Abstract
This work evaluates the hypothesis that proteins with an identical supersecondary structure (SSS) share a unique set of residues—SSS-determining residues—even though they may belong to different protein families and have very low sequence similarities. This hypothesis was tested on two groups of sandwich-like proteins (SPs). Proteins in each group have an identical SSS, but their sequence similarity is below the “twilight zone.” To find the SSS-determining residues specific to each group, a unique structure-based algorithm of multiple sequences alignment was developed. The units of alignment are individual strands and loops rather than whole sequences. The algorithm is based on the alignment of residues that form hydrogen bonds between corresponding strands. Structure-based alignment revealed that 30–35% of the positions in the sequences in each group of proteins are “conserved positions” occupied either by hydrophobic-only or hydrophilic-only residues. Moreover, each group of SPs is characterized by a unique set of SSS-determining residues found at the conserved positions. The set of SSS-determining residues has very high sensitivity and specificity for identifying proteins with a corresponding SSS: It is an “amino acid tag” that brands a sequence as having a particular SSS. Thus, the sets of SSS-determining residues can be used to classify proteins and to predict the SSS of a query amino acid sequence.
Keywords: protein prediction, sequence pattern recognition, sequence/structure relation, structure-based sequence alignment
A fundamental principle that governs the sequence–structure relation of proteins states that the native structure of a protein is determined by its amino acid sequence (1, 2). This principle implies that similar sequences encode similar structures. The idea that sequence similarity translates into structural similarity underlies most modern high-accuracy algorithms of structure prediction (3–10). It was shown that proteins tend to share similar 3D structures when their sequence identity exceeds 30% (11). This is an important observation because it provides the threshold for structure prediction and also suggests that a relatively small number of residues in a sequence are critical to structure formation, whereas others play a relatively minor structural role. Thus, even though each residue makes some contribution to 3D structure formation, the relative weights of the contributions vary greatly. Residues conserved across all proteins with a similar 3D structure presumably make a crucial contribution to structure stability.
The goal of this research was to find the residues that play an essential role in supersecondary structure formation (SSS). The reason for focusing on the relation between primary sequence and SSS, rather than on the usually considered relation between sequence and tertiary structure, is that the definition of SSS identity is much more rigorous than the semiquantitative notion of 3D structure similarity. For example, beta sandwich proteins are said to have an identical SSS if they have the same number of strands and the same order (arrangement) of strands in each of their 2 beta sheets. It is important to note that proteins with an identical SSS may differ markedly in the number and composition of residues within strands and loops and that their sequence similarity may be below the “twilight zone.”
This work evaluates the hypothesis that proteins with an identical SSS share a unique set of SSS-determining residues. The residues at conserved positions will be referred to collectively as “SSS-determining residues” because they are presumably determining SSS formation. To prove the hypothesis of uniqueness of SSS-determining residue sets, it is necessary to demonstrate that even markedly dissimilar sequences with the same SSS share the same SSS-determining residues and that this set of residues is not present in sequences with a different SSS. If the hypothesis is true, knowledge of SSS-determining residues would enable one to distinguish sequences of proteins with a particular SSS from all others.
Comparison of sequences and identification of conserved positions require a multiple sequence alignments procedure. The most widely used alignment algorithms, such as PSI-BLAST or HMM, use the dynamic approach to examine numerous variants of alignments and to estimate the number of conserved positions (12, 13). However, when it comes to very low similarities between sequences (less than 10–15% sequence identity), applications of these methods are very complicated and limited (14, 15). It was shown that the PSI-BLAST human-controlled procedure varied for different protein superfamilies and cannot detect all subtle relations between proteins (14).
For proteins with large diversity, structure-based sequence alignment is usually applied instead (16–18). The advantage of using structural data for purposes of alignment is that structure is less susceptible to change than sequence during evolution. On the other hand, comparison of structures is more difficult than that of sequences, because the criteria of assessing structure similarity are not as well defined (19).
