Abstract
Identification of splice sites is essential for the expression of most eukaryotic genes, allowing accurate splicing of pre-mRNAs. The splice sites are recognized by the splicing machinery based on sequences within the pre-mRNA. Here, we show that the exon sequences at the splice junctions play a significant, previously unrecognized role in the selection of 3′ splice sites during the second step of splicing. The influence of the exon sequences was enhanced by the Prp18 mutant Prp18ΔCR, and the strength of an exon sequence in Prp18ΔCR splicing predicted its effect in wild-type splicing. Analysis of the kinetics of splicing in vitro demonstrated that 3′ splice sites were chosen competitively during the second step, likely at the same time as exon ligation. In wild-type yeast, splice site selection for two genes studied was altered by point mutations in their exon bases, affecting splicing fidelity and alternative splicing. Finally, we note that the degeneracy of the genetic code allows competing 3′ splice sites to be eliminated from coding regions, and we suggest that the evolution of the splicing signals and the genetic code are connected.
Keywords: pre-mRNA splicing, Prp18, splice site selection
Identification of splice sites is necessary both for ensuring the fidelity of pre-mRNA splicing and for the regulation of alternative splicing. Mutations that impair the recognition of the splice sites are responsible for a significant fraction of genetic diseases (1). Splice sites are defined by sequence elements in the pre-mRNA that are recognized by many splicing factors (2). The 3′ splice site is usually the first AG downstream of the branchpoint and is immediately preceded by a pyrimidine or an A (3, 4). A polypyrimidine tract is often present upstream of the splice site. The influence of each element varies and not all are required (5). Recognition of elements in the 3′ splice site region is required early in splicing in metazoans but not in yeast (6).
Identification of the 3′ splice site for exon ligation appears to occur in the second step, and these events are well studied in yeast. After the first transesterification, the RNA helicase Prp16 rearranges the spliceosome (7). Prp18, Slu7, and Prp22 join the spliceosome, which then rapidly ligates the exons (8–10). The U2, U5, and U6 snRNAs and the Prp8 protein, which are required for the first step of splicing, also participate directly in the second step (11, 12).
The 3′ splice site is identified by various mechanisms. The distance between the branch point and the 3′ splice site is important, with distant sites dependent on their polypyrimidine tracts (13, 14). The G's at the 5′ and 3′ ends of the intron interact (13, 15), and base −3 at the 3′ site interacts with base +3 at the 5′ site and the U6 snRNA (16). Prp8 is involved in recognition of both the polypyrimidine tract and the AG dinucleotide at the 3′ splice site (12, 17). Mutation of Slu7 impairs the use of distant sites (18, 19). Analyses of in vivo splicing suggest that the 3′ splice site recognition events occur during the second step, but this conclusion is not well established.
Prp18 stabilizes the interaction of the ends of the exons with loop 1 of the U5 snRNA (20, 21). This function is lost in the Prp18ΔCR protein that lacks a conserved 24-amino acid region (22). In splicing with Prp18ΔCR, the second step is slowed, and its rate depends critically on the exon bases at both the 5′ and 3′ splice junctions. In general, exon sequences that can base-pair more stably to loop 1 of the U5 snRNA facilitate the second step of Prp18ΔCR splicing. The exon sequences have a minimal effect on wild-type splicing (20). Prp18 likely acts together with Prp8 to stabilize interaction of the exons with U5 (23, 24).
In this study we assessed the role of exon sequences at the splice junctions in 3′ splice site selection. We found that the exon sequences are important in both model and natural substrates, suggesting that their role is generally significant, and that their influence is evident only during the second step of splicing.
Results
Splicing of Model Substrates in Vivo.
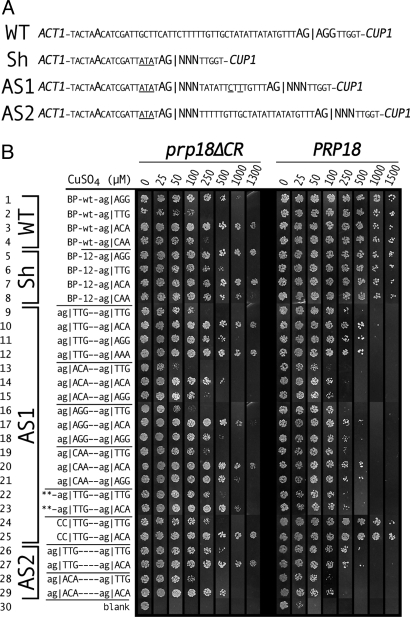
To assess the effect of exon sequences on 3′ splice site selection in yeast, we made ACT1-CUP1 reporters with two 3′ splice sites (Fig. 1A). We placed one 3′ splice site 14 bases from the branch site in an ACT1-like sequence that is used efficiently (19). In the substrate AS1 the second 3′ splice site was placed 19 bases downstream of the first site in a slightly altered ACT1 context. In this model substrate, we found that the splice sites were chosen at similar levels. Only the mRNA resulting from splicing to the downstream 3′ splice site can be translated into Act1-Cup1 fusion protein. Act1-Cup1 protein production is quantitatively measured by copper resistance (25). We used ag|ACA and ag|TTG as examples of strong and weak exon sequences (the | symbol denotes the splice junction), based on their strengths in Prp18ΔCR splicing (20).
