Abstract
Perhaps the simplest of biological timing systems, bacteriophage holins accumulate during the phage morphogenesis period and then trigger to permeabilize the cytoplasmic membrane with lethal holes; thus, terminating the infection cycle. Canonical holins form very large holes that allow nonspecific release of fully-folded proteins, but a recently discovered class of holins, the pinholins, make much smaller holes, or pinholes, that serve only to depolarize the membrane. Here, we interrogate the structure of the prototype pinholin by negative-stain transmission electron-microscopy, cysteine-accessibility, and chemical cross-linking, as well as by computational approaches. Together, the results suggest that the pinholin forms symmetric heptameric structures with the hydrophilic surface of one transmembrane domain lining the surface of a central channel ≈15 Å in diameter. The structural model also suggests a rationale for the prehole state of the pinholin, the persistence of which defines the duration of the viral latent period, and for the sensitivity of the holin timing system to the energized state of the membrane.
Keywords: glycine zipper, holin, lysis, SAR domain, thiol modification
The most frequent cytocidal event in the biosphere is the holin-mediated permeabilization of the bacterial membrane that terminates the bacteriophage infection cycle (1). At an allele-specific time, the holin triggers to disrupt the cytoplasmic membrane by the formation of nonspecific holes, the first step in a three-step lytic process involving three other phage proteins, the endolysin and Rz-Rz1, the components of the spanin complex (1, 2). Besides deenergizing the membrane and bringing macromolecular synthesis to a halt, hole-formation allows the destruction of the murein layer by the endolysin, which in turn allows the disruption of the outer membrane by the spanin complex. The entire process is finished within seconds of triggering (3). The canonical holins, such as that of phage λ, form very large holes that allow fully-folded endolysins to pass through the membrane to attack the murein. These holes are nonspecific, allowing passage of unrelated endolysins (4), and large enough for 500-kDa proteins (5).
Recently, an alternative and remarkably different class of holin-endolysin systems has emerged (6–8). This class, represented by the lambdoid bacteriophage 21, utilizes endolysins having novel N-terminal secretory signals called signal-anchor release (SAR) domains. SAR sequences initially act as signal-anchor domains and promote the integration of proteins into the cytoplasmic membrane with type II, N-in, C-out topology (6, 9). Importantly, despite the fact that the muralytic domain of the SAR endolysins is secreted to the periplasm, these enzymes are catalytically inactive in their membrane-tethered state, which prevents premature lysis that would curtail the cytoplasmic assembly of progeny virions. Maintenance of the tethered, inactive state of the endolysin depends on the energized state of the bilayer, and release into the periplasm is quantitative when the holin triggers to disrupt the membrane. Thus, for SAR endolysins, the holin protein need only produce lesions large enough to allow the passage of ions through and depolarization of the cytoplasmic membrane. Indeed, unlike lesions formed by the λ holin, lesions formed by the phage 21 holin did not allow the passage of either λ endolysin or a periplasmic marker-tagged GFP protein (8). The term “pinholin” has been proposed to differentiate the small-hole (pinhole) forming character of the phage 21 holin from the canonical holins that form large, nonspecific holes. A focus on the molecular basis of pinholin function seems warranted, not only because of the intriguing functional differences with the canonical holins, but also because a homolog of the S21 pinholin has been shown to be involved in the programmed release of the Shiga-like toxin by enterohemorrhagic Escherichia coli O157:H7 (10, 11).
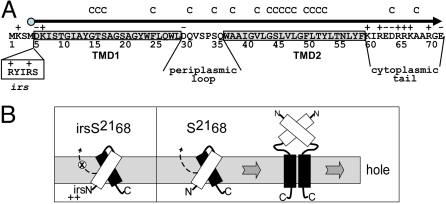
Like the λ holin gene S, the phage 21 holin gene, S21 encodes two proteins, S2171 and S2168, by virtue of alternate translational starts (Fig. 1A); the longer gene product, like that of lambda, has inhibitory character, and thus, is an antiholin (12). Analysis of S2168, an allele that produces only the holin, has shown that despite its small size of 68 residues, the product has two functional domains corresponding to the two predicted transmembrane domains (TMDs) (Fig. 1A). TMD1 is not only dispensable for hole-formation and lysis, but in fact, is itself a SAR domain that must exit the bilayer in order for TMD2 to be competent for hole-formation (Fig. 1B) (7). This property is most dramatically illustrated by the modified allele irsS2168, which encodes the S2168 with the irs epitope, containing two positively-charged residues, added to the N terminus. Because the extra charges prevent TMD1 from exiting the membrane, the irsS2168 protein has an absolute lysis-defective phenotype (7). In this article, we present biochemical, ultrastructural, and computational studies of the elements of S2168 that are involved in the membrane lesion. The results are discussed in terms of a model for the structure of the pinhole and the implications for its temporally-scheduled formation.
Fig. 1.
Features of S2168. (A) Sequence of the S21 reading frame. The 71 residues encoded by the S21 gene are shown, with the S2168 reading frame, which begins with Met4, depicted by the arrowhead line. TMD1 and TMD2 are indicated by shaded boxes, and the regions corresponding to the periplasmic loop and cytoplasmic tail are so labeled. Residues altered to Cys for this study are indicated by C above the sequence. The irs epitope inserted after Met-4 in the irsS2168 variant is shown in the clear box. (B) Model for pinhole formation by S2168. TMD1 (white) is initially in the membrane, bound to TMD2 (black). When TMD1 exits the membrane, TMD2 is able to dimerize and then oligomerize, in the pathway to pinhole formation. TMD1 undergoes homotypic interactions in the periplasm that facilitate, but are not necessary, for pinhole formation. To the left, the inhibitory effect of the irs epitope, with its two positive charges, on the release of TMD1 from the membrane is depicted. Modified from Park et al. (7) with permission.
