low-mass SNOs. Stereochemistry of SNO donors, interactions between donors and their substrates, and the conformational state of target proteins are shown to represent additional determinants of specificity. The multifactorial requirements for S-nitrosylation highlight the limitations of interpretations based on proteomic analyses that rely on a single type of NO donor or physiological stimulus and which remove proteins from natural environments rich in cofactors that may influence SNO reactivity.
For many of the proteins analyzed in this study, thiol accessibility to SNO appears to be a critical determinant of reactivity. The facile reaction of alkyl SNOs with C397 of Uba4p is consistent with the proposed role of this residue in mediating sulfane sulfur (S0) transfer between protein Cys thiols (42) and suggests that it is solvent-exposed. On the other hand, the selective reactivity of PASNO with Ych1p suggests that the active site prefers aryl versus alkyl side chains (to the exclusion of CysNO), consistent with the enzyme's tyrosine phosphatase activity. Less efficient S-nitrosylation of Qns1p and Yuh1p by GSNO versus CysNO, and the preferential inhibition of NaAD+-bound Qns1p by l- versus d-CysNO, also appear to reflect the ability of these small molecules to access enzyme active sites, which have evolved to bind specific substrates. Other similar examples have recently been uncovered, including the preferential reactivity of GSNO (versus CysNO) with E. coli OxyR (43), and the stereoselective inhibition of T-type calcium channels by l- versus d-CysNO (44). NaAD+-dependent S-nitrosylation of Qns1p points further to the importance of allostery in control of S-nitrosylation, and it is notable that S-nitrosylation of hemoglobin (45), serum albumin (46), tissue transglutaminase (47), and the ryanodine receptor (48) are also modulated allosterically (by O2, fatty acid, Ca2+ and Ca2+-calmodulin, respectively). More generally, allosteric effectors are shown to have profound consequences on efficiency of S-nitrosylation and thus may be underappreciated determinants of SNO reactivity.
Microarray-based proteomic screens of S-nitrosylation may reveal SNO-reactivity patterns for large classes of proteins. The human genome alone encodes for ≈100 Cys-dependent DUBs that, at least in structurally-characterized examples, have catalytic Cys residues at the N termini of α-helices (49). However, the low reactivity of yeast DUBs is consistent with the selective accessibility of their active sites to substrates containing ubiquitin or ubiquitin-like proteins, and, in fact, none of the catalytic Cys residues of mammalian DUBs have been identified as SNO targets [UCH-L1, a ubiquitin C-terminal hydrolase, is S-nitrosylated at a noncatalytic Cys (23, 50)]. Thus, whereas SNOs appear to regulate numerous enzymes in the ubiquitylation pathway (2), DUBs appear to be resistant to SNO-based regulation. Similarly, >100 human Cys-dependent protein tyrosine phosphatases (PTPs; including Cdc25A-C) have been identified that are defined by a Cys-containing catalytic loop (51, 52), and although mammalian PTPs are increasingly recognized as SNO targets (Table S2A), only one HCX5R motif-containing yeast phosphatase (Ymr1p) was detected among the most SNO-reactive proteins on the microarray. Further study is therefore needed to define the reactivity determinants of this large enzyme class. In addition, the DJ-1 family, including DJ-1 itself (53) and yeast Hsp31p (54), appears to define a class of proteins whose active sites are readily oxidized but are apparently inaccessible to low-mass SNOs.
