Protein folding has run the gamut from complexity, to simplicity, and in a way, back to complexity. In the beginning, there were the slow and complex folding mechanisms of multidomain proteins involving multiple parallel or sequential intermediates en route to the native state (1). Then followed small proteins that can fold by a rapid two-state mechanism (2). Finally, when the single leftover activation barrier was reduced as much as possible through protein engineering, energy landscape roughness was revealed in folding kinetics (3). Landscape roughness temporarily traps very fast folders as they navigate many pathways toward the native state (4). The article by Noé et al. (5) in this issue of PNAS shows how molecular dynamics simulations can be used very efficiently to create a roadmap of the pathways on which a protein explores the energy landscape.
The feat is accomplished through a clever process of stitching pieces of the map together from simulations that each probe is just a small patch of the energy landscape. Noé et al. (5) deftly pulled together a new approach based on decomposing the state space of a protein into a mosaic of substates that are used to combine and correctly weight the independent simulations. The idea is based on Markov modeling, which can avoid picking a reaction coordinate a priori (see refs. 6–10 and references in ref. 5.). The new aspect of Noé et al.'s work is that it shows how to reconstruct the equilibrium ensemble of folding pathways from the piecewise kinetic information and coarse-grain it onto a few major free-energy minima to visualize the results.
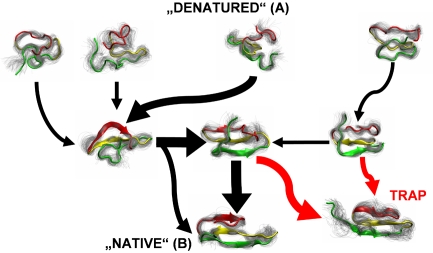
The specific energy landscape mapped out by Noé et al. (5) is that of the Pin WW domain, a fast folding three-stranded β-sheet with two turns and a small hydrophobic core (11). On their map of essential pathways (Fig. 1 shows a simplified version), many roads lead from A to B: unfolded states A have a wide range of structures and travel to the native basin B by a number of sequential and parallel paths. On these paths, the protein takes brief pauses in non-native structural ensembles, which one can think of as very short-lived intermediates. Some of these paths lead into cul-de-sacs (traps), others lead to the folded structure more or less directly, and yet others move proteins in parallel like an efficient superhighway. None lead unfolded WW domain quite straight from A to B.
Fig. 1.
A simplified version of the map of pathways on the energy landscape of Pin WW domain considered by Noé et al. (5). Representative native (B), unfolded (A), intermediate, and trap structures are shown. The thickness of the arrows represents the probability of pathways. Serial, parallel, and cul-de-sac pathways are shown. A full quantitative version of the energy landscape map is given in ref. 5.
In experiments however, the Pin WW domain is an apparent two-state folder (11). Albeit a fast folder, it still takes on average 80 μs at 42 °C to find its way to the native state (11). Even at the higher temperature conditions simulated by Noé et al. (5), it still takes ≈15 μs. That means the main folding barrier is large enough to dominate folding kinetics. The protein climbs from the unfolded state to a transition state where turn 1 forms in the rate-limiting step, then drops toward the native state. Noé et al.'s map (5) shows that ≈70% of the protein form turn 2 rapidly and then turn 1. A respectable 30% proceed in reverse. Experiment and simulation are not necessarily at odds: turn 2 could form rapidly either before or after the rate-limiting step involving turn 1, depending on the road taken. Also, when a protein folds slowly, the minority route might be difficult to detect in an experimental free-energy analysis dominated by a large barrier.
Faster variants of the Pin WW domain such as FiP35 have been made to reveal more of those minority routes seen by Noé et al. (5): when the barrier for formation of turn 1 is lowered sufficiently by re-engineering the amino acid sequence, a fast “molecular phase” appears in the kinetics (12). The molecular phase is caused by rapid diffusion in and out of small valleys on the rough energy landscape. It becomes visible because very fast-folding proteins get to spend a good deal of time all over the free-energy landscape, instead of being stuck in A or B most of the time. Multitrajectory simulations of FiP35 folding directly support multiple paths on a rough free-energy landscape (13). In the faster-folding FiP35 WW domain, turn 1 has been optimized to the point where it can compete with turn 2 in the race to native turn structure.
The rapidly sampled paths on Noé et al.'s (5) roadmap of the energy landscape are in many ways just like the kinetic pathways that connect the metastable intermediates of large proteins, albeit on a much faster time scale. The difference in time scales nicely illustrates the hierarchical nature of the energy landscape (14). When one engineers away the large roadblocks, smaller ones remain to take their place, until we finally reach the thermal energy scale RT, on which all interconversion becomes highly efficient (15, 16).
It should therefore not surprise us that very similar sequences can lead to completely different folds (17) or that a certain protein fold, upon mutation, can be reached through completely different folding mechanisms (18). A “folding mechanism” is simply a free-energy path more prevalent than others because it offers a slightly lower free energy on the way from A to B. Folding populations are exponentially sensitive to free energy thanks to the Boltzmann factor e−ΔG/RT, so a very small difference in free-energy ΔG between two paths is enough for one of them to carry most of the population (18). Yet higher-energy paths remain on the landscape, and mutation through natural selection or protein engineering can bring them down in free energy. Thus, a rough-energy landscape with several alternative paths allows evolution to change protein function, and sometimes even a protein fold, while providing robust routing toward a folded state (16).
