Abstract
M2 is a double-stranded RNA (dsRNA) element occurring in the hypovirulent isolate Rhs 1A1 of the plant pathogenic basidiomycete Rhizoctonia solani. Rhs 1A1 originated as a sector of the virulent field isolate Rhs 1AP, which contains no detectable amount of the M2 dsRNA. The complete sequence (3,570 bp) of the M2 dsRNA has been determined. A 6.9-kbp segment of total DNA from either Rhs 1A1 or Rhs 1AP hybridizes with an M2-specific cDNA probe. The sequences of M2 dsRNA and of PCR products generated from Rhs 1A1 total DNA were found to be identical. Thus this report describes a fungal host containing full-length DNA copies of a dsRNA element. A major portion of the M2 dsRNA is located in the cytoplasm, whereas a smaller amount is found in mitochondria. Based on either the universal or the mitochondrial genetic code of filamentous fungi, one strand of M2 encodes a putative protein of 754 amino acids. The resulting polypeptide has all four motifs of a dsRNA viral RNA-dependent RNA polymerase (RDRP) and is phylogenetically related to the RDRP of a mitochondrial dsRNA associated with hypovirulence in strain NB631 of Cryphonectria parasitica, incitant of chestnut blight. This polypeptide also has significant sequence similarity with two domains of a pentafunctional polypeptide, which catalyzes the five central steps of the shikimate pathway in yeast and filamentous fungi.
Keywords: RNA-dependent RNA polymerase
In the last three decades, fungal double-stranded RNA (dsRNA) elements have been the subject of considerable research because of their potential adverse effects on plant pathogenic fungi, and the prospects of utilizing them in biocontrol schemes against the host fungus (1–3). dsRNAs have been associated with cytoplasmic hypovirulence (4) or virulence (5) in Rhizoctonia solani. We have shown that dsRNAs are ubiquitous in natural R. solani populations that include isolates covering a wide range of virulence, including hypovirulence (6). Subsequent surveys conducted in Japan (7), Florida (8), and Louisiana (9) confirmed our findings. We have also shown that the conflicting reports on the role of dsRNAs in R. solani could be attributed, at least in part, to the high degree of genetic diversity among dsRNA elements occurring in natural populations of the pathogen (10, 11). More importantly, we have presented several lines of indirect evidence suggesting that specific dsRNA elements might be involved in up- or down-regulation of virulence in R. solani (10–14).
Recently, we described a genetic model wherein new dsRNAs appear or existing dsRNAs disappear from a given genotype (Rhs 1AP) with concomitant changes in virulence (15). Rhs 1AP is a virulent culture, member of anastomosis group 3, which is the major cause of the rhizoctonia disease syndrome of potato in North America (16, 17). Rhs 1A1 originated as a sector of Rhs 1AP and contains, in addition to the two dsRNAs of Rhs 1AP (L2 and M1), three genetically distinct dsRNAs (L1, M2, and S1). Molecular sizes of L1, L2, M1, M2, and S1 dsRNAs were estimated to be 25, 23, 6.4, 3.6, and 1.2 kbp, respectively (15). Rhs 1A1 is hypovirulent and has been shown to be an effective biocontrol agent against virulent R. solani in the field. It also has plant growth-promoting properties (18, 19).
To understand the nature and role of dsRNA in R. solani, we have constructed cDNA clones of the above dsRNAs for sequencing and transfection studies (15). In this paper, we report that the M2 element is phylogenetically related to a hypovirulence-associated mitochondrial dsRNA from the ascomycete Cryphonectria parasitica. It also has significant sequence similarity with polypeptides of known cellular functions. More importantly, M2-related DNA sequences occur in both the M2-containing culture Rhs 1A1 and the parental Rhs 1AP. This is a feature not reported to date for other fungal dsRNA genetic elements.
MATERIALS AND METHODS
Cloning and sequencing.