Therefore, for comparison of sequences of beta proteins that share the same SSS but belong to different superfamilies and have slight relations, a unique SSS-based multisequence alignment algorithm was developed. Two main features of this algorithm are that (i) units of alignment are individual strands and loops rather than whole sequences and (ii) the alignment of strands is based on the residues that form interstrand hydrogen bonds. The proposed approach makes it possible to align sequences with very low similarity and variable lengths, which would not have been possible using the extant alignment techniques.
The objects of our investigation are 2 groups of sandwich-like proteins (SPs), each defined by a single SSS. Proteins with an identical SSS may differ widely in length and residue content of strands and loops. The alignment algorithm allowed us to identify conserved positions and to describe sets of SSS-determining residues for each of the 2 different sandwich-like SSSs. Each of the 2 SSSs was shown to be characterized by a unique set of SSS-determining residues that is not found in proteins with different SSSs. Thus, each set of SSS-determining residues is a highly sensitive and specific marker for its respective SSS, and hence makes it possible to predict the SSS of a query sequence for which no prior structural information is available.
Results
SSSs of SPs.
In the structural classification of proteins (SCOP) and CATH databases, SPs are defined as 2 beta sheets packed face to face (20, 21). SSSs of these proteins can be rigorously defined by specifying the number and order (arrangement) of strands in each of their 2 beta sheets. Any SPs with the same number and order of strands (in the same orientation) in each beta sheet share the same SSS (Fig. 1). The ability to define the SSS strictly made it possible for us to develop a unique structural classification of SPs, which classifies these proteins in accordance with their SSS (22). Every variant of the SSS of SPs is shown in the publicly accessible “SSS database” (http://binfs.umdnj.edu/sssdb), together with a list of all protein structures that are described by the given SSS variant.
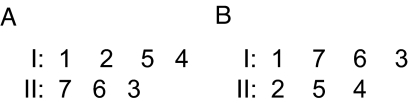
Fig. 1.
Schematic representation of the arrangement of strands in the 2 SSSs. The numbers designate the strands that make up sheets I and II. (A) The SSSs are formed by proteins of Superfamily Ig (Family: C1-set domains), Superfamily E-set domains (Family: Class II viral fusion proteins C-terminal domain), and Superfamily Fibronectin type III (Family: Fibronectin type III) (see motif 2E in SSS database). (B) The SSSs are formed by proteins of Superfamily Ig (Family: I-set domains), Superfamily E-set domains (Families: E-set domains of sugar-using enzymes and Internalin Ig-like domain), Superfamily E-set domains (Family: Internalin), and Superfamily Cadherin (Family: Cadherin) (see motif 3D in SSS database).
Definition of What Constitutes a “Conserved Position” for Purposes of Sequence Alignment of Proteins with Identical SSSs but Dissimilar Sequences.
The goal of a sequence alignment is to maximize the number of conserved positions occupied by identical or chemically similar residues in all aligned sequences. In this research, residue similarity is defined based on whether the residues are hydrophobic or hydrophilic. The reason for selecting hydrophobicity/hydrophilicity as the criterion of conserved positions is because the critical importance of distribution of hydrophobic and hydrophilic amino acids in defining the secondary structures has been demonstrated in a number of studies (23–28). It is therefore plausible to assume that distribution of hydrophobic and hydrophilic residues is largely responsible for SSS as well. Preliminary analysis of residue conservation in SP sequences revealed that residues V, I, L, M, F, W, and C are usually interchangeable at the hydrophobic positions, whereas residues Q, N, E, D, R, K, H, T, S, G, and P are interchangeable at the hydrophilic positions. Thus, a position was classified as “conserved hydrophobic” or “conserved hydrophilic” if all, or almost all, residues found in this position belong either to the hydrophobic or the hydrophilic group. Two residues, A and Y, were found with roughly equal frequency in both hydrophobic conserved positions in strands and in the hydrophilic conserved positions in loops. Therefore, for the purposes of identification of conserved positions in SPs, these 2 residues were considered as hydrophilic if found in loops and as hydrophobic if found in strands.