Results
Detergent-Solubilized S2168 Forms Channel-Like Structures.
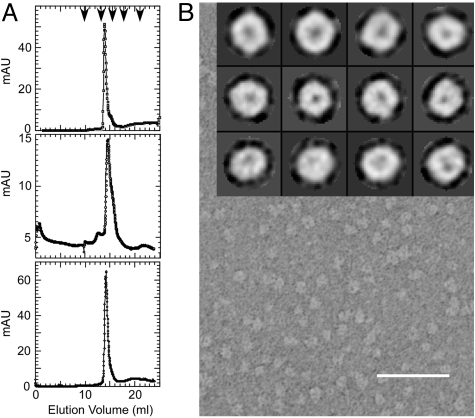
After purification in n-dodecyl-β-D-Maltopyranoside (DDM), S2168 was found to form oligomers with an apparent size of 104 kDa in size-exclusion chromatography (Fig. 2A; Fig. S1). This size corresponds to complexes with 4–7 S2168 molecules, based on a range of ≈100–140 for the aggregation number of DDM (13–16). Electron microscopy of negatively-stained S2168 purified in DDM revealed small doughnut-shaped particles that were reasonably monodisperse (Fig. 2B). Reference-free classification of the S2168 oligomers resulted in roughly spherical class averages that measured between 75 and 80 Å in diameter, with a central stain-filled cavity of ≈15 Å (see top row in Fig. 2B Inset). Some of these classes displayed a roughly hexagonal shape, although no symmetry had been imposed. Examination of structures formed by S2168ΔTMD1 and by lysis-defective irsS2168 resulted in particles with similar class averages. This result was not surprising for irsS2168, because the membrane barrier that restrains the tagged TMD1 is removed by detergent. We suggest that the similarity in the negative-stain images between the oligomer of the full-length holin and that of S2168ΔTMD1 reflects positioning of TMD1 in oligomeric bundles stacked over the pinhole (Fig. 1B).
Fig. 2.
In vitro characterization of his-tagged S2168 and its derivatives. (A) Gel filtration of S2168his and derivatives. Purified protein was applied to a Superdex 200 column and analyzed as described in Materials and Methods. (Top) S2168his; (Middle) S2168ΔTMD1his; (Bottom) irsS2168his. Arrowheads indicate the protein standards, from left to right: 670, 158, 44, 17, and 1.35 kDa. (B) Electron micrograph of negatively stained S2168his purified in DDM. (Scale bar, 500 Å.) (Inset) Single particle averages for S2168his (top row), S2168ΔTMD1his (middle row), and irsS2168his (bottom row). Each single box is 148 × 148 Å.
Hole-Forming Ability of S2168 Alleles.
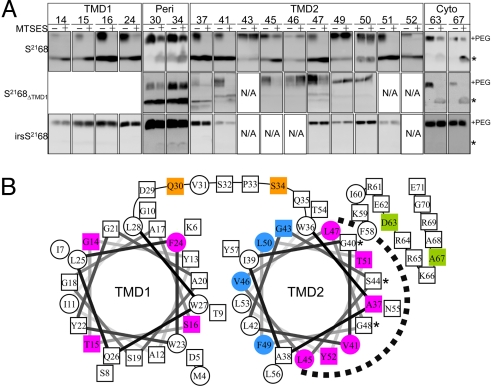
The negative stain images of the purified pinholin suggest an oligomeric structure with a small channel, consistent with its biological function. To probe the structure of the pinholin in the membrane, we used a variation of cysteine-scanning-accessibility. This approach involves treating whole cells expressing the desired S2168 single-cysteine mutant allele with membrane-impermeant thiol reagent (2-sulfonatoethyl) methanethiosulfonate (MTSES). After quenching the reaction, cells are solubilized with SDS/urea, and the extent of MTSES modification is assessed by derivatizing the remaining free cysteines with a high molecular mass (5 kDa) derivative of methoxy-PEG maleimide. For the hole-forming S21 alleles, residues in both the periplasm and cytoplasm should be modified by MTSES, given a luminal diameter of at least the 6 Å needed for passage of the reagent through the bilayer (17). We first examined the accessibility of cysteines located in either the periplasmic loop or the cytoplasmic domain of S2168. As can be seen in Fig. 3A, positions in the periplasmic loop of S2168 (positions 30 and 34) show partial protection, as can be seen from comparing the unmodified species (asterisk) with or without MTSES, or by comparing the PEGylated species (+PEG), with or without MTSES. (The intensities of the unmodified and PEGylated forms cannot be directly compared with each other because of inherent differences in the efficiency of blot transfer and immunostaining.) For both irsS2168 and S2168ΔTMD1, the protection is much less efficient, possibly due to a difference in the membrane proximity of these residues in the different contexts. That is, in the WT protein, the periplasmic location of TMD1 may displace the loop residues from the bilayer. Importantly, the protection of the cytoplasmic C-terminal positions (63 and 67) is efficient for both the hole-forming alleles, S2168 and S2168ΔTMD1, but undetectable for the lysis-defective irsS2168. Also, membrane depolarization by 2,4-dinitrophenol (DNP) has no effect on the irsS2168 C-terminal inaccessibility to MTSES (Fig. S2). Together with previous findings, we conclude that (i) the cytoplasmic membrane of cells expressing irsS2168 retains its impermeability to MTSES, and (ii) the lumen of pinholes formed by both S2168 and S2168ΔTMD1 is large enough to permit the diffusion of MTSES into the cytoplasm.