Our data show that structural motifs such as the Cys at the N terminus of a helix (N-Cap Cys) are highly variable with regard to SNO-reactivity, and are therefore unlikely to be universal motifs for S-nitrosylation. Nonetheless, the N-Cap Cys motif might serve as a useful SNO-site predictor: N termini of helices in proteins are often solvent-exposed (55), and charge stabilization by the helix dipole may lower the pKa of the N-Cap Cys by approximately two pH units (56, 57) [a potentially greater contribution to pKa than a proximal basic (e.g., His) residue (58)]. Notably, the catalytic cysteines of rhodanese/Cdc25 phosphatases and PTPs are near the cationic side of a helix (Fig. S4A), and the helix dipole is often suggested as a modulator of Cys thiol reactivity in these enzymes. In addition to the catalytic thiols of Cys-dependent hydrolases [e.g., papain (59), CTP synthetase (35), Qns1p and Yuh1p (this work)], C32 and C62 of human thioredoxin (60), C12 of β-tubulin (23), and C149 of GAPDH (23)–known S-nitrosylation sites—are all at the N termini of α-helices. Nonetheless, statistical significance for this motif among SNO-sites is yet to be demonstrated.
Despite occasional false-positive SNO-protein identifications and the lack of complete proteome coverage by commercially available microarrays, our study highlights a number of advantages of this high-throughput methodology for studies of S-nitrosylation, including: (i) the facile identification of SNO-proteins (without the need for mass spectrometry); (ii) the ability to perform relative quantification of SNO-reactivity across a proteome; (iii) the ability to assess the effects of multiple donors and cofactors; (iv) the lack of bias with regard to endogenous protein abundance. With regard to the latter factor, it is worth noting that neither SNO-MK2 nor SNO-Cdk5 (Fig. 2C) have been previously identified by MS-based analyses of SNO-treated RAW246.7 macrophages or brain extracts, respectively (23, 50, 61). Inasmuch as the in vitro targets of SNOs (e.g., in tissue extracts) have been shown to significantly overlap the sets of proteins that are S-nitrosylated by endogenous NO (21), protein microarrays present a tractable methodology for deconvoluting the multiple pathways for S-nitrosylation and denitrosylation that govern steady-state SNO levels in vivo (12, 61, 62).
Materials and Methods
Materials and methods of syntheses of S-nitrososthiols are provided in SI Materials and Methods
Detection of Protein S-Nitrosylation on Protein Microarrays.
Invitrogen yeast and human Protoarrays for kinase substrate identification (KSI) were treated with S-nitrosothiols and assayed for protein S-nitrosylation by using a modified biotin switch protocol as described in SI Materials and Methods. Raw microarray data can be accessed at the National Center for Biotechnology Information Gene Expression Omnibus web site (www.ncbi.nlm.nih.gov/geo/).
Cloning and Purification of Yeast Proteins.
Yeast proteins were cloned from yeast genomic DNA, expressed as GST- or 6xHis-tagged fusion proteins in E. coli and purified as described in SI Materials and Methods.
Assay of S-Nitrosylation in Cell Lysates and in Purified Proteins.
Protein S-nitrosylation was measured in cell lysates and in purified proteins by using the biotin switch technique or Hg-coupled photolysis chemiluminescence as described in SI Materials and Methods. Mass spectrometry was used in the identification of SNO-sites within purified protein as described in SI Materials and Methods.
Assay of Enzyme Activities.
Rhodanese, tyrosine phosphatase, NAD+ synthetase, and ubiquitin C-terminal hydrolase activities were assayed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Charles Brenner (University of Iowa) for providing Qns1p plasmid, Greg Michaud (Invitrogen) for advice and for supplying blank microarray slides, the Duke Proteomics Core and Will Thompson for MS analysis, Douglas Hess and Erin Whalen for helpful suggestions during the course of our investigation, and Erica Washington for technical assistance. This work was supported by National Institutes of Health Grants P01-HL075443, R01-HL096673, R01-HL059130, and U19-ES012496.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900729106/DCSupplemental.