Fig. 1.
Copper resistances of reporter yeast with mutant act1-CUP1 plasmids with two 3′ splice sites. (A) Sequences of ACT1-CUP1 plasmids showing the region near the 3′ splice site, emphasizing the branch point and 3′ splice sites. Wild-type ACT1-CUP1 (WT), the shortened Sh substrate, and the alternative splicing substrates AS1 and AS2 are shown. Exon bases shown as NNN were mutated. Base differences between the new plasmids and ACT1 are underlined. (B) Copper resistances conferred on prp18ΔCR (Left) and PRP18-wt (Right) yeast by plasmids based on WT ACT1-CUP1 (1–4), Sh (5–8), AS1 (9–25), or AS2 (26–29). The 3′ splice site(s) is (are) shown at the Left. In plasmids 22 and 23, the last four bases of exon1 were changed from TCTG to AAAA. The figure is a composite of plates from a single experiment with prp18ΔCR yeast and a single experiment with PRP18 yeast. The original colors were individually adjusted for conversion to black and white.
Individually, both 3′ splice sites in AS1 were used efficiently in wild-type yeast and remained sensitive to exon sequence in prp18ΔCR yeast (Fig. 1B, 1–8, 24, 25). In WT ACT1, splicing was sensitive to exon sequence in prp18ΔCR yeast (Fig. 1B Left, 1–4), as reported in ref. 20. Splicing to the upstream site was assayed using the Sh substrate (Fig. 1 A and B, 5–8), and splicing to the downstream site, by blocking the upstream site in AS1 (Fig. 1B, 24–25). In prp18ΔCR yeast, the exon sequences were influential at both sites, but moving the 3′ splice site closer to the branch site improved splicing of weaker substrates (comparing Fig. 1B Left, 1–4 with 5–8 and 24–25), consistent with earlier work (26). In wild-type yeast, the upstream and downstream 3′ sites were used efficiently independent of the exon sequence (Fig. 1B Right), as expected (20).
In prp18ΔCR yeast, the exon sequences at the 3′ splice junctions had a very large effect on alternative splice site choice (Fig. 1B 9–21 and Fig. S1). The example substrates (9) ag|TTG−ag|TTG, (10) ag|TTG–ag|ACA, (13) ag|ACA–ag|TTG, and (14) ag|ACA–ag|ACA conferred copper resistances of 0.1, 1.3, 0.04, and 0.4 mM. With either upstream site, changing the downstream site from weak to strong increased copper resistance by ≈10-fold. With either downstream site, changing the upstream site from weak to strong reduced resistance by 3- to 4-fold, giving an overall maximum change of 30-fold in copper resistance (comparing 10 and 13).
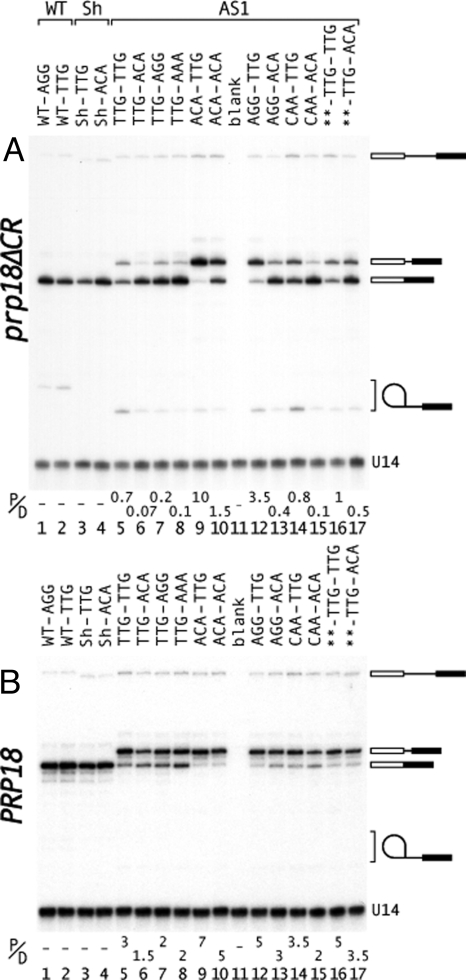
The changes in splice site selection were shown directly by primer extension (Fig. 2A). Altering the downstream splice site from |TTG to |ACA led to both more splicing downstream and less splicing upstream (comparing lanes 5 with 6, and 9 with 10 in Fig. 2A). Likewise, changing the upstream site from |TTG to |ACA shifted splicing to the upstream site (comparing lanes 5 with 9, and 6 with 10 in Fig. 2A). Combining changes at both sites caused a nearly quantitative switch in splicing (Fig. 2A, lanes 6 and 9). The primer extension results showed that the copper resistance is faithfully reporting splicing to the downstream site. The ratio of proximally to distally spliced mRNA (P/D in Fig. 2) is an indicator of relative splicing efficiency (18). For prp18ΔCR yeast (Fig. 2A), the ratio varied by 100-fold and correlated well with copper resistance when splicing efficiency was high but less well when splicing efficiency was low (comparing, for example, lanes 5 and 10 in Fig. 2A). Quantitative interpretation of the in vivo results is complicated by the relative instability of the proximal mRNA due to nonsense-mediated decay (Fig. S2). We interpret the ratio cautiously as an indicator of relative splicing rates.