Fig. 3.
MTSES protection analysis of S2168. (A) Whole cells carrying plasmids encoding single-Cys substitutions of S2168 or its derivatives were induced, treated with the nonpermeant thiol reagent MTSES, subjected to organic denaturation and delipidation, treated with PEG-maleimide, and analyzed by immunoblot, as described in Materials and Methods. The asterisk and “+PEG” indicate the position of the unmodified monomer and the PEGylated species, respectively. MTSES protection is indicated by an MTSES-dependent decrease in the PEGylated species and increase in the unmodified species. NA, not available; Peri, periplasmic loop; Cyto, cytoplasmic tail. (B) MTSES-protected positions in TMD2 map to its most hydrophilic face. Helical projections of TMD1 and TMD2 are shown, with hydrophobic residues as circles and hydroxylated, and hydrophilic residues, as well as Ala and Gly, as squares. Magenta and blue indicate transmembrane positions of S2168 that do or do not show protection by MTSES. Also shown are the positions in the periplasmic loop (orange) and cytoplasmic tail (green) that show MTSES protection. The dashed arc indicates the deduced luminal face of TMD2. Asterisks, GxxxSxxxG motif in TMD2.
Identification of the TMD2 Residues Lining the S2168 Channel.
Because MTSES penetrates the lesion formed by S2168, it was feasible to use the cysteine-scanning-accessibility method outlined above to determine which face of the TMD2 helix faces the lumen of the pinhole. Single cysteines were engineered along the length of TMD2 helices encoded by S2168, irsS2168, and S2168ΔTMD1 (Fig. 1A). With one exception, S2168S44C, none of the mutants changed function. MTSES did not react with any of irsS2168 proteins carrying cysteine substitutions in TMD2, which is consistent with its inability to form holes in the cytoplasmic membrane (Fig. 3A; Fig. S2). By contrast, in both S2168 and S2168ΔTMD1, the same positions lining the hydrophilic face of TMD2 were found to be accessible to MTSES (Fig. 3 A and B), suggesting that these residues face the lumen of the pinhole. The behavior of the lysis-defective mutant S2168S44C confirms the linkage between MTSES-sensitivity and lethal pinhole formation. Although located in the hydrophilic face, S2168S44C lost MTSES-accessibility, even when the membrane was depolarized (Figs. S2 and S3). However, when coexpressed with S2168, triggering occurred very early and it became MTSES-accessible (Fig. S3).
SAR Domain of S2168 Is in the Periplasm.
Next, cysteine was substituted for four residues which would occupy the different helical faces of TMD1. In the S2168 context, all four positions were found to be accessible to MTSES (Fig. 3A). These results extend our previous results that, during the pathway to pinhole-formation, TMD1 becomes protease-sensitive in spheroplasts (7), and indicate that it becomes completely relocated to the aqueous milieu of the periplasm, rather than just to the external face of the cytoplasmic membrane. In contrast, the same positions in the nonlethal irs-tagged derivative were completely insensitive to MTSES, consistent with its retention in the membrane.
Cross-Linking of the Pinholin in the Membrane and in Detergent.
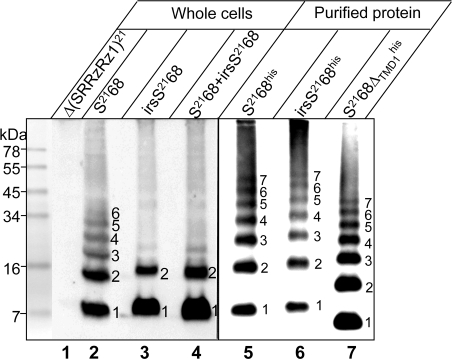
To identify the oligomeric states of S2168 and its derivatives, whole cells expressing S2168 or irsS2168 were treated with the membrane-permeable amine-specific cross-linker, dithiobis[succinimidyl propionate] (DSP) and analyzed by immunoblot using anti-S21 antibodies. For S2168, a cross-linked ladder up to at least hexameric order was obtained (Fig. 4, lane 2). In contrast, oligomerization appeared to be blocked at the dimer stage in cells expressing the nonlethal irsS2168 (lane 3). Also, we have previously shown that the presence of the irs epitope on S2168 not only renders the protein nonfunctional, but also converts it to an antiholin, which blocks the hole-forming activity of S2168 (7). Accordingly, when S2168 and irsS2168 were coexpressed, the formation of cross-linked oligomers of S2168 above the dimeric species was inhibited (lane 4). These data demonstrate that the cross-linking ladder generated from treating whole cells can be correlated with the lytic function of the pinholins present. The DSP cross-linking pattern for purified S2168 in detergent was similar to that obtained from the membrane-embedded protein, except that, including the monomer, seven species were clearly identifiable (Fig. 4, lane 5), each with an Mr corresponding to an integral multiple of the Mr of the monomer (Fig. S4). The same multimer ladder, shifted for the reduced molecular mass of the monomer, was seen for the other lytic allele, S2168ΔTMD1, as well as for the nonlytic irsS2168, which, in the absence of the membrane, has no barrier to pinhole formation (Fig. 4, lanes 6 and 7).