References
- 1.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 3.Inoue K, et al. Nitrosothiol formation catalyzed by ceruloplasmin. Implication for cytoprotective mechanism in vivo. J Biol Chem. 1999;274:27069–27075. doi: 10.1074/jbc.274.38.27069. [DOI] [PubMed] [Google Scholar]
- 4.Angelo M, Singel DJ, Stamler JS. An S-nitrosothiol (SNO) synthase function of hemoglobin that utilizes nitrite as a substrate. Proc Natl Acad Sci USA. 2006;103:8366–8371. doi: 10.1073/pnas.0600942103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minning DM, et al. Ascaris haemoglobin is a nitric oxide-activated ‘deoxygenase’. Nature. 1999;401:497–502. doi: 10.1038/46822. [DOI] [PubMed] [Google Scholar]
- 6.Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature. 2001;409:622–626. doi: 10.1038/35054560. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat Chem Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 8.Whalen EJ, et al. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 9.Ozawa K, et al. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, et al. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 12.Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. A genetic analysis of nitrosative stress. Biochemistry. 2009;48:792–799. doi: 10.1021/bi801813n. [DOI] [PubMed] [Google Scholar]
- 13.de Jesus-Berrios M, et al. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–1968. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Stroeher UH, et al. A pneumococcal MerR-like regulator and S-nitrosoglutathione reductase are required for systemic virulence. J Infect Dis. 2007;196:1820–1826. doi: 10.1086/523107. [DOI] [PubMed] [Google Scholar]
- 15.Lipton AJ, et al. S-nitrosothiols signal the ventilatory response to hypoxia. Nature. 2001;413:171–174. doi: 10.1038/35093117. [DOI] [PubMed] [Google Scholar]
- 16.Hogg N, Broniowska KA, Novalija J, Kettenhofen NJ, Novalija E. Role of S-nitrosothiol transport in the cardioprotective effects of S-nitrosocysteine in rat hearts. Free Radical Biol Med. 2007;43:1086–1094. doi: 10.1016/j.freeradbiomed.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Stamler JS, Lamas S, Fang FC. Nitrosylation. The prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 18.Derakhshan B, Hao G, Gross SS. Balancing reactivity against selectivity: The evolution of protein S-nitrosylation as an effector of cell signaling by nitric oxide. Cardiovasc Res. 2007;75:210–219. doi: 10.1016/j.cardiores.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess DT, Matsumoto A, Nudelman R, Stamler JS. S-nitrosylation: Spectrum and specificity. Nat Cell Biol. 2001;3:E46–E49. doi: 10.1038/35055152. [DOI] [PubMed] [Google Scholar]
- 20.Greco TM, et al. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 22.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radical Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung LA, Snyder M. Proteome chips for whole-organism assays. Nat Rev Mol Cell Biol. 2006;7:617–622. doi: 10.1038/nrm1941. [DOI] [PubMed] [Google Scholar]
- 25.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 27.Saccharomyces. Genome Database, www.yeastgenome.org.
- 28.Michaud GA, et al. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003;21:1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 29.Lefievre L, et al. Human spermatozoa contain multiple targets for protein S-nitrosylation: An alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics. 2007;7:3066–3084. doi: 10.1002/pmic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bordo D, Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep. 2002;3:741–746. doi: 10.1093/embo-reports/kvf150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz J, et al. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry. 2008;47:6479–6489. doi: 10.1021/bi800477u. [DOI] [PubMed] [Google Scholar]
- 33.Bryant T, Williams DLH, Ali MHH, Stedman G. Nitrosothiosulfate ion (S2O3NO−) as a nitrosating species. J Chem Soc Perkin Trans. 1986;2:193–196. [Google Scholar]