Fig. 2.
Analysis of alternative 3′ splicing in vivo. Primer extension products of RNA isolated from (A) prp18ΔCR or (B) wild-type yeast are displayed. The exon sequences for substrates based on WT ACT1-CUP1 (lanes 1, 2), Sh (lanes 3, 4), and AS1 (lanes 5–17) are shown at the top (substrates are diagrammed in Fig. 1A). The first three exon bases after each 3′ splice site are shown at the top of each lane. In the pre-mRNAs in lanes 16 and 17, TCTG| at the end of exon1 was replaced with AAAA|. The positions of the pre-mRNA, two alternatively spliced mRNAs, lariat intermediate, and U14 standard are indicated. The lariat intermediate in lanes 3 and 4 comigrated with U14. The ratio of proximally to distally spliced mRNA (P/D) is shown below each lane (average of two measurements).
We were surprised to find that the exon sequences affected splicing of alternative splicing substrates in wild-type yeast, even though the sequences have no effect on wild-type, single-site splicing (Fig. 1B Right, 1–8) (20). The illustrative substrates 9, 10, 13, and 14 conferred resistance to 0.4, 0.8, 0.08, and 0.15 mM copper. Changing the downstream site changed copper resistance twofold, and changing the upstream site, fivefold, giving up to a 10-fold change in copper resistance (comparing 10 and 13 in Fig. 1B Right). The observed effects are in the same direction in prp18ΔCR and wt yeast, but the effects are larger in the mutant. Primer extension showed that the changes in copper resistance were caused by splicing changes (Fig. 2B, lanes 5, 6, 9, and 10). The small increases in downstream splicing on changing |TTG to |ACA (comparing lanes 5 with 6, and 9 with 10, in Fig. 2B) were commensurate with the twofold increases in copper resistance (Fig. 1B, 9, 10, 13, and 14). Strengthening the upstream site caused a more pronounced shift to upstream splicing (Fig. 2B, lanes 5, 6, 9, and 10). The ratio of proximal to distal splicing varied by fivefold in wild-type yeast and correlated well with copper resistance. Overall, the results show that the exon sequences have a significant effect when there is a choice between 3′ splice sites.
The effects of several exon junction sequences were examined. In both wild-type and prp18ΔCR yeast the ordering of strengths of the upstream splice sites, |ACA > |AGG > |CAA ≥ TTG (comparing 9–21 in Fig. 1B), was in accord with that found for single-site splicing in prp18ΔCR yeast (20). Primer extension results (Fig. 2B, lanes 12–15) were consistent with the copper-resistance measurements. At the downstream site, the ordering of strengths, |ACA=|AAA > |AGG=|GAA ≥ |CAA > |TTG (Fig. 1B and Fig. S1), was generally in accord with previous work, with the exception that |CAA was stronger than expected at the downstream site. The overall consistency of our results with expectations based on single-site strengths argues strongly that the changes in splice site choice we observe are dictated by the strength of the adjacent exon sequences. The observation that the exon sequences shift splice site choice in wild-type yeast in the same direction as in prp18ΔCR yeast argues that the mechanistic basis for the shifting is the same in wild-type and prp18ΔCR yeast.
We previously showed that superior 5′ splice sites could compensate for weak 3′ splice sites (20). We replaced the exon sequence at the 5′ splice site with AAAA|gt in the AS1 substrates ag|TTG–ag|TTG and ag|TTG–ag|ACA (Fig. 1B, 22 and 23). AAAA| increased upstream splicing (and reduced downstream splicing) in ag|TTG–ag|ACA (comparing 23 with 10 in Fig. 1B, and lanes 17 and 6 in Fig. 2 A and B) by improving splicing to the upstream |TTG site. With AAAA|gt, the difference between the substrates 22 and 23 is lessened, compared with 9 and 10 (Fig. 1B). The results are consistent with the notion that the relative strengths of the exon sequences determines which site is chosen. The results show that the 5′ splice site can affect the choice of 3′ splice sites.
We tested alternative splicing in the AS2 substrate (Fig. 1A), in which the downstream 3′ splice site was placed at the wild-type distance from the branch point in an ACT1 context. The effects of exon sequence in the AS2 substrate (Fig. 1B, 26–29) were similar to, but slightly smaller than, those in AS1 (see Fig. S1).
Alternative Splicing in Vitro.