Fig. 4.
DSP cross-linking reveals pinholin oligomerization in vivo and in vitro. (Left) Whole cells carrying prophage λQ21Δ(SRRzRz1)21 only (lane 1), or prophage λQ21Δ(SRRzRz1)21 with either plasmid pS2168 (lane 2), pirsS2168* (lane 3), or prophage λS2168 with the plasmid pirsS2168* (lane 4), as indicated, were induced, treated with the cross-linker DSP, and analyzed by SDS/PAGE and immunoblot, as described in Materials and Methods. (Right) Protein purified from expression of alleles encoding his-tagged S2168 or its derivatives, as indicated, was treated with DSP and analyzed by immunoblot. Oligomeric state indicated for each cross-linked species.
Pinhole Arrangement Is Suggested by Computational Modeling.
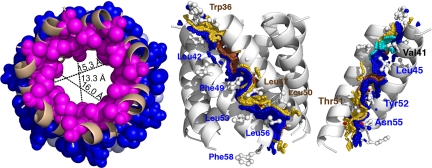
Because its lethality drastically limits the level of protein expression, the pinhole is currently not amenable to high-resolution structural analysis by crystallography or NMR. However, the simplicity of S2168ΔTMD1, comprising little more than a single transmembrane helix, and the fact that it forms holes with the same lumen-facing residues as the full-length holin, suggested a computational approach could be used to complement the biochemical and ultrastructural investigations. Therefore, a sequence corresponding to residues 36–58 of TMD2 was used in a simulated annealing search for closed N-mers, varying N from 5 to 7, and the resulting structures evaluated according to two general criteria. First, the distribution of MTSES-insensitive residues (43, 46, 49, and 50), which should be facing the lipid, and MTSES-sensitive residues (37, 41, 45, 47, 51, and 52), which should be facing the lumen. Second, the size of the predicted transmembrane pore, which must be large enough to allow both passage of MTSES through the membrane and allow the chemistry of MTSES labeling without steric clash. The structures obtained for n = 5–7 all gave lipid-lumen distributions that were compatible with the experimental results, with the exception of Leu47, which, although MTSES-sensitive, was always lipid-exposed, not lumen-facing. However, Leu47 lipid exposure was somewhat attenuated in the heptameric structure (Fig. 5), in that only the delta carbons were lipid-exposed in this bundle, and the remaining atoms of the side chain (e.g., Cβ and Cγ) were involved in helix–helix interactions that are juxtapositioned to the lumen surface of the pinhole. The pore-size criterion significantly favored the heptamer model, where the diameter of the luminal cavity is ≈15.3 Å at Val41, constricting to 13.3 Å at Leu45 in the center, and opening up to 16.0 Å at Thr51 (Fig. 5). These dimensions are significantly larger than in the hexamer (7–13 Å) and pentamer (≈6 Å) (Fig. S5). Because MTSES occupies an ≈12 × 6 Å cylindrical space (18), only the hexamer and heptamer structures are compatible with MTSES permeation, and the hexameric lumen would likely put unacceptable steric constraints on MTSES reactivity toward luminal thiols. Together, the simulated annealing computations favor a model for the functional pinhole based on a heptameric bundle of TMD2 helices, consistent with the results obtained with treating the purified pinholes with the cross-linker DSP (Fig. 4).
Fig. 5.
Computational model for the heptameric pinhole. (Left) Top down view of n = 7 pinhole model with pore distances shown at various depths. MTSES- sensitive and insensitive positions are shown in magenta and blue, respectively. (Center) Side view of n = 7 pinhole model showing the helix–helix interaction contact surface. Contact surface A containing the glycine zipper is shown in gold and labeled in brown; the Gly40, Ser44, and Gly48 contact surface is colored brown. Contact surface B is shown and labeled in blue. (Right) A luminal view of the contact surface, with five helices removed for clarity. Contact surfaces are colored as in Center, except that Val41 is shown in cyan.
Using the energy function in the CHI software suite (see Materials and Methods for details), we identified 20 residues involved in helix–helix interactions in the heptamer, which, by occluded surface analysis (19, 20), led to the assignment of two interaction surfaces could for each helix: surface A, comprising residues W36A37xxG40V41xG43S44xxL47G48xL50T51xxT54; and surface B, comprising residues A38xxV41L42xxL45xxxF49xxY52L53xN55L56xF58 (Fig. 5). Val41 has a unique role in the pinholin, because it has significant interactions on the inner surface of the pore in both the A and B faces. Interestingly, the core of surface A is formed by a triplet of small residues, G40xxxS44xxxG48, which makes a sterically unhindered pocket accommodating the side chains of the bulky residues L45, F49, and Y52. The heptamer structure also displays three interhelical hydrogen bonds between Ser or Thr side chains and backbone carbonyls on different helices: (i) Ser44/Leu45; (ii) Thr51/Tyr52; and (iii) Thr54/Leu56.
Discussion
The pinholins are a new class of holins that forms small holes using only a single TMD (7). Interest in the prototype pinholin, S2168, was piqued by the simplicity of its primary structure, by evidence indicating that its N-terminal TMD was required to exit the bilayer for triggering to occur, and by its near identity with the holin of the prophage 933w, which controls release of Stx toxin during E. coli O157:H7 infections (Fig. S6) (10, 11). Here, we have used structural, biochemical, and computational studies to interrogate the structure of the lethal pinholin lesion, both in the membrane and purified in detergent.