Set of SSS-Determining Residues for the SSS Shown in Fig. 1A.
According to our analysis of SSSs, as presented in the SSS database, there are 601 SPs with the SSS shown in Fig. 1A. The SCOP classification assigns these proteins to 3 superfamilies and 3 families. Sequences from different superfamilies are strongly dissimilar. For example, for structures 1c5c and 1f42, the European Molecular Biology Open Software Suite (EMBOSS) Needle program for the pairwise sequence global alignment (29) shows 4.5% identity and 7.1% similarity.
Step 1: Selection of Representative Proteins and Their Sequence Alignment.
The selection of representatives is based on SCOP structural classification. The smallest unit in this hierarchical classification is “species.” Proteins from 3 different families with the SSS shown in Fig. 1A belong to 14 different species. For purposes of SSS-based sequence alignments, 10 random sequences from 10 different species were chosen as a “learning set.” The alignment revealed the 30 conserved positions shown in Table S1 (in Supporting Information), of which 19 were hydrophilic and 11 were hydrophobic. Residues at these conserved positions will be referred to as “SSS-determining residues” because they presumably are largely responsible for determining the SSS. SSS-determining residues are shown in Table 1. The syntax of Table 1 is almost identical to that of PROSITE patterns (30). Table 1 also contains information regarding which secondary structure unit any given conservative position is located in (Table 1, top row).
Table 1.
SSS-determining residues for the SSS shown in Fig. 1A
Column “Strand,” SSS-determining residues for the given strand; column “Loop,” SSS-determining residues for the loop between strands.
The residues at lines a1 are obtained from the alignment of the learning set of sequences. The augmented sets of the SSS-determining residues are shown at lines a2. The expressions X and 3X show that the distances between 2 consecutive conserved positions are always the same in all proteins with the same SSS (e.g., 1 residue, 2 residues). The expression ″(d,r)″ X indicates that the minimum number of residues between 2 consecutive conserved positions is ″d″ residues and the maximum number of residues between 2 consecutive conserved positions is ″r″ residues.
Step 2: Testing Specificity and Sensitivity of SSS-Determining Residues.
The goal of this step is to determine whether the set of the SSS-determining residues represents the characteristic fingerprint of all proteins with the given SSS. If this particular set of SSS-determining residues (Table 1, line a1) is highly specific and sensitive for these proteins, scanning the SCOP database that contains sequences of 71,786 diverse structures using this set of residues would lead to the detection of all, or almost all, the proteins with this SSS and none, or few, proteins with a different SSS.
The set of residues obtained in step 1 was input into the EMBOSS/Preg program (29) and used to search the SCOP database. This test revealed 304 of the 601 proteins (“true positives”) and no “false positives.” Thus, the set of residues in Table 1, line a1, is highly specific for the SSS in Fig. 1A but not very sensitive: It identified less than 60% of sequences with the SSS in question. It is therefore probable that the learning set used to derive the residue pattern, which consisted of just 10 sequences, is not sufficiently representative of the wide diversity of sequences with the SSS from Fig. 1A. Therefore, in the next step of the algorithm, the residue content at individual conserved positions was gradually extended so as to increase the sensitivity of the set.
Step 3: Refining the Definition of SSS-Determining Residues.
To obtain an augmented set of SSS-determining residues, the following procedure was suggested. Residues were added step by step to conserved hydrophobic and hydrophilic positions, respectively. At each step, a set of residues is input into the EMBOSS/Preg program and used to rescan the SCOP database to determine whether an “extra” residue changes the specificity of the set. If an additional true-positive sequence is detected, the extra residue is added to the “waiting list” of allowed residues at the given conserved position. After all conserved positions are tested, all residues from the waiting list are added to the conserved positions. Then an augmented set of residues is input into the EMBOSS/Preg program and used to rescan the SCOP database to test the specificity of the set.