Supercomputers are now making many multimicrosecond molecular dynamics possible in explicit solvent for the full protein chain (13). Such simulations involve hundreds of thousands of atoms (19). Millisecond trajectories are now a fait accompli (20), enough to directly map out folding routes of small, very fast-folding proteins with a single trajectory. The utility of patching together free-energy landscapes, however, remains intact even with such powerful trajectories. Although an analysis of the fastest folders is now possible without stitching, the slower wild-type Pin WW domain tackled by Noé et al. (5) is presently out of reach at physiological temperature. It will be within reach soon, but then other, larger, and more complex proteins will take its place and reveal their folding maps to statistical mechanicians and simulators.
Experimental fast folding studies will continue to play a role in refining molecular dynamics force fields as the basis for energy landscape mapping. Fast kinetic data provide benchmarks complementary to the small-molecule thermodynamic data that force fields are currently based on. After all, if we want to predict the dynamics of a large molecule over long time scales, what better calibration than an experiment measuring the dynamics of such a molecule? A key problem that remains to be solved for direct comparison of experiment and simulation: both experimentalists and simulators must strive to find a common ground among folding observables. Q, the number of native contacts, will not be easy to obtain experimentally, nor is a circular dichroism spectrum easily computable except for aromatic amino acid couplets. The time scales and sizes of computed and measured systems now overlap. The observables need to do the same.
When a protein folds slowly, the minority route might be difficult to detect.
Structure prediction of novel protein folds by computational folding kinetics is just around the corner. The recipe is simple: model the unfolded polypeptide on the computer and let it run long enough to refold and unfold several times. Perhaps the main reason no such prediction has been turned in to contests such as CASP (Critical Assessment of Techniques for Protein Structure Prediction) yet (21) is simply that the sequence alone does not guarantee a folding time within the means of current computer power. A modified version of the contest could be useful to entice simulators: supply the sequence and a measured folding relaxation time, so the ab initio simulator can decide whether the sequence lies within available computational means. As a bonus, the computed folding time can be compared with measurement. Eventually, such computations will be fast enough so the folding time can be predicted along with the structure.
Footnotes
The author declares no conflict of interest.
See companion article on page 19011.
References
- 1.Ikai A, Tanford C. Kinetic evidence for incorrectly folded intermediate states in the refolding of denatured proteins. Nature. 1971;230:100–102. doi: 10.1038/230100a0. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SE, Fersht AR. Folding of chymotrypsin inhibitor-2. I. Evidence for a two-state transition. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- 3.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 4.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins Struct Funct Genet. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 5.Noé F, Schütte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Natl Acad Sci USA. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chodera JD, Singhal N, Pande VS, Dill KA, Swope WC. Automatic discovery of metastable states for the construction of Markov models of macromolecular conformational dynamics. J Chem Phys. 2007;126:15501. doi: 10.1063/1.2714538. [DOI] [PubMed] [Google Scholar]
- 7.Singhal N, Snow CD, Pande VS. Using path sampling to build better Markovian state models: Predicting the folding rate and mechanism of a tryptophan zipper beta hairpin. J Chem Phys. 2004;121:415–425. doi: 10.1063/1.1738647. [DOI] [PubMed] [Google Scholar]
- 8.Chodera JD, Dill KA, Swope WC, Pitera JW. Constructing master equation models of protein folding and dynamics from atomistic simulation. Protein Sci. 2004;13(Suppl 1):101–102. [Google Scholar]
- 9.Berezhkovskii A, Hummer G, Szabo A. Reactive flux and folding pathways in network models of coarse-grained protein dynamics. J Chem Phys. 2009;130:205102. doi: 10.1063/1.3139063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noé F, Fischer S. Transition networks for modeling the kinetics of conformational change in macromolecules. Curr Opin Struct Biol. 2008;18:154–162. doi: 10.1016/j.sbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Jäger M, Nguyen H, Crane J, Kelly J, Gruebele M. The folding mechanism of a β-sheet: The WW domain. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 12.Liu F, et al. An experimental survey of the transition between two-state and downhill protein folding scenarios. Proc Natl Acad Sci USA. 2008;105:2369–2374. doi: 10.1073/pnas.0711908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ensign DL, Pande VS. The Fip35 WW domain folds with structural and mechanistic heterogeneity in molecular dynamics simulations. Biophys J. 2009;96:L53–L55. doi: 10.1016/j.bpj.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 15.Kubelka J, Chiu TK, Davies DR, Eaton WA, Hofrichter J. Sub-microsecond protein folding. J Mol Biol. 2006;359:546–553. doi: 10.1016/j.jmb.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 16.Gruebele M. Downhill protein folding: Evolution meets physics. Comptes Rendus Biol. 2005;328:701–712. doi: 10.1016/j.crvi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Alexander PA, He Y, Chen Y, Orban J. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SJ, Matsumura Y, Dumont C, Kihara H, Gruebele M. Slowing down downhill folding: A three-probe study. Biophys J. 2009;97:295–302. doi: 10.1016/j.bpj.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shea J, Brooks CL. From folding theories to folding proteins: A review and assessment of simulation studies of protein folding and unfolding. Annu Rev Phys Chem. 2001;52:499–535. doi: 10.1146/annurev.physchem.52.1.499. [DOI] [PubMed] [Google Scholar]
- 20.Klepeis JL, Lindorff-Larsen K, Dror RO, Shaw DE. Long-time scale molecular dynamics simulations of protein structure and function. Curr Opin Struct Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Moult J, et al. Critical assessment of methods of protein structure prediction: Round VII. Proteins Struct Funct Bioinform. 2007;69:3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]