Topological Dynamics and Pinhole Formation in the Membrane.
The picture that TMD1 exits the membrane during the hole-formation pathway has been strongly reinforced here, with the finding that in S2168, which triggers, but not in irsS2168, which does not, residues all around the TMD1 helix become sensitive to MTSES attack in whole cells (Fig. 3A). Also, it was shown that coupled to this topological change in TMD1, positions along one surface of TMD2 and in the C-terminal cytoplasmic tail become accessible to MTSES (Fig. 3A). Not unexpectedly, the surface of S2168 that becomes solvent-exposed is its most hydrophilic surface (Fig. 3B). Although the arc of accessibility begins and ends with Leu residues (L45, L47), all but one (V41) of the other residues on that face are either polar (N55), hydroxylated (S44, T51, and Y52), or of neutral hydrophobicity (A37, G40, and G48). Importantly, protection for most of the lumen-facing residues approaches completion (Fig. 3A), indicating that essentially all of the S2168 protein, which is present in ≈6,400 copies per cell at the time of triggering, based on quantitative immunoblotting with purified S2168 as a standard (Fig. S7), is involved in pinholes. Also, the DSP-cross-linking data clearly indicate that oligomeric complexes of S2168 up to at least hexamer order are present in the membrane after triggering (Fig. 4). In contrast, if TMD1 is prevented from exiting the membrane by the irs tag, thus, blocking pinhole formation, DSP-cross-linking indicates that the protein cannot oligomerize beyond a dimer. Thus, the pinhole formation system seems to be set up for an all-or-nothing response that suddenly forms ≈900 pinholes. Although we do not know the ion flux the pinholes would support, they must be sufficiently large to permit efficient MTSES reaction with luminal thiols, which would certainly require at least 1-nm diameter (18). Because single colicin channels of ≈0.8-nm diameter can depolarize an E. coli cell in a few minutes (21, 22), it seems likely that >900 heptameric pinholes is greatly in excess of what is needed for depolarization within seconds. We suggest that this result reflects the biological mandate for a pinholin to effect instantaneous and complete deenergization of the membrane at the programmed time; thus, causing quantitative activation of the prelocalized SAR endolysins and rapid lysis. It would be counterproductive to form pinholes gradually, which would presumably reduce ATP-generation and the biosynthetic capacity of the infected cell in parallel.
Structure of the Pinhole.
The pinholes are too small to visualize in membranes where triggering has occurred, so our only direct structural information in the biological context comes from the thiol accessibility and cross-linking data described above. However, the pinhole can be purified in DDM as a holo-heptameric complex (Figs. 2 and 4). Also, searches using simulated annealing of TMD2 generated a heptameric model for the pinhole that is consistent with all of these results. Importantly, one of the two heterotypic interaction surfaces in the heptameric model contains the sequence G40xxxS44xxxG48. The motif appears to be critical for pinhole formation, because one of the single Cys substitutions used in this study, S44C, does not trigger. Also, similar motifs are conserved in TMD2 in other holins likely to be pinholins, some not detectably related to S2168 (Fig. S6). Such “glycine-zipper” motifs featuring Gly and other residues with small side chains at intervals of 3 amino acids underlie the heterotypic interactions between TMDs in the other well-studied homo-oligomeric channel proteins: KcsA (tetramer), MscL (pentamer), VacA (hexamer), and MscS (heptamer) (23). In each of these cases, the homo-oligomerizing helix using this motif is the sole TMD that lines the pore, and, as in our model for the heptameric pinhole, the interaction is heterotypic, with the motif embedded in one face that interacts with a different face of the next helix. Last, it must be noted that the GxxxGxxxG motif was first identified in the homotypic dimer interaction surface between the TMDs of glycophorin (24), and has recently been shown to mediate homotypic dimerization of the TMD of myelin protein zero (MPZ), both in membranes and in detergent (25).
Implications for the Prehole State and Holin Triggering.
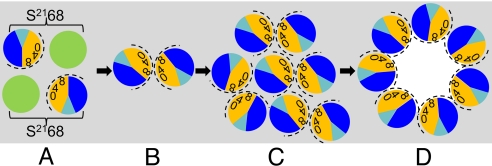
Together, the MTSES-protection patterns, chemical cross-linking data, and computational results suggest a general model for the hole-formation pathway. In this perspective, the pinholins accumulate as dimers, randomly dispersed in the membrane; these dimers are inactive because TMD1 is still in the bilayer (Figs. 1B and 6). We have shown that, in a population of pinholins, TMD1 gradually exits the membrane, with the proportion increasing as the triggering time approaches (7). The release of TMD1 leads to formation of the active dimers (Figs. 1B and 6), with only TMD2 in the bilayer; obviously, the ΔTMD1 protein would accumulate directly in this conformation. In the active dimer, we suppose that the hydrophilic face of TMD2 is sequestered against itself with the G40xxxS44xxxG48 motif supporting a homotypic interface. When a critical concentration of active dimers is reached, formation of 2D aggregates begins (Fig. 6). There is precedent for this model with bacteriorhodopsin (BR). BR accumulates as a monomer in the cytoplasmic membrane until a critical concentration (Cr) is reached, after which the free BR concentration remains constant and the hexagonally-packed BR lattice accumulates. In the pinholin aggregates, triggering would then result from a change in how TMD2 interacts with itself, shifting from the homotypic interaction defining the dimer to the heterotypic A:B interaction, also dependent on the zipper motif. The driving force for this conversion could be hydration of the luminal-side chains, which, in the heptameric model presented here, are mostly hydrophilic.