The augmented set of SSS-determining residues is presented in Table 1, line a2. When the search was carried out with the augmented residue set, it yielded 573 true-positive sequences out of a total of 601 sequences and no false-positive sequences.
Step 4: The Set of SSS-Determining Residues with a Single Mismatch Position.
To identify additional true positives, scans of the database were carried out using the set of SSS-determining residues shown in Table 1, line a2, but with 1 permitted mismatch: In each scan, the content of 1 of the 30 conserved positions was left unspecified (e.g., any residue was allowed). These 30 additional scans revealed additional 18 true-positive sequences but no false-positive sequences.
Furthermore, it was shown that 6 sequences with the SSS shown in Fig. 1A, which were not detected using augmented sets with 1 mismatched position, have 2 mismatching positions.
The very high sensitivity and 100% specificity of the SSS-determining residues suggest an important conclusion: substitution of a hydrophilic residue for a hydrophobic residue, or vice versa, in residues with the same SSS is allowed at just 1–2 conserved positions.
Set of SSS-Determining Residues for the SSS Shown in Fig. 1B.
The SSS database contains 58 protein structures with the SSS presented in Fig. 1B. In the SCOP database, these proteins are assigned to 3 superfamilies, 4 families, and 11 species (Table S2, legend in Supporting Information). There is a very low similarity of sequences from different families.
Step 1: Selection of Representative Proteins and Sequence Alignment.
Six sequences from 6 species were randomly selected as a learning set (Table S2 in Supporting Information). The alignment revealed 31 hydrophobic and hydrophilic conserved positions. The residue content at each conserved position is shown in Table 2, line a1. These residues comprise the initial set of SSS-determining residues.
Table 2.
SSS-determining residues for the SSSs shown in Fig. 1B
See legend for Table 1. Two augmented sets of the SSS-determining residues are shown at lines a2′ and a2″.
Step 2: Testing Specificity and Sensitivity of SSS-Determining Residues.
Using the EMBOSS/Preg program to scan all sequences in the SCOP databank with the set of residues in Table 2, line a1, disclosed 12 true positives of 58 sequences and no false-positive sequences. Thus, the original set of residues obtained from the analysis of a few representative sequences has low specificity.
Step 3: Refining the Definition of SSS-Determining Residues.
The additional set of SSS-determining residues was obtained in the same way as for proteins with the SSS shown in Fig. 1A. However, the addition of different residues to the list of allowed residues at the conserved positions resulted in an augmented set that had low specificity: The augmented set picked up a number of false-positive sequences. To overcome this problem, the initial set of residues from step 1 was divided into 2 subsets; then, for every subset of residues, the procedure of the expansion of the allowed residue content at the conserved positions was performed independently (Table 2, lines a2′ and a2′′). The augmented subset of SSS-determining residues identified 9 true-positive sequences, and the second augmented subset revealed 18 true-positive sequences.
Step 4: The Set of SSS-Determining Residues with a Single Mismatch Position.
Two subsets were tested independently, allowing for a single mismatch. When the SCOP databank was scanned with augmented subsets and a mismatch at any single conserved position, there were 6 additional true-positive sequences identified with one subset, 25 additional true-positive sequences identified with another subset, and no false-positive sequences. Thus, the combined search with both augmented subsets (with 1 mismatch allowed) had 100% specificity and selectivity: It identified all structures with the SSS shown in Fig. 1B and no structures with any other SSS.
Discussion
Characterization and classification of all existing SSS of SPs and working out of the rules pertaining to the organization of secondary structure units are detailed in our previous publication (22). This paper deals with the next stage of analysis: determination of specific sequence characteristics common to all proteins with a given SSS. We found out fuzzy rules (grammars) that determine the relation between sequence and SSS. The algorithm of alignment is the algorithm of extraction of these rules. The main concept here is the conserved position (the key position), which is defined with some uncertainty (fuzzy position).