- 34.Massiere F, Badet-Denisot MA. The mechanism of glutamine-dependent amidotransferases. Cell Mol Life Sci. 1998;54:205–222. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun O, Knipp M, Chesnov S, Vasak M. Specific reactions of S-nitrosothiols with cysteine hydrolases: A comparative study between dimethylargininase-1 and CTP synthetase. Protein Sci. 2007;16:1522–1534. doi: 10.1110/ps.062718507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojcik M, Seidle HF, Bieganowski P, Brenner C. Glutamine-dependent NAD+ synthetase. How a two-domain, three-substrate enzyme avoids waste. J Biol Chem. 2006;281:33395–33402. doi: 10.1074/jbc.M607111200. [DOI] [PubMed] [Google Scholar]
- 37.Mouilleron S, Golinelli-Pimpaneau B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr Opin Struct Biol. 2007;17:653–664. doi: 10.1016/j.sbi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Skoneczna A, Micialkiewicz A, Skoneczny M. Saccharomyces cerevisiae Hsp31p, a stress response protein conferring protection against reactive oxygen species. Free Radical Biol Med. 2007;42:1409–1420. doi: 10.1016/j.freeradbiomed.2007.01.042. [DOI] [PubMed] [Google Scholar]
- 39.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Que LG, Yang Z, Stamler JS, Lugogo NL, Kraft M. S-nitrosoglutathione reductase: An important regulator in human asthma. Am J Respir Crit Care Med. 2009;180:226–231. doi: 10.1164/rccm.200901-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 42.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SO, et al. OxyR: A molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 44.Joksovic PM, Doctor A, Gaston B, Todorovic SM. Functional regulation of T-type calcium channels by S-nitrosothiols in the rat thalamus. J Neurophysiol. 2007;97:2712–2721. doi: 10.1152/jn.00926.2006. [DOI] [PubMed] [Google Scholar]
- 45.McMahon TJ, Stone AE, Bonaventura J, Singel DJ, Stamler JS. Functional coupling of oxygen binding and vasoactivity in S-nitrosohemoglobin. J Biol Chem. 2000;275:16738–16745. doi: 10.1074/jbc.M000532200. [DOI] [PubMed] [Google Scholar]
- 46.Ishima Y, et al. Effects of endogenous ligands on the biological role of human serum albumin in S-nitrosylation. Biochem Biophy Res Comm. 2007;364:790–795. doi: 10.1016/j.bbrc.2007.10.094. [DOI] [PubMed] [Google Scholar]
- 47.Lai TS, et al. Calcium regulates S-nitrosylation, denitrosylation, and activity of tissue transglutaminase. Biochemistry. 2001;40:4904–4910. doi: 10.1021/bi002321t. [DOI] [PubMed] [Google Scholar]
- 48.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci USA. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3:697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- 50.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alonso A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 52.Andersen JN, et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito G, Ariga H, Nakagawa Y, Iwatsubo T. Roles of distinct cysteine residues in S-nitrosylation and dimerization of DJ-1. Biochem Biophys Res Comm. 2006;339:667–672. doi: 10.1016/j.bbrc.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 54.Wilson MA, St Amour CV, Collins JL, Ringe D, Petsko GA. The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily. Proc Natl Acad Sci USA. 2004;101:1531–1536. doi: 10.1073/pnas.0308089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doig AJ, MacArthur MW, Stapley BJ, Thornton JM. Structures of N-termini of helices in proteins. Protein Sci. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model alpha-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J Mol Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 57.Roos G, Loverix S, Geerlings P. Origin of the pKa perturbation of N-terminal cysteine in alpha- and 3(10)-helices: A computational DFT study. J Phys Chem B. 2006;110:557–562. doi: 10.1021/jp0549780. [DOI] [PubMed] [Google Scholar]
- 58.Miranda JJ. Position-dependent interactions between cysteine residues and the helix dipole. Protein Sci. 2003;12:73–81. doi: 10.1110/ps.0224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xian M, Chen X, Liu Z, Wang K, Wang PG. Inhibition of papain by S-nitrosothiols. Formation of mixed disulfides. J Biol Chem. 2000;275:20467–20473. doi: 10.1074/jbc.M001054200. [DOI] [PubMed] [Google Scholar]
- 60.Weichsel A, Brailey JL, Montfort WR. Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry. 2007;46:1219–1227. doi: 10.1021/bi061878r. [DOI] [PubMed] [Google Scholar]
- 61.Forrester MT, et al. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.