Fig. 6.
Model for the pinhole formation pathway. View is top-down from periplasm; gray, lipid. TMD1 and TMD2 are shown as green and sectored circles, respectively. In TMD2, orange and dark blue represent A and B interaction faces (Fig. 5), with 0, 4, and 8 indicating the helical positions of the G40, S44, and G48 residues, respectively. The face accessible to MTSES in the pinhole is indicated by the dashed arc. (A) Inactive dimers, with bracket linking cognate TMD1 and TMD2; one possible orientation is shown. (B) Active dimers, after escape of TMD1 from membrane; homotypic interface. (C) Aggregate of active dimers; actual number of dimers is likely to be much larger. (D) Heptameric pinholes after triggering; heterotypic interface.
In this pathway, the prehole conformation is essential for pinhole formation, rather than just an inactive state. It should be noted that the S44C substitution is inactive in hole-formation, but can be recruited into pinholes by functional S2168 (Fig. S3). We suggest that S44C disrupts the prehole active dimer by poisoning the homotypic interface, as it should in the central position of the GxxxSxxxG motif, but can be accommodated in the heterotypic interface. Thus, the mutant protein would be incorporated into holes by being a proximal monomeric bystander when the concerted triggering event occurs.
This model does not directly address the sensitivity to the energized state of the membrane, which might oppose the hydration step, stabilize the GxxxSxxxG interaction, or destabilize the heterotypic interhelical interactions in the pinhole. In this regard, the helices of the modeled heptamer are aligned at an angle of 34° to the normal, a geometry that might be strongly affected by the presence of the membrane potential (Fig. 5). The sensitivity to the proton motive force (pmf) is a key component of the biological function of the pinholin, and indeed all holins, because it confers “all or nothing” character. In our rationale, the spontaneous conversion of a single aggregate of pinholin dimers to a pinhole leads immediately to pmf collapse, assuring coordinated holin triggering throughout the whole cell. In any case, the structural and biological model presented here provides a framework for future experiments to examine the working parts of this simple, universal, and ancient biological timing system.
Materials and Methods
General Methods.
Bacterial culture growth, plasmids and prophage construction, and general DNA manipulations are described in SI Materials and Methods. In general, S2168 was expressed from plasmid pS2168, which carries the modified phage 21 lysis gene cassette, genes (SRRzRz1)21 (26) under its native promoter pR′21. The other lysis genes were inactivated by nonsense mutations, so that only S2168 is expressed. Similarly, the irsS2168 and S2168ΔTMD1 alleles are expressed from plasmids pirsS2168 and pS2168ΔTMD1, respectively, which are of the same construction as pS2168, except that the promoter is pR′ from phage lambda. The pS2168 is transactivated by the phage 21 late gene activator, Q21, provided by the thermal-inducible prophage λQ21Δ(SRRzRz1)21. Both pirsS2168 and pS2168ΔTMD1 are transactivated by lambda Q, provided by the plasmid pQ, a low-copy plasmid in which Q is under the control of an IPTG-arabinose inducible promoter (27). For experiments requiring coexpression of the WT S2168 with certain derivative alleles, prophage λS2168 was used to provide one S2168 allele and Q21, which trans-activates plasmids pS2168a or plasmid pirsS2168*. Single-cysteine substitutions were inserted in corresponding plasmids by site-directed mutagenesis. Bacterial strains, prophages and plasmids are listed in Table S1. Structures of key plasmids and phages are in Fig. S8.
Purification and Gel Filtration Chromatography of S2168his and Its Variants.
For all experiments involving purified protein, alleles encoding his-tagged S21 gene products were used, with the sequence GGH6GG inserted between residues 66 and 67 (7). The presence of the oligohistidine tag had no effect on function or oligomerization (Fig. S9). S2168his, S2168ΔTMD1his, and irsS2168his were purified from an induced culture of E. coli C43(DE3) (28), carrying the plasmid pETS2168his, pETS2168ΔTMD1his, and pETirsS2168his, respectively. Induction, detergent extraction, and purification of the his-tagged proteins by immobilized metal affinity chromatography have been described (29) and can be found in SI Materials and Methods. S2168his and its variants were eluted in buffer containing 20 mM Tris, pH 7.9/150 mM NaCl/500 mM imidazole/0.1% (wt/vol) DDM. Purified protein samples were analyzed by gel filtration using a Superdex 200 10/300 GL column (Amersham Pharmacia) on an AKTA FPLC workstation (Amersham Pharmacia) (SI Materials and Methods). Protein standards were purchased from Bio-Rad and used according to the manufacturer's instructions.
Electron Microscopy.
Purified protein at 0.5 μM was applied to freshly glow-discharged formvar-carbon coated grids and stained with 2% (wt/vol) aqueous uranyl acetate according to the method of Valentine et al. (30). Micrographs were recorded on a JEOL 1200EX TEM operating at 100 KV, with a calibrated magnification of 48,600×, and scanned on a Leafscan 45 microdensitometer to a final sampling size of 4.12 Å per pixel at the specimen level. ≈900 particles for S2168his, 1,100 particles for S2168ΔTMD1his, and 710 particles for irsS2168his were selected manually, low-pass filtered to remove high-frequencies beyond 10 Å, and subjected to iterative, reference-free classification using the refine2d.py command in the EMAN (31) software package.