It is shown that each of 2 groups of SSSs examined in this work is described by a unique set of conserved hydrophobic and hydrophilic positions, whose residues are decisive for formation of the respective SSSs. This finding implies that not only does amino acid sequence determine protein structure, as shown by Anfinsen over 50 years ago (1, 2), but that there is a way to find residue content at critical positions from SSSs. Thus, the relation between protein sequence and structure is reciprocal.
The residues at the conserved positions are referred to as SSS-determining residues, because their presence is required for the sequence to assume a particular SSS. Thus, residues in sequences may be conceptually divided into 2 groups: a relatively small set of SSS-determining residues and a larger group of all other “supporting” residues. Mutation of SSS-determining residues is generally limited to residues that belong to the same group, either hydrophilic or hydrophobic. By contrast, mutations of supporting residues are much more permissive and interchange of a hydrophobic amino acid for a hydrophilic amino acid, and vice versa, is common.
The concept of structure-determining and supporting residues may help to explain the various exceptions to the rule that more than 30% sequence identity results in structure similarity. Exceptions occur in either direction: sequences with very low residue similarity can have very similar structures (31–33), whereas others with very high sequence similarity can have completely dissimilar 3D structures (34). Assuming the decisive role of just a few key residues for structure formation, we can explain why very similar sequences are structurally dissimilar by positing that they do not share the same set of structure-determining residues. Conversely, even widely dissimilar sequences will fold into similar structures if they contain the same set of structure-determining residues. When comparing sequences with respect to their structure-determining and supporting residues, 4 scenarios are possible:
Both SSS-determining and supporting residues are similar. These proteins have a high degree of overall sequence homology and similar SSS (and, most likely, a similar 3D structure as well).
Sequences share the same set of SSS-determining residues, although among supporting residues, there is a large degree of variability. Proteins have a low total sequence similarity but identical SSS; however, a significant variability in their 3D structure is possible, given the differences in lengths and conformations of loops and strands. In this work, proteins of this kind were studied: those with widely dissimilar sequences attributable to high variability among supporting residues but identical SSS because of the presence of the same SSS-determining residues.
There is little, if any, overlap among SSS-determining residues and high variability among supporting residues. These proteins have very low total sequence similarity and, most likely, different SSSs as well.
There is little, if any, overlap among SSS-determining residues but a high degree of similarity among supporting residues. These proteins are likely to belong to different protein folds despite the high degree of sequence similarities.
A case in point is demonstrated by 2 proteins with 88% sequence identity and yet entirely different tertiary structures: a 3-α–helix fold and a α/β-fold (34). This example illustrates the idea that the fold can be encoded by only 7 amino acids, which constitute just 12% of the sequence. Presumably 7 residues in common between these 2 proteins are the SSS-determining residues, whereas the remaining 49 residues (the supporting residues) “provide a relatively neutral sequence background” (34).
Methods
The Structure-Based Algorithm for Sequence Alignment of Proteins with the Same SSS but Widely Dissimilar Sequences.
The essential feature of the algorithm is that the alignment procedure is performed separately for a set of strands in a beta sheet and for loops rather than the entire sequence. All necessary information about secondary structure and hydrogen bond contacts can be obtained from the SSS database.
Two Rules of Alignment of Residues in Strands.
The alignment of corresponding strands is based on the alignment of the residues that form hydrogen bond contacts between strands in beta sheets. The rules of alignment of residues within a strand are discussed in the next sections.
Rule 1.
If the main chain atoms of residue a and residue a′ form an H-bond in one protein and residue b forms an H-bond with residue b′ in another protein, if a is aligned with b and both are assigned the same position index, a′ will be aligned with b′ and both residues will have a common position index as well.
This rule can be illustrated by the example of structures A and B shown in Fig. 2. Residue a1 in strand 1 of structure A forms an interstrand hydrogen bond with residue a′1 in strand B. There is an analogous pair of residues in structure B, residues b1 and b′1, which forms hydrogen bond contacts between strands 1 and 2. If we align residue a1 with residue b1, rule 1 dictates that residues a′1 and b′1 will also be aligned with each other.