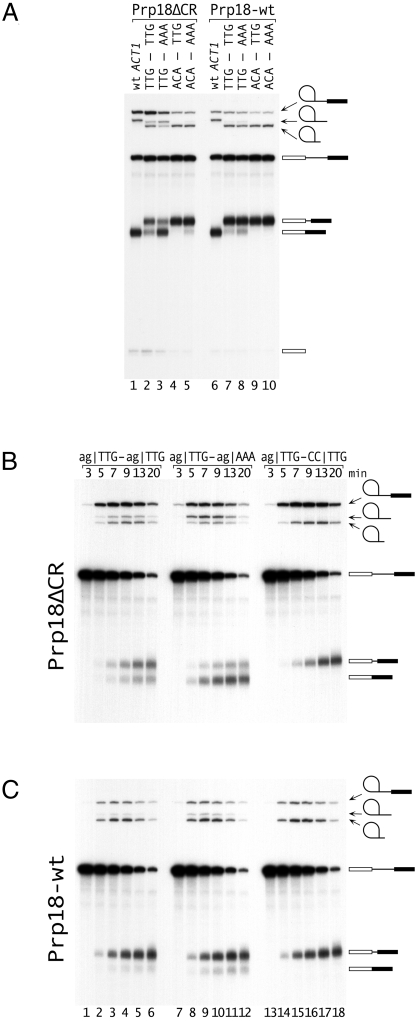
We assayed the splicing of representative AS1 pre-mRNAs in vitro. In Prp18ΔCR splicing (Fig. 3A, lanes 1–5), strengthening the downstream site from |TTG to |AAA resulted in more downstream splicing with either |TTG or |ACA as the upstream site (comparing lanes 2 and 3, or 4 and 5). The 4-fold shift in the ratio of proximal to distal splicing in lanes 2 and 3 was similar to the 6-fold shift seen in vivo (Fig. 2A lanes 5 and 8). Strengthening the upstream sites from |TTG to |ACA shifted splicing sharply to the upstream site (comparing lanes 2 and 4, or 3 and 5). Exchanging the strong and weak exon sequences changed the ratio of proximal to distal splicing by at least 50-fold (lanes 3 and 4). In wild-type splicing (Fig. 3A, lanes 6–10), the effects were smaller; strengthening the downstream site from |TTG to |ACA increased downstream splicing approximately twofold (lanes 7 and 8). Placing |ACA at the upstream site shifted essentially all of the splicing to the that site (lanes 9 and 10). Overall, the in vitro results were similar to the in vivo results. The proximal to distal ratios were higher in vitro. This difference might result from slow (relative to splicing in vivo) assembly of Slu7, Prp18, and/or Prp22 onto the substrate, which is expected to favor the upstream site (9, 19, 26), or from the relative instability of the (untranslatable) proximal mRNA in vivo.
Fig. 3.
Analysis of alternative 3′ splicing in vitro. (A) Splicing of four substrates with alternative 3′ splice sites and wt ACT1 was assayed using extract from a PRP18-knockout strain supplemented with either Prp18ΔCR or Prp18-wt protein. The sequences of the three exon bases at each 3′ splice site are shown at the top; the substrates were based on AS1 (Fig. 1A). The positions of the pre-mRNA, lariat-intron and exon1 intermediates, released lariats (long and short), and alternatively spliced mRNAs are indicated. Splicing was carried out for 15′. (B and C). Kinetics of splicing of three substrates with (B) Prp18ΔCR and (C) Prp18-wt proteins. The 3′ splice sites of the AS1-based substrates are shown at the top of B. In the substrate in lanes 13–18, the downstream AG was mutated to CC. The positions of the RNAs are indicated.
We analyzed the kinetics of splicing of pre-mRNAs with ag|TTG as the upstream site and ag|TTG, ag|AAA, or cc|TTG as the downstream site (Fig. 3 B and C). In Prp18ΔCR splicing (Fig. 3B), splicing of ag|TTG–cc|TTG (lanes 13–18) was slowest, accumulated the most intermediates and yielded the most proximal mRNA. The ag|TTG–ag|AAA pre-mRNA (lanes 7–12) was spliced fastest accumulating the least intermediates and giving the least proximal mRNA. We calculated second-step rate constants (8, 20, 27) using one rate constant to describe the second step for each product (18) (Fig. S3). In Prp18ΔCR splicing (Fig. 3B), upstream splicing had the same rate for all three substrates whereas the downstream rates changed depending on the exon sequence (Table 1). This result implies that the 3′ splice site was chosen competitively during the second step; the reduction in the amount of proximal product was accounted for by more rapid splicing to the distal site. The ratio of the two mRNAs was approximately constant over the reaction course, consistent with a single kinetic step. The wild-type splicing results (Fig. 3C) support the idea of competition between splice sites, although the numbers were less compelling because of the dominance of proximal splicing.
Table 1.
Second-step rate constants
| Prp18 | Splice site | ACT1 | ag|TTG–cc|TTG | ag|TTG–ag|TTG | ag|TTG–ag|ACA |
|---|---|---|---|---|---|
| Wild-type | prox. | 1 | 1.0 | 1.1 | 1.0 |
| dist. | – | 0 | 0.18 | 0.34 | |
| Prp18ΔCR | prox. | 0.23 | 0.13 | 0.13 | 0.12 |
| dist. | – | 0 | 0.06 | 0.22 |
Rate constants for the second step of splicing. The rate constant is the ratio of the rate of formation of mRNA to the amount of intermediate (Fig. S3) (20, 27). Values were calculated from replicates of the experiments in Figures 3B and 3C (20) and were normalized to ACT1 wild-type splicing. Measurements were reproducible to within 10%. prox., proximal; dist., distal.
The primer extension analysis strongly supports the idea that the 3′ splice sites are chosen in a competition that is affected by the exon sequences. In all of the sets of substrates shown in Fig. 2, the amount of product at the upstream site is affected by exon bases at the downstream site, and vice versa. Only a competition can explain the changes in splicing to one site when the other site is altered (14). Previous work showed that the efficiency of splicing in prp18ΔCR yeast is affected by exon bases at the splice junctions (20). However, models in which splice site selection occurs independent of exon sequence, followed by a second step whose efficiency depends on exon sequence are not consistent with the results presented here. In any model consistent with our results, the exon sequences must be involved in the selection of 3′ splice sites, not just in the efficiency of the second step. The in vitro results are explained by a model with a single kinetic step, but more complicated models cannot be excluded.