Construction of a cDNA library of the M2 dsRNA has been described previously (15). Sequencing of the M2 specific cDNA clones was carried out by primer walking from both sides of two nearly full-length clones, M2–31 (bases 63–3,556, Fig. 1) and M2–53 (bases 170–3,557), using the dideoxy chain termination method (20) and the Sequenase kit, version 2 (United States Biochemical-Amersham). Initial sequence information was verified by incorporating dITP in the sequencing protocol. In addition, many regions of the cDNA clones were sequenced from both strands and verified by cycle sequencing of cloned DNA PCR or reverse transcription (RT)–PCR products with an automated DNA sequencer (Applied Biosystems, ABI373A). The terminal sequences were derived by sequencing of the M2 dsRNA using an avian myeloblastosis virus reverse transcriptase sequencing kit (United States Biochemical-Amersham), and verified by sequencing cDNA clones obtained by amplification of cDNA copies of the M2 dsRNA ends using the 5′ Rapid Amplification of cDNA Ends kit (Life Technologies, Gaithersburg, MD). The presence of circular or oligomeric forms of M2 was indicated by inverse RT-PCR (21) of a dsRNA-enriched sample from Rhs 1A1. The two primers used for inverse RT-PCR, P14 (5′-CTCACGTAATAGAACCTCCA) and P15 (5′-GTCCGATCTTAACCTCCATA), were obtained from the terminal sequences of clone M2–31. Sequencing methods for the M2-specific PCR products from Rhs 1A1 total DNA were as described above and a detailed account of sequencing strategy of the same is given in the result section. Sequence analysis was carried out using the pcgene 6.5 program (IntelliGenetics), and the Wisconsin GCG nucleic acid sequence analysis software package.
Figure 1.
cDNA sequence of the sense-strand of the M2 dsRNA with the major ORF (ORF A) according to either the universal or the mitochondrial codon usage in filamentous fungi.
Subcellular Fractionation.
For determining subcellular localization of dsRNA, nuclei were purified as described by Gealt et al. (22). Mitochondria were purified using the procedure of Specht et al. (23) as modified by Syminis (24). Fluorescent staining of the nuclear fraction using acridine orange (22), and the mitochondrial fraction using rhodamine-123 (25) were used to tentatively identify the respective fractions, and ensure freedom from contaminating organelles. The postmitochondrial supernatant was used to obtain the membrane fraction (30,000 × g, 30 min per pellet), the ribosomal/virus particle fraction (120,000 × g, 3 hr per pellet), and the cytosolic fraction (postribosomal supernatant). Before lysis and phenol extraction, nuclei and mitochondria were treated with pancreatic RNase, in the absence of NaCl, to eliminate inadvertent, external contamination with dsRNA. Subcellular nucleic acid samples were treated with pancreatic DNase I, and RNase in 0.3 M NaCl to remove DNA and single-stranded RNA (ssRNA), respectively. RNA from the above fractions was analyzed by Northern blot hybridization as described (15).
Southern and Northern Blot Hybridizations.
Genomic DNA was extracted according to Sambrook et al. (26). Extraction of total RNA from Rhs 1AP or Rhs 1A1 was according Logemann et al. (27) with minor modifications. 32P-labeled transcripts of both polarities, and of comparable specific activity, were generated from a linearized M2–31 clone using T7 and T3 RNA polymerase, respectively. Southern and Northern blot analyses were carried out following standard protocols (26).
RESULTS
Analysis of M2 dsRNA Sequence.
Sequencing showed that the M2 dsRNA has 3,570 nucleotides (Fig. 1), and is relatively enriched in AU (57%). The sequence of M2 specific DNA from Rhs 1A1 was identical to that shown in Fig. 1. Two potential hairpin loops are centered at positions 3,165 and 3,552 near the 3′-end of the sense strand (Fig. 2). Moreover, the two ends of the sense strand could fold into a 10-bp panhandle, thus forming a pseudocircular structure, leaving the terminal U unpaired (Fig. 2). Inverted repeats 10–13 bases long begin at bases 2 and 3,561; 337 and 3,163; 1,044 and 2,852; 1,860 and 3,020; 2,019 and 2,563; and 2,575 and 2822. Palindromic repeats of 10–16 bases are centered at bases 621, 1541, 2,220, 2,651, 2,865, 2,873, 3,091, and 3,438.
Figure 2.
Stem-loops at the 3′-end and panhandle formation at the termini of the M2 dsRNA sense-strand. Base positions are shown in parenthesis. The 3,561th C base of the stem-loop could potentially pair with the 11th G base to form a 10-bp panhandle.
Coding Frame and Codon Usage Analysis.
We scanned both strands of the M2 sequence for the presence of termination codons using the universal genetic code and the mitochondrial code for filamentous fungi, which utilizes the universal termination codon UGA as a tryptophan codon (28). Using either genetic code, one strand was shown to have a major ORF (ORF A) of 754 aa in reading frame 2 (bases 422–2,683, Fig. 1). It was designated as sense-strand. Another ORF of 95 aa (bases 2,743–3,027) is also located on this strand in frame 1. The complementary strand could potentially encode polypeptides of 89, 93, and 100 aa (bases 2,052–2,351, 2,365–2,631, and 2,836–3,114). ORF A has a preference of A or U over G or C at the third or wobble position. A, U, G, and C at the wobble position correspond to 30.7%, 30.3%, 20.0%, and 18.8%, respectively. This trend is in accordance with codon usage in the mitochondria of higher fungi (29).
The C-terminal part of ORF A contains the consensus motif (GDD) (30) and the three conserved regions A, B, and D, of RNA-dependent RNA polymerase (RDRP) (Fig. 3). ORF A has a significant sequence similarity with a protein encoded by a hypovirulence-associated mitochondrial dsRNA of Cryphonectria parasitica (ref. 31 and Fig. 3). Furthermore, this polypeptide possesses three transmembrane helices at positions 413–434, 468–486, and 552–571, respectively (32).
Figure 3.
A portion of the alignment of the putative polypeptide of ORF A of the M2 dsRNA and a corresponding region of the RDRP of a mitochondrial dsRNA from Crephonectria parasitica strain NB631 (NB). —, RDRP domains (31) (domains a–d). The consensus amino acids of RDRPs are in boldface. ∗, Identical amino acids; ⋅, conserved amino acids.
Cellular Nucleic Acid Analysis.
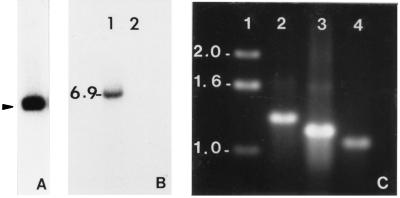
For Southern blot analyses, total genomic DNA was fractionated after digestion with BamHI, which does not have a recognition site within the full-length M2 cDNA sequence and hybridized with oligo-primed, 32P-labeled DNA probes. A restriction DNA fragment of 6.9-kbp hybridized with the M2-specific probe in both Rhs 1A1 (Fig. 4B, lane 1), which contains M2 dsRNA and Rhs 1AP (data not shown), which possesses no detectable amount of M2 (15).
Figure 4.
Southern blots and PCR of total DNA from Rhs 1A1 revealing the presence of a DNA copy of the M2 dsRNA. Autoradiographs of blots of BamHI digests from M2 dsRNA clone M2–31 (A; 16 hr exposure), total DNA from the R. solani isolate Rhs 1A1 (B, lane 1; 48 hr exposure), and total DNA of Nicotiana tabacum (B, lane 2). The blots were hybridized with a 32P-labeled cDNA clone (M2–31) of M2. C shows the ethidium bromide-stained DNA size standards (lane 1), and PCR products amplified with primer combinations encompassing bases 1–1316 (lane 2), 1263–2474 (lane 3), and 2435- 3570 (lane 4) of the M2 sequence. Numbers on the left indicate size (kbp), and arrow shows the position of the recombinant plasmid carrying clone no. 31. A control PCR reaction with tobacco DNA and M2 specific primers did not give rise to any product (data not shown).
The M2-related 6.9-kbp restriction fragment was extremely recalcitrant to cloning, yielding deleted and rearranged inserts. Thus, we amplified the complete M2-specific DNA sequence into three overlapping fragments (bases 1–1,316, 1,263–2,474, and 2,435–3,570) with 20–22 nucleotide primers (Fig. 4C, lanes 2–4), using total DNA from Rhs 1A1 treated with RNase (RNase Plus, 5 Prime → 3 Prime). Sequencing of the three PCR products showed that they were identical to respective M2 cDNA regions. We ruled out the possibility of contamination of the fungal total DNA by the M2 cDNA clones (nos. 31 and 53) by designing primer pairs complementary to the two termini of the M2 dsRNA (bases 1–22 and 3,550–3,570, respectively). The sequence of the former primer is absent from the cDNA clones, and that of the latter has a 6-bp or 7-bp overlap with cDNA clones no. 31 and no. 53, respectively. The above primer combinations gave rise to M2-specific PCR products of the expected size. In addition, we included a PCR reaction with a M2-specific primer (bases 63–82) and a T3 sequencing primer that is present in both clones in similar orientation. This primer combination gave rise to the expected M2-specific PCR product only when the M2 cDNA clone (plasmid) was spiked into the total R. solani DNA in the PCR reaction mixture (data not shown). A control PCR of tobacco DNA with M2 specific primers did not give rise to any product (data not shown).
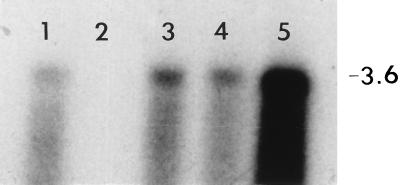
Cellular fractionation data showed that the largest amount of M2 dsRNA was in the cytosol, and a smaller amount was associated with nuclease-treated mitochondrial fractions (Fig. 5). Nuclei contained no detectable amount of the M2 dsRNA.
Figure 5.
Autoradiograph of a dsRNA Northern blot of subcellular fractions from the R. solani isolate Rhs 1A1. The blot includes mitochondrial (lane 1), nuclear (lane 2), membrane (lane 3), high-speed pellet (lane 4), and high-speed supernatant (lane 5) fractions prepared as described in the text. The blot was hybridized with a probe similar to that described in Fig. 4. The position of the 3.6-kbp M2 dsRNA is indicated on the right.
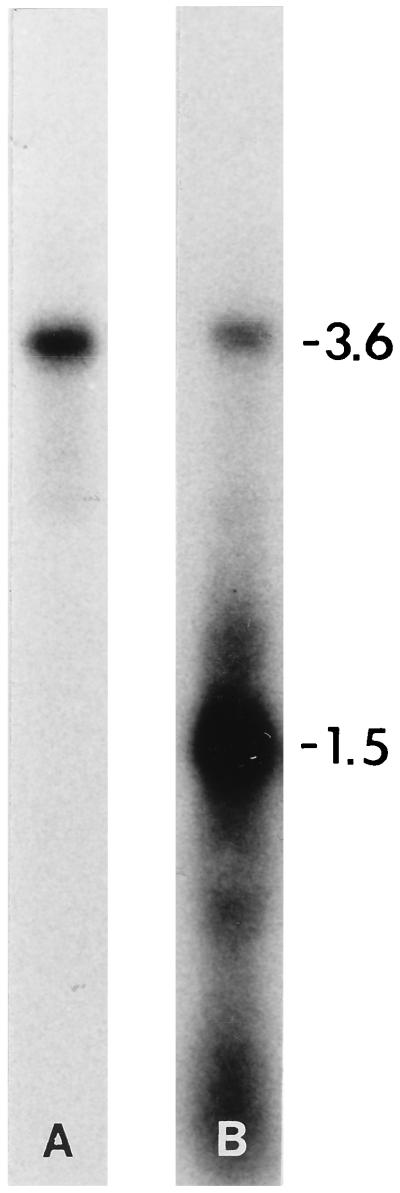
Similar to the results obtained with the oligo-primed DNA-probe of mixed polarity (15), complementary-sense RNA probes hybridized with the 3.6-kbp M2 dsRNA and a ssRNA (Fig. 6B) that migrated to 1.5-kbp position in a nondenaturing gel. In a previous report we have shown that this ssRNA is 3.6 kb and is potentially a full-length transcript of the M2 dsRNA (15). In contrast, the sense RNA probes hybridized with the M2 dsRNA only (Fig. 6A). Total RNA from Rhs 1AP did not hybridize with M2 RNA probe of either polarity (data not shown).
Figure 6.
Northern blot hybridization analyses of total RNA from the R. solani isolate Rhs 1A1 electrophoresed in a nondenaturing gel. Blots were hybridized with sense polarity (A) and complementary-sense polarity (B), 32P-labeled ssRNA probes transcribed in vitro from a 3.4-kbp cDNA clone (M2–31) of M2 dsRNA. Positions of M2 dsRNA (3.6 kbp) and 3.6-kb ssRNA, which migrates to a position corresponding to 1.5 kbp (based on the 1-kbp DNA ladder) (15), are indicated on the right. Total RNA from Rhs 1AP did not hybridize with M2-specific RNA probes of either polarity (data not shown).
A dsRNA-enriched fraction, treated with RNase-free DNase (Promega) and subjected to inverse RT-PCR, gave rise to a PCR product of 584 bp. Cloning and sequencing of this DNA showed a covalent attachment of the two M2 dsRNA termini, thus suggesting the presence of a concatemer or circular dsRNA form of the M2 element in Rhs 1A1 (15).
DISCUSSION
The M2 element is distinct from known dsRNAs found in filamentous plant pathogenic fungi for the following reasons. It is one of three dsRNAs that are apparently suppressed in the virulent culture Rhs 1AP (14), but replicate prolifically in the sector-derived, hypovirulent subculture Rhs 1A1 (15). Transmission of M2 via hyphal anastomosis coincides with a decrease in virulence. Moreover, the degree of virulence reduction appears to be directly proportional to the titer of M2-related dsRNA (14). The existence of a complete copy of M2 in DNA form in both Rhs 1AP and Rhs 1A1 is one of the most distinctive attributes of this dsRNA element. In contrast with the C. parasitica mitochondrial dsRNA (31), M2 occurs in both the cytosol and within mitochondria or nuclease-resistant entities cofractionating with mitochondria (Fig. 5). Northern hybridization analysis had shown the presence of M2-related RNAs that migrate more slowly than the M2 dsRNA (15). This information, in conjunction with the fact that the sequence of the inverse RT-PCR showed a covalent joining of the two M2 dsRNA termini, suggests that replication of M2 involves circular and/or concatemeric structures. Similar to the NB631 mitochondrial dsRNA from C. parasitica (31), M2 has a full-length transcript (Fig. 6 and ref. 15). Whereas the polarity of the NB631 dsRNA transcript is not known, the M2 transcript has a sense-polarity (Fig. 6).
The two 3′-terminal hairpins (Fig. 2) are quite similar in structure and relative position to those occurring on dsRNA of the yeast killer system and shown to be involved in transcription, replication, and assembly of the respective dsRNA (33). Inverted repeats, such as the M2 terminal inverted repeat (Fig. 2), are common in reoviruses (34). Xu et al. (35) demonstrated that any alteration in the 3′-terminal inverted repeat affects the in vitro translational efficiency, 5′-terminal domain nuclease (T1) sensitivity of a genome segment (S8), and packaging of wound tumor virus.
RNA viruses evolve rapidly, and this results in considerable sequence divergence even among related viruses (36). The only gene common to all dsRNA viruses is their RDRP, which shows homology of short stretches of conserved aa or motifs (37, 38). In this respect, it is significant that a stretch of M2 dsRNA has a 72% sequence identity (data not shown) with a region (bases 1,416–1,503) of a hypovirulence-associated, mitochondrial dsRNA from C. parasitica strain NB631 (31). A more extensive sequence similarity (36% identities and 33% conservative substitutions in a region of 287 amino acids) was observed when all four RDRP motifs (30) of the ORF A polypeptide were aligned with those of the RDRP gene of the NB631 dsRNA (Fig. 3).
A stretch of ORF A (190–517 aa) is phylogenetically related to two domains of the pentafunctional polypeptide AROM from yeast (Saccharomyces cerevisiae) that is a mosaic of five monofunctional domains, and carries out steps 2 to 6 of the shikimic acid pathway (39). These two domains correspond to the 3-dehydroquinase (AroD) and shikimate dehydrogenase (AroE). In a region of 343 amino acids, the ORF A putative protein has 21% identities, and 43% conserved substitutions with the AroD and AroE domains of the AROM protein (Fig. 7). Interestingly, R. solani cultures produce phenylacetic acid, a catabolite of the aromatic amino acid phenylalanine in R. solani (40) and other fungi (41). Phenylacetic acid is capable of producing the same disease syndrome on potato as the pathogen itself (13, 42). The amount of phenylacetic acid produced by Rhs 1A1 is significantly lower than that produced by virulent isolates of anatomosis group 3 when grown on defined media (13, 15). We have also shown that the phenylacetic acid-producing capacity of Rhs 1A1 is fully restored when phenylalanine is added to media (data not shown). The above data, in conjunction with the fact that M2 replicates only in Rhs 1A1, suggests that the M2-encoded ORF A polypeptide might interfere with specific steps of the shikimate pathway resulting in reduced levels of phenylalanine. This inference is further supported by the following facts. The shikimate pathway shares two metabolites (3-dehydroquinate and dehydroshikimate) with the quinate pathway that leads to the production of protocatechuate from quinate in many fungi (43). Transcription of the eight-gene cluster of the quinate pathway is positively or negatively regulated by the qutA or qutR gene products, respectively. The QUTA protein (transcription activator) is phylogenetically related to the two N-terminal domains of the pentafunctional AROM protein, dehydroquinate synthase and 5-enolpyruvyl-shikimate-3-phosphate synthase. The QUTR protein (transcription repressor) is phylogenetically related to the extreme C terminus of 5-enolpyruvyl-shikimate-3-phosphate synthase and the three C-terminal domains shikimate kinase, dehydroquinase, and shikimate dehydrogenase (43). The overall sequence similarity of the above two regulatory proteins to the AROM protein is similar to the relatedness between the ORF A protein of the M2 dsRNA element and the respective domains of the AROM protein. We propose that the ORF A protein of M2 acts as a truncated, inactive repressor by binding to the quinate pathway activator but still allowing transcription of the quinate pathway gene cluster. This would divert 3-dehydroquinate and dehydroshikimate from the shikimate to the quinate pathway, thus causing a significant decrease in aromatic amino acid synthesis (44). This hypothesis is further supported by experimental evidence showing that Neurospora crassa mutants possessing an inactive repressor of the quinate pathway exhibit constitutive expression of the genes of this pathway (45).
Figure 7.
A portion of the alignment of the putative polypeptide of ORF A of M2 dsRNA with the Aro D (aa 1,146–1,356), and Aro E (aa 1,357–1,411) domains of the pentafunctional polypeptide (Sc) shown to be involved in the shikimate pathway in S. cerevisiae.
Acknowledgments
This work was supported in part by the U.S. Department of Agriculture/National Research Initiative Competitive Grants Program Grant 95–37303-2410, and the Maine Agriculture and Forestry Experiment Station.
ABBREVIATIONS
- dsRNA
double-stranded RNA
- ssRNA
single-stranded RNA
- RDRP
RNA-dependent RNA polymerase
- RT
reverse transcription
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U51331).
References
- 1.Van Alfen N K, Jaynes R A, Anagnostakis S L, Day P R. Science. 1975;189:890–891. doi: 10.1126/science.189.4206.890. [DOI] [PubMed] [Google Scholar]
- 2.Nuss D L, Koltin Y. Annu Rev Phytopathol. 1990;28:37–58. doi: 10.1146/annurev.py.28.090190.000345. [DOI] [PubMed] [Google Scholar]
- 3.Nuss D L. Microbiol Rev. 1992;56:561–576. doi: 10.1128/mr.56.4.561-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castanho B, Butler E E, Shepherd R J. Phytopathology. 1978;68:1515–1519. [Google Scholar]
- 5.Finkler A, Koltin Y, Barash I, Sneh B, Pozinak B. J Gen Virol. 1985;66:1221–1232. [Google Scholar]
- 6.Zanzinger D H, Bandy B P, Tavantzis S M. J Gen Virol. 1984;65:1601–1605. [Google Scholar]
- 7.Hyakumachi M, Sumino A, Ueda I, Shikada E. Annu Phytopathol Soc Jpn. 1985;51:372–373. [Google Scholar]
- 8.Washington J R, Martin F N. Phytopathology. 1991;81:1162. (abstr.). [Google Scholar]
- 9.Kousik C S, Snow J P, Valverde R A. Phytopathology. 1994;84:44–49. [Google Scholar]
- 10.Bharathan N, Tavantzis S M. Phytopathology. 1990;80:631–635. [Google Scholar]
- 11.Bharathan N, Tavantzis S M. Phytopathology. 1991;81:411–415. [Google Scholar]
- 12.Tavantzis S M. In: Advances in Potato Pest Biology and Management. Zehnder G W, Powelson M L, Jansson R K, Raman K V, editors. St. Paul, MN: Am. Phytopathol. Soc. Press; 1995. pp. 565–579. [Google Scholar]
- 13.Tavantzis S M, Lakshman D K. In: Pathogenesis and Host Specificity in Plant Diseases, Vol. III, Viruses and Viroids. Singh R P, Singh U S, Kohmoto K, editors. Kidlington, U.K.: Pergamon/Elsevier Science; 1995. pp. 249–267. [Google Scholar]
- 14.Jian J, Lakshman D K, Tavantzis S M. Mol Plant–Microbe Interact. 1997;10:1002–1009. doi: 10.1094/MPMI.1998.11.7.601. [DOI] [PubMed] [Google Scholar]
- 15.Lakshman D K, Tavantzis S M. Phytopathology. 1994;84:633–640. [Google Scholar]
- 16.Bandy B P, Leach S S, Tavantzis S M. Plant Dis. 1988;72:596–598. [Google Scholar]
- 17.Carling D E, Leiner R H, Westphale P C. Am Potato J. 1989;66:693–701. [Google Scholar]
- 18.Tavantzis S M. In: Innovations in Pest Management. Engelstad S, Coli W M, Carlson J L, editors. Amherst: Univ. of Mass. Press; 1988. pp. 44–46. [Google Scholar]
- 19.Bandy B P, Tavantzis S M. Am Potato J. 1990;67:189–199. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochman H, Gerber A S, Hartl D L. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gealt M A, Sheir-Neiss G, Morris N R. J Gen Microbiol. 1976;94:204–210. doi: 10.1099/00221287-94-1-204. [DOI] [PubMed] [Google Scholar]
- 23.Specht C A, Dirusso C C, Novotny C P, Ulrich R C A. Anal Biochem. 1982;119:158–163. doi: 10.1016/0003-2697(82)90680-7. [DOI] [PubMed] [Google Scholar]
- 24.Syminis T E. Extrachromosomal DNA in Rhizoctonia solani. Orono: Univ. of Maine; 1989. [Google Scholar]
- 25.Johnson L V, Walsh M L, Chen L B. Proc Natl Acad Sci USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 28.Fox T D. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- 29.Aota, S., Gojobori, T., Ishibashi, F., Maneyama, T. & Ikemura, T. (1988) Nucleic Acids Res. 16 (Suppl.), R315–R402. [DOI] [PMC free article] [PubMed]
- 