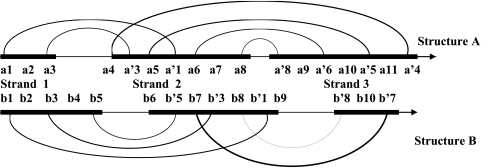
Fig. 2.
Beta sheets with 3 strands in structures A and B. The strands are schematically shown by bold lines. The thin line shows the loops between the strands. The hydrogen bonds between residues are indicated by arcs.
Rule 2.
No gaps are allowed within strands: consecutive residues in a strand are always assigned consecutive position indices.
From these 2 rules, it follows that if residue a1 in Fig. 2 is aligned with residue b1, the immediately downstream residues a2 and a3 in strand 1 of structure A must be aligned with residues b2 and b3 in strand 1 of structure B. Likewise, residues a5 and a′3 in strand 2 of structure A must be aligned with residues b8 and b′3 in strand 2 of structure B. Thus, after initial alignment of a pair of H-bond–forming residues is made, one can systematically invoke the 2 rules to align all residues unambiguously in a beta sheet, as illustrated for residues of strands 1, 2, and 3 in Fig. 3.
Fig. 3.
Sequence alignment is based on hydrogen bond contacts. (A) In this variant, the procedure starts with the alignment of residues a1 and b1. (B) In this variant, the initial selection of residues for alignment is residues a1 and b3.
It is clear from this discussion that alignment of residues depends on the initial choice of H-bonded residues that serve as a “nucleus” of alignment in our approach. Let us consider all possible strand alignments in the beta sheet of structures A and B. In variant 1, shown in Fig. 3A, the initial pair of H-bonded residues, which will serve as a nucleus of alignment, are residues a1 and b1. In variant 2, shown in Fig. 3B, the initial choices are residues a1 and b3. (Note that alignment of residues a1 and b2 is not allowed, because residue a1 is involved in hydrogen bonding, whereas residue b2 is not.) Usually, strands are connected by 2–4 hydrogen bonds in a beta sheet; thus, the total number of possible variants is quite limited—just 2–4 variants per beta sheet. All these possible variants of alignment of strands need to be considered. The “optimal variant” of alignment is the variant that affords the greatest number of conserved positions.
Alignment of Residues in Loops.
The multiple sequence alignment is performed independently for each loop. All sequences in proteins that correspond to loops between strand 1 and 2 are aligned among themselves, and the same procedure is then followed for loops between strands 2 and 3, and so forth. Because conformation of loops may be very variable in different proteins, no structural data are used for loop alignment and multiple sequence alignment of loops was carried out by hand to generate gaps in sequences.
Supplementary Material
Acknowledgments.
We thank Drs. M. Shibata, A. Gorban', A. Koonin, and A. Finkelstein for critical comments and discussions and the Gabriella and Paul Rosenbaum Foundation for continuous encouragement of the research project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909714106/DCSupplemental.