Cysteine Modification.
Cultures were grown to A550 = 0.4, induced with IPTG/arabinose or by a thermal shift, as appropriate, and aerated past the time of holin triggering or, in the case of nonlethal S2168 alleles, for 50 min. Cells corresponding to 0.25 A550 units were collected by centrifugation, washed twice with 1 mL PB (50 mM phosphate buffer, pH 7), and then resuspended in 0.25 mL of PB. Each sample was divided into two 125 μL aliquots. To one, 10 mM MTSES (Anatrace) was added and to the other, an equivalent amount of water was added. After 30 min at room temperature, 50 mM L-cysteine was added to quench any unreacted MTSES. After 10 min, the cells were diluted by the addition of 0.75 mL PB, collected by centrifugation, washed twice with 1 mL PB, resuspended in 100 μL PB, and extracted with of 750 μL chloroform:methanol:water (1:4:1). After incubation on ice for 30 min, the denatured and delipidated proteins were collected by centrifugation at 13,000 × g for 5 min at 4 °C. The protein pellets were washed once with 400 μL 95% methanol and resuspended in 100 μL PEGylation buffer [10M urea/1% SDS/1 mM EDTA/0.6 M Tris, pH 7; adapted from Lu and Deutsch (32)]. Fifty μL of each sample was transferred to a clean tube and treated with 0.2 mM mPEG-maleimide (Creative Biochem) for 30 min at room temperature, and then precipitated with 1 mL cold ethanol. After overnight at −20 °C, proteins were collected by centrifugation at 13,000 × g for 15 min at 4 °C. The pellets were air-dried and resuspended in sample loading buffer for analysis by SDS/PAGE (33). For experiments involving depolarization of the membrane, 1 mM DNP was added into the culture at the time of harvesting and used to supplement PB in each step until quenching with L-cysteine.
Chemical Cross-Linking.
A culture volume corresponding to one A550 unit was harvested, washed and resuspended in 640 μL of PBS (PBS, containing 0.1 M sodium phosphate/0.15 M NaCl, pH 7.2), and treated with 2 mM DSP (Pierce) at room temperature for 30 min. The reaction was quenched by the addition of 20 mM Tris, pH 7.5, and the proteins were collected by precipitation with trichloroacetic acid for analysis by SDS/PAGE. For the DSP cross-linking of purified S2168his or its variants, 1 pmol of each protein in 0.1% DDM was used. The reaction was mixed with 2× sample loading buffer directly after quenching, boiled for 5 min, and analyzed by SDS/PAGE.
SDS/PAGE and Western Blotting.
SDS/PAGE and Western blotting was performed as described (7), and can be found in SI Materials and Methods; 10% Tris-Tricine gels were used to separate protein samples. An antibody raised in rabbit against the S21 C-terminal peptide KIREDRRKAARGE was used as primary antibody to detect S21 protein variants (12). Horseradish peroxidase-conjugated goat-anti-rabbit secondary antibody was from Pierce.
Computational Methods.
A 24-residue sequence corresponding to residues 36–58 of the S21 TMD2 (WAAIGVLGSLVLGFLTYLTNLYF; Fig. 1A) was used to compute oligomeric pinholin structures using CHI. The simulated annealing search calculation protocols have previously been described in detail (34, 35), except that the CHI suite of scripts uses CNS to implement the molecular dynamics. The OPLS topology and parameter sets were used with all polar hydrogens included (36). CHI uses an implicit membrane solvent with a dielectric constant = 2.
Conformations of TMD2 oligomers ranging from n = 4–7 were independently generated in different computational searches. Each oligomeric state was symmetrically searched with a helix rotation angle, α, ranging from 0° to 360°, and a sampling step size of 5°, starting with both left- (Ω = 45°) and right-handed crossing angles (Ω = −45°). For each oligomer, four trials were carried out for each rotation and crossing angle conformation using simulated annealing of all atomic coordinates during which rotation- and crossing-angles were free to vary. The resulting 576 structures were analyzed to determine the final rotation and crossing angles. Clusters of low-energy structures were calculated by determining the frequency of structures appearing in a particular region of interaction space using a cutoff of 1.0-Å rmsd and a minimum requirement of 10 structures to define a cluster. An average structure for each cluster was calculated and evaluated for consistency with the experimental data on the MTSES sensitivity and pore size from permeability.
The A- and B- helix–helix interaction surfaces were identified for the experimentally compatible oligomeric structures by calculating the distribution of interchain interactions using the occluded surface algorithm (19, 20). The occluded surface can be thought of as the contacting molecular surface between two helices and is represented in Fig. 5 as a dot surface. Hydrogen bonding interactions were calculated using HBPLUS (37).
Supplementary Material
Acknowledgments.
This work was supported by the Public Health Service Grant GM27099 (to R.Y.), National Science Foundation Grant MCB0423807 (to K.G.F.), the Robert A. Welch Foundation, and the Program for Membrane Structure and Function at Texas A&M University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907941106/DCSupplemental.