Effects of Exon Base Changes in Natural pre-mRNAs.
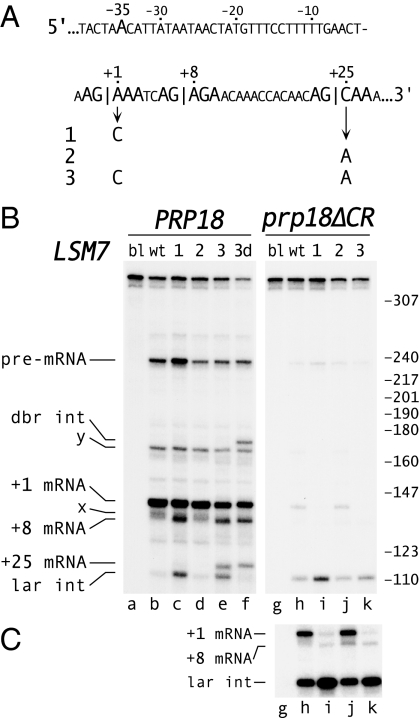
We studied the effects of exon bases on splicing of two natural substrates, LSM7 and REC102. In LSM7 (Fig. 4A) the correct 3′ splice site at +1, aag|AAA, has a strong exon sequence but has an uncommon “a” at base −3. The downstream site at +8 has a strong exon sequence, and 1 of 9 pre-mRNAs in a large-scale cDNA sequencing project was spliced at this site (28). A second potential downstream site at +25 has the weak exon sequence |CAA. We weakened the authentic 3′ splice site by changing base +1 from A to C (Fig. 4A, mutants 1 and 3) and strengthened the +25 site by changing base +25 from C to A (mutants 2 and 3), thereby exchanging the strong |AAA and weak |CAA exon sequences.
Fig. 4.
Effect of exon bases at the splice junctions on fidelity of LSM7 splicing. (A) Sequence of the 3′ splice site region of LSM7, emphasizing the branch site at −35 and two downstream AG dinucleotides. Three LSM7 mutants are shown. (B and C). Primer extension analysis of splicing of wt LSM7 and the three mutants in PRP18 ΔUPF1 and prp18ΔCR ΔUPF1 yeast. Alleles of LSM7 are indicated at the top. B shows a single, uniformly adjusted exposure, and C shows a longer exposure of the prp18ΔCR samples from a separate gel. The positions of the pre-mRNA and mRNAs (numbers denote the first base of exon 2 in the spliced product) are shown at the Left. The positions of the lariat intermediate and debranched intermediate are indicated. To identify the primer extension products, RT-PCR products (using the original primer and a primer at the 5′ end of the RNA) were purified from a polyacrylamide gel and sequenced. The lariat intermediate was identified by treating the RNA from mutant 3 with Dbr1 protein before primer extension, leading to a loss of lariat intermediate and the appearance of a new band of the expected size (lane f). The lengths of the primer extension products agree with their calculated values. The identities of the bands “x” and “y” are unknown; RT-PCR results argue that “x” is distinct from +8 spliced product.
We analyzed the splicing of wild-type and mutant LSM7 in wild-type yeast (Fig. 4B). Primer extension (Fig. 4B, lane b) and RT-PCR/sequencing analysis showed splicing only to the authentic +1 site. Mutation of the A at +1 to C (Fig. 4A, mutant 1) resulted in some splicing to the +8 site and a trace of splicing to the +25 site (Fig. 4B, lane c). Strengthening the +25 site from |CAA to |AAA did not have any effect by itself (Fig. 4 A and B, mutant 2), but when combined with weakening of the +1 site, this mutation resulted in considerable splicing to the +25 site (mutant 3). In mutant 3, 40% of the splicing was to the +8 and +25 sites, which were used approximately equally (Fig. 4B, lane e). The second-step rates, estimated from the ratios of mRNA to intermediate (Fig. 4B), were consistent with notion that the 3′ splice sites were in competition.
In prp18ΔCR yeast, the second step of splicing of LSM7 was slow (Figs. 4 B and C, lane h). Weakening the +1 site greatly reduced wild-type splicing and resulted in very limited splicing to the +8 site (Figs. 4 B and C, lane i). Strengthening the +25 site (lane k) resulted in a trace of splicing to that site, detectable by RT-PCR (Fig. S4), but activation of this site was less than in wild-type yeast. Levels of all mRNAs are depressed in prp18ΔCR yeast (20, 21); our model substrates show that we can interpret splicing of pre-mRNAs independent of this reduction.