References
- 1.Sela M, White FH, Jr, Anfinsen CB. Reductive cleavage of disulfide bridges in ribonuclease. Science. 1957;125:691–692. doi: 10.1126/science.125.3250.691. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen C. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Bowie JU, Luthy R, Eisenberg DA. Method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 4.Wallner B, Elofsson A. All are not equal: A benchmark of different homology modeling programs. Protein Sci. 2005;14:1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalton J, Jackson R. An evaluation of automated homology modelling methods at low target template sequence similarity. Bioinformatics. 2007;23:1901–1908. doi: 10.1093/bioinformatics/btm262. [DOI] [PubMed] [Google Scholar]
- 6.Misura K, Chivian D, Rohl CA, Kim DE, Baker D. Physically realistic homology models built with ROSETTA can be more accurate than their templates. Proc Natl Acad Sci USA. 2006;103:5361–5366. doi: 10.1073/pnas.0509355103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayeem A, Sitkoff D, Krystek S., Jr A comparative study of available software for high-accuracy homology modeling: From sequence alignments to structural models. Protein Sci. 2006;15:808–824. doi: 10.1110/ps.051892906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopp J, Schwede T. Automated protein structure homology modeling: A progress report. Pharmacogenomics J. 2004;5:405–416. doi: 10.1517/14622416.5.4.405. [DOI] [PubMed] [Google Scholar]
- 9.Xiang Z. Advances in homology protein structure modeling. Curr Protein Pept Sci. 2006;7:217–227. doi: 10.2174/138920306777452312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moult J. A decade of CASP: Progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15:285–289. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Gunalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol. 2006;16:172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durbin R, Eddy SR, Krogh A, Mitchison G. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge, UK: Cambridge Univ Press; 1999. [Google Scholar]
- 14.Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 15.Hill EE, Morea M, Chothia C. Sequence conservation in families whose members have little or no sequence similarity: The four-helical cytokines and cytochromes. J Mol Biol. 2002;322:205–233. doi: 10.1016/s0022-2836(02)00653-8. [DOI] [PubMed] [Google Scholar]
- 16.Konagurthu A, Whisstock J, Stuckey P, Lesk A. MUSTANG: A multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 17.Yang AS, Honig B. An integrated approach to the analysis and modeling of protein sequences and structures. III. A comparative study of sequence conservation in protein structural families using multiple structural alignments. J Mol Biol. 2000;301:691–711. doi: 10.1006/jmbi.2000.3975. [DOI] [PubMed] [Google Scholar]
- 18.Kim C, Lee B. Accuracy of structure-based sequence alignment of automatic methods. BMC Bioinformatics. 2007;8:355–372. doi: 10.1186/1471-2105-8-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye Y, Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]
- 20.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 21.Orengo CA, et al. CATH—A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 22.Chiang Y-S, Gelfand TI, Kister AE, Gelfand IM. New classification of supersecondary structures of sandwich-like proteins uncovers strict patterns of strand assemblage. Proteins. 2007;68:915–921. doi: 10.1002/prot.21473. [DOI] [PubMed] [Google Scholar]
- 23.Silverman BD. Underlying hydrophobic sequence periodicity of protein tertiary structure. J Biomol Struct Dyn. 2005;22:411–423. doi: 10.1080/07391102.2005.10507013. [DOI] [PubMed] [Google Scholar]
- 24.Xiong H, Buckwalter BL, Shieh H-M, Hecht MH. Periodicity of polar and nonpolar amino acids is the major determinant of secondary structure in self-assembling oligomeric peptides. Proc Natl Acad Sci USA. 1995;92:6349–6353. doi: 10.1073/pnas.92.14.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eudes R, Le Tuan K, Delettré J, Mornon JP, Callebaut I. A generalized analysis of hydrophobic and loop clusters within globular protein sequences. BMC Struct Biol. 2007;7:2–24. doi: 10.1186/1472-6807-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandel-Gutfreund Y, Gregoret LM. On the significance of alternating patterns of polar and non-polar residues in beta-strands. J Mol Biol. 2002;323:453–461. doi: 10.1016/s0022-2836(02)00973-7. [DOI] [PubMed] [Google Scholar]
- 27.Woodcock W, Mornon J-P, Henrissat B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992;5:629–635. doi: 10.1093/protein/5.7.629. [DOI] [PubMed] [Google Scholar]
- 28.Avbelj F, Fele L. Role of main-chain electrostatics, hydrophobic effect and side-chain conformational entropy in determining the secondary structure of proteins. J Mol Biol. 1998;279:665–684. doi: 10.1006/jmbi.1998.1792. [DOI] [PubMed] [Google Scholar]
- 29.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 30.Sigrist CJA, et al. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 31.Chothia C, Lesk A. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devos D, Valencia A. Practical limits of function prediction. Proteins. 2000;41(1):98–107. [PubMed] [Google Scholar]
- 33.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol. 2003;333(4):863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 34.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci USA. 2007;104:11963–11968. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.