References
- 1.Young R, Wang IN. Phage Lysis. In: Calendar R, editor. The Bacteriophages. 2nd Ed. Oxford: Oxford Univ Press; pp. 104–126. [Google Scholar]
- 2.Berry J, Summer EJ, Struck DK, Young R. The final step in the phage infection cycle: The Rz and Rz1 lysis proteins link the inner and outer membranes. Mol Microbiol. 2008;70:341–351. doi: 10.1111/j.1365-2958.2008.06408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young R. Bacteriophage holins: Deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. [PubMed] [Google Scholar]
- 4.Bläsi U, Young R. Two beginnings for a single purpose: The dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang IN, Deaton J, Young R. Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J Bacteriol. 2003;185:779–787. doi: 10.1128/JB.185.3.779-787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M, Struck DK, Deaton J, Wang IN, Young R. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc Natl Acad Sci USA. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park T, Struck DK, Deaton JF, Young R. Topological dynamics of holins in programmed bacterial lysis. Proc Natl Acad Sci USA. 2006;103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park T, Struck DK, Dankenbring CA, Young R. The pinholin of lambdoid phage 21: Control of lysis by membrane depolarization. J Bacteriol. 2007;189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu M, et al. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- 10.Wagner PL, et al. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol Microbiol. 2002;44:957–970. doi: 10.1046/j.1365-2958.2002.02950.x. [DOI] [PubMed] [Google Scholar]
- 11.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barenboim M, Chang CY, dib Hajj F, Young R. Characterization of the dual start motif of a class II holin gene. Mol Microbiol. 1999;32:715–727. doi: 10.1046/j.1365-2958.1999.01385.x. [DOI] [PubMed] [Google Scholar]
- 13.Rosevear P, VanAken T, Baxter J, Ferguson-Miller S. Alkyl glycoside detergents: A simpler synthesis and their effects on kinetic and physical properties of cytochrome c oxidase. Biochemistry. 1980;19:4108–4115. doi: 10.1021/bi00558a032. [DOI] [PubMed] [Google Scholar]
- 14.VanAken T, Foxall-VanAken S, Castleman S, Ferguson-Miller S. Alkyl glycoside detergents: Synthesis and applications to the study of membrane proteins. Methods Enzymol. 1986;125:27–35. doi: 10.1016/s0076-6879(86)25005-3. [DOI] [PubMed] [Google Scholar]
- 15.Aoudia M, Zana R. Aggregation behavior of sugar surfactants in aqueous solutions: Effects of temperature and the addition of nonionic polymers. J Colloid Interface Sci. 1998;206:158–167. doi: 10.1006/jcis.1998.5627. [DOI] [PubMed] [Google Scholar]
- 16.Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan RS, et al. The yeast mitochondrial citrate transport protein. Probing the secondary structure of transmembrane domain iv and identification of residues that likely comprise a portion of the citrate translocation pathway. J Biol Chem. 2000;275:12009–12016. doi: 10.1074/jbc.275.16.12009. [DOI] [PubMed] [Google Scholar]
- 19.Pattabiraman N, Ward KB, Fleming PJ. Occluded molecular surface: Analysis of protein packing. J Mol Recognit. 1995;8:334–344. doi: 10.1002/jmr.300080603. [DOI] [PubMed] [Google Scholar]
- 20.Fleming KG, Engelman DM. Computation and mutagenesis suggest a structure for the synaptobrevin transmembrane dimer. Proteins. 2001;45:313–317. doi: 10.1002/prot.1151. [DOI] [PubMed] [Google Scholar]
- 21.Schein SJ, Kagan BL, Finkelstein A. Colicin K acts by forming voltage-dependent channels in phospholipid bilayer membranes. Nature. 1978;276:159–163. doi: 10.1038/276159a0. [DOI] [PubMed] [Google Scholar]
- 22.Davidson VL, Brunden KR, Cramer WA, Cohen FS. Studies on the mechanism of action of channel-forming colicins using artificial membranes. J Membrane Biol. 1984;79:105–118. doi: 10.1007/BF01872115. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, et al. Transmembrane glycine zippers: Physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 25.Plotkowski ML, et al. Transmembrane domain of myelin protein zero can form dimers: Possible implications for myelin construction. Biochemistry. 2007;46:12164–12173. doi: 10.1021/bi701066h. [DOI] [PubMed] [Google Scholar]
- 26.Bonovich MT, Young R. Dual start motif in two lambdoid S genes unrelated to lambda S. J Bacteriol. 1991;173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gründling A, Manson MD, Young R. Holins kill without warning. Proc Natl Acad Sci USA. 2001;98:9348–9352. doi: 10.1073/pnas.151247598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 29.Savva CG, et al. The holin of bacteriophage lambda forms rings with large diameter. Mol Microbiol. 2008;69:784–793. doi: 10.1111/j.1365-2958.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valentine RC, Shapiro BM, Stadtman ER. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry. 1968;7:2143–2152. doi: 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- 31.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Deutsch C. Pegylation: A method for assessing topological accessibilities in Kv1.3. Biochemistry. 2001;40:13288–13301. doi: 10.1021/bi0107647. [DOI] [PubMed] [Google Scholar]
- 33.Tran TA, Struck DK, Young R. The T4 RI antiholin has an N-terminal signal anchor release domain that targets it for degradation by DegP. J Bacteriol. 2007;189:7618–7625. doi: 10.1128/JB.00854-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Arkin IT, Engelman DM, Brunger AT. Computational searching and mutagenesis suggest a structure for the pentameric transmembrane domain of phospholamban. Nat Struct Biol. 1995;2:154–162. doi: 10.1038/nsb0295-154. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Engelman DM, Brunger AT. Improved prediction for the structure of the dimeric transmembrane domain of glycophorin A obtained through global searching. Proteins. 1996;26:257–261. doi: 10.1002/(SICI)1097-0134(199611)26:3<257::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 37.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.