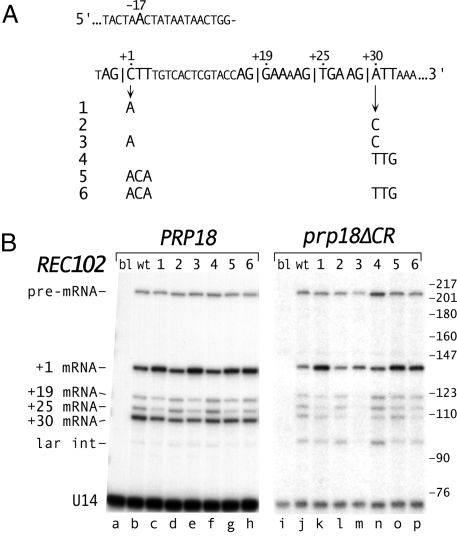
Splicing of the REC102 pre-mRNA uses alternative 3′ splice sites (29, 30). Fig. 5A shows the 4 AG dinucleotides that could be 3′ splice sites. Only the +30 site allows translation of the REC102 mRNA. We strengthened the weak upstream (+1) site, |CTT, and weakened the relatively strong downstream (+30) site, |ATT (Fig. 5A).
Fig. 5.
Effect of exon bases at the splice junctions on REC102 splice site choice. (A) Sequence of REC102 including its 3′ splice sites and branch site. Six mutations at the exon junctions are shown. (B) Primer extension analysis of splicing of REC102. The mutants of REC102 (blank, wt and 1–6) and the PRP18 ΔUPF1 (lanes a–h) and prp18ΔCR ΔUPF1 (lanes i–p) yeast are indicated at the top. The positions of the pre-mRNA, 4 mRNAs (numbers denote the first base of exon 2 in the spliced product), and lariat intermediate are indicated. Primer extension products were annealed to an oligo within exon1 and cut with HpaII before electorphoresis to facilitate separation (Fig. S5). To identify the primer extension products, RT-PCR products (using the original primer and a primer in exon 1) were purified from denaturing polyacrylamide gels and sequenced. The lariat intermediate was identified by treating the RNA from prp18ΔCR yeast with Dbr1 before primer extension (Fig. S5). The lengths of the primer extension products agree with their calculated values. B was assembled from two separate primer extension experiments. In direct comparisons, the amounts of spliced products in prp18ΔCR yeast were two- to fourfold less than in wild-type yeast.
We assayed splicing of wild-type and mutant REC102 by primer extension in both PRP18-wt and prp18ΔCR yeast (Fig. 5B). All 4 potential 3′ splice sites were used in wild-type yeast (Fig. 5B, lane b) with the +1 and +30 sites used approximately equally, and the +19 and +25 sites, ≈5-fold less. Strengthening the +1 site (mutants 1 and 5) increased its utilization at the expense of the other sites (Fig. 5B, lanes c and g). Weakening the +30 site (mutants 2 and 4) reduced its usage somewhat (lanes d and f). Combining the mutations (mutants 3 and 6, lanes e and h) gave the largest shifts in splicing, increasing the ratio of +1 to +30 site usage by threefold. The effects of exon sequences were larger in prp18ΔCR yeast than in wild-type yeast. The +1 site was used preferentially with wild-type REC102 (lane j), and strengthening it increased its usage two- to threefold (lanes k and o). Weakening the +30 site reduced its usage up to threefold (lanes l and n). Combining the mutations (mutants 3 and 6) shifted splicing fivefold (lane p) to >10-fold (lane m) to the +1 site.
Discussion
Our experiments show that the selection of the 3′ splice site is significantly influenced by exon bases at the splice junctions. The choice of splice sites is made competitively during the second step, and the exon sequences play an important role in selection in natural substrates. Previous work showed that exon sequences at both splice junctions significantly affect the rate of the second step of splicing with the mutant Prp18ΔCR protein but have almost no effect on the rate of wild-type splicing (20). Here, in model pre-mRNAs with two 3′ splice sites, we found that the choice of the 3′ splice site in prp18ΔCR splicing was governed by the exon bases at the 3′ splice junction, with nearly quantitative switches in splice site choice with the strongest and weakest exon sequences. The strengths of exon sequences in alternative splicing and in single-site prp18ΔCR splicing were well-correlated, consistent with a common mechanism for both processes. Surprisingly, the exon sequences had a significant effect on 3′ splice site selection by wild-type spliceosomes, changing the ratio of alternatively chosen sites in model substrates severalfold. Analysis of splicing kinetics in vitro showed that the 3′ splice site was chosen in competition during the second step. The dependence of the choice on Prp18 strongly suggests that the choice is coincident with exon ligation. In wild-type yeast, the exon sequences affected splice site choice in natural pre-mRNAs. Mutation of the exon bases at the splice junctions altered the splicing of the pre-mRNAs of LSM7 and REC102, suggesting a general role for these bases in splice site selection.
Exon Sequences and Splice Site Choice.
The exon bases at the splice junctions are an important determinant of 3′ splice site choice, affecting both splicing fidelity and alternative splicing. The activation of the downstream splice sites in LSM7 by the mutations in the exon bases demonstrates the importance of the exon sequences in ensuring the fidelity of LSM7 splicing. Many transcripts will have a potential 3′ splice site—a YAG or AAG—within 25 bases of the correct 3′ splice site. Splicing fidelity is likely ensured by a combination of 3′ splice site determinants, including a pyrimidine tract, the −3 base, and the distance from the branch site (4, 13, 14, 16). For LSM7, point mutations in the exon bases shifted splicing to a site that lacks a pyrimidine tract and is distant (60 bases) from the branch site, suggesting that the exon bases play an important and general role in ensuring the fidelity of splice site choice.
The REC102 results show that exon bases are important in determining alternative 3′ splice site choice. Alternative splicing is uncommon in S. cerevisiae but is common in higher eukaryotes. Despite differences in yeast and metazoan splicing, yeast Prp18 can replace human Prp18 in splicing in vitro (31), suggesting that 3′ splice site identification mechanisms have been conserved. In metazoans, the 3′ splice site region is identified before the first step of splicing, but the 3′ splice site for exon joining is chosen after the first step (2, 5). Exon sequences could affect this second-step selection of 3′ splice sites. Our results could apply to cases where alternative 3′ splice sites are chosen using the same branch site (32), including the closely spaced 3′ splice sites that occur often (33).
Mechanism of Choosing the 3′ Splice Site.
Our results show that the 3′ splice site is chosen competitively during the second step of splicing. The sensitivity of the outcome to Prp18 argues that the choice is made during the Prp18-dependent, ATP-independent stage of the second step during which the second transesterification occurs (8). After the first step of splicing, ATP hydrolysis by Prp16 leads to protection of the 3′ splice site (7). We think that the ATP hydrolysis activates the spliceosome, permitting binding of Slu7, Prp18, and Prp22, and that this second-step spliceosome selects the 3′ splice site and carries out the reaction (similar to the scheme of (9, 10)). This model has a single kinetic step for identifying the 3′ splice site and carrying out the reaction. Our results are inconsistent with mechanisms in which the spliceosome selects a 3′ splice site before binding of Prp18, with Prp18 needed only to facilitate the transesterification. Previous work on 3′ splice site selection in yeast argued against directional scanning mechanisms and in favor of competition (13, 14), in agreement with our conclusions.
Our work shows that Prp18 can influence splice site selection. Wild-type Prp18 mutes the response to different exon sequences, which can cause a 100-fold shift in 3′ splice site choice in prp18ΔCR yeast. The second-step proteins Slu7 and Prp8 also affect 3′ splice site selection in that mutants are sensitive to the branchpoint-3′ splice site distance or the presence of a polypyrimidine tract (17, 18, 34). Slu7, Prp8, Prp22, and Prp18 act in concert during the transesterification stage of the second step (9, 19, 23, 26), and this stage appears to be critical for defining the 3′ splice site.
Competing 3′ Splice Sites and the Genetic Code.
We suggest that the competition between splice sites during the second step may have been a force in evolution. Any AG near the correct splice site can be a competitor because no other bases are required for a 3′ splice site (although GAG is strongly disfavored (3, 4, 16)). Problems would be expected if the encoded protein's function demanded an amino acid encoded by AGN or NAG near a 3′ splice site. However, all such AG-containing codons have substitutes that lack an AG dinucleotide. For NAG, NAA is always a synonymous codon, and this third position degeneracy is typical in the genetic code. For AGN, it is unusual that there are substitute codons; AGY is Ser, which is also encoded by UCN, and AGR is Arg, which is also encoded by CGN. (An AG dinucleotide is required in the rare case in which a selenocysteine UGA codon is followed by a GNN codon, resulting in a weak GAG 3′ splice site.) Thus, an AG dinucleotide is never required for coding, and we suggest that splicing and/or the genetic code may have evolved to avoid potential difficulties with 3′ splice site selection using this degeneracy of the genetic code. Only one other amino acid, Leu, is encoded by different initial bases (CUN and UUR), making CU another avoidable dinucleotide.
The genetic code is thought to have evolved to minimize replication and translation errors (35), and it continues to evolve slowly (36). It is not known when pre-mRNA splicing developed relative to the fixing of the genetic code, but several arguments favor its presence early in evolution (37, 38). We can redirect a modern spliceosome to an incorrect site. We surmise that an ancient spliceosome could be more easily mis-directed, and that this difficulty could have provided sufficient selective pressure to affect the evolution of the genetic code. Alternatively, splicing could have evolved to use AG. The spliceosome can, for example, use AC as the 3′ splice site (15), and the dominant use of AG might reflect evolution to nonconflicting signals.
Materials and Methods
Yeast Strains.
The copper-reporter strain YLC10 (prp18::KANr) was described in ref. 20. UPF1-knockout strains were made by recovering the upf1::KANr fragment from BY4741 (YMR080C::KANr) (Research Genetics) by PCR, transforming it either into DB120 (PRP18/prp18::URA3) (derived from W303 (a/α leu2-3,112 his3-11,15 trp1-1 can1-100 ade2-1 ura3-1)) and dissecting to obtain DB127 (MATα upf1::KANr prp18::URA3), or into W303a to obtain DB129 (upf1::KANr).
Plasmids.
Plasmids bearing act1-CUP1 genes with two 3′ splice sites were made in pGAC14 (25) by replacing the ClaI-KpnI fragment that spans the region with inserts made from oligonucleotides. LSM7 and REC102 were recovered by PCR and cloned into the single-copy vector pYX132 (Novagen) in which transcription is driven by the TPI1 promoter (39). Two additional mutations were made in LSM7, A165G and A166C (numbered relative to the first coding base in the gene), and in REC102, A301C and A302C. These mutations are translationally silent and did not detectably affect splicing of the wild-type substrates. These bases were the 3′ termini of the primers to prevent the wild-type mRNAs from interfering in the extension. Mutations in the LSM7 and REC102 were made by QuikChange Mutatgenesis (Stratagene).
Splicing Assays.
Protocols for measuring copper-resistance and for primer extension, using the ACT1-CUP1 and U14 primers from Siatecka et al. (40), were described in ref. 20. LSM7 and REC102 were tested in DB129 and in DB127 transformed with pRS315-prp18ΔCR. The LSM7 results were similar in wild-type and ΔUPF1 yeast, although the +8 mRNA (Fig. 4A) is not translatable. For REC102, there was an increase in the relative amounts of the +1, +19, and +25 mRNAs (Fig. 5A) compared with the in-frame +30 mRNA in the ΔUPF1 yeast, and we used ΔUPF1 yeast for analysis of LSM7 and REC102 splicing. Primer for LSM7 complemented bases 165–191, and, for REC102, bases 301–322. REC102 cDNAs were treated with RNase A and RNase H, annealed to oligonucleotide complementary to bases 105–127, and digested with HpaII before electrophoresis (Fig. S5). Lariat intermediates were identified by treating the RNA from one mutant with Dbr1 protein (from B. Schwer, Weill Cornell Medical College, New York) (41) before primer extension.
For in vitro splicing, act1 genes were cloned into pDKSa2, and splicing was assayed at 32 °C, as described in ref. 20. We used |AAA as the strong downstream site in vitro because |ACA affected the first step of splicing. For Fig. 3 B and C, aliquots from single reactions were withdrawn at the times indicated at the top of B.
Supplementary Material
Acknowledgments.
We thank Beate Schwer for helpful comments during this work, Michelle Hastings for comments on the manuscript, and Dagmar Bačíková (Uniformed Services University of the Health Sciences, Bethesda) for construction of UPF1 yeast strains. This work was supported by National Institutes of Health Grant GM57267 (to D.S.H.), and by Uniformed Services University of the Health Sciences Grant C017HO (to D.S.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907948106/DCSupplemental.
References
- 1.Lopez-Bigas N, Audit B, Ouzounis C, Parra G, Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:710–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Sheth N, et al. Comprehensive splice-site analysis using comparative genomics. Nucl Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spingola M, Grate L, Haussler D, M Ares J. Genome-wise bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore MJ. Intron recognition comes of AGe. Nat Struc Biol. 2000;7:14–16. doi: 10.1038/71207. [DOI] [PubMed] [Google Scholar]
- 6.Rymond B, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 7.Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horowitz DS, Abelson J. Stages in the second reaction of pre-mRNA splicing: The final step is ATP independent. Genes Dev. 1993;7:320–329. doi: 10.1101/gad.7.2.320. [DOI] [PubMed] [Google Scholar]
- 9.Schwer B, Gross CH. Prp22, a DExH RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James SA, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umen JG, Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 12.Grainger RJ, Beggs JD. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luukkonen BGM, Séraphin B. The role of the branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- 15.Parker R, Siliciano PG. Evidence for an essential non-Watson–Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 16.Collins CA, Guthrie C. Genetic interactions between the 5′ and 3′ splice site consensus sequences and U6 snRNA during the second catalytic step of pre-mRNA splicing. RNA. 2001;7:1845–1854. [PMC free article] [PubMed] [Google Scholar]
- 17.Umen JG, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 18.Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 1992;6:2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- 19.Brys A, Schwer B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 20.Crotti LB, Bačíková D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bačíková D, Horowitz DS. Genetic and functional interaction of evolutionarily conserved regions of the Prp18 protein and the U5 snRNA. Mol Cell Biol. 2005;25:2107–2116. doi: 10.1128/MCB.25.6.2107-2116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bačíková D, Horowitz DS. Mutational analysis identifies two separable roles of the Saccharomyces cerevisiae splicing factor Prp18. RNA. 2002;8:1280–1293. doi: 10.1017/s1355838202023099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronova A, Bačíková D, Crotti LB, Horowitz DS, Schwer B. Functional interactions between Prp8, Prp18, Slu7, and U5 snRNA during the second step of pre-mRNA splicing. RNA. 2007;13:1437–1444. doi: 10.1261/rna.572807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teigelkamp S, Newman AJ, Beggs JD. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Schwer B. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucl Acids Res. 1997;25:2146–2152. doi: 10.1093/nar/25.11.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horowitz DS, Abelson J. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of splicing in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2959–2970. doi: 10.1128/mcb.13.5.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miura F, et al. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–486. doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horowitz DS, Krainer AR. A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev. 1997;11:139–151. doi: 10.1101/gad.11.1.139. [DOI] [PubMed] [Google Scholar]
- 32.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcárcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 33.Hiller M, Platzer M. Widespread and subtle: Alternative splicing at short-distance tandem sites. Trends Genet. 2008;24:246–255. doi: 10.1016/j.tig.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402:207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 35.Crick FHC. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 36.Santos MA, Moura G, Massey SE, Tuite MF. Driving change: The evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Ann Rev Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- 38.Roy SW, Gilbert W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat Rev Gen. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 39.Scott EW, Allison HE, Baker HV. Characterization of TPI gene expression in isogeneic wild-type and gcr1-deletion mutant strains of Saccharomyces cerevisiae. Nucl Acids Res. 1990;18:7099–7107. doi: 10.1093/nar/18.23.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucl Acids Res. 2005;33:6349–6360. doi: 10.1093/nar/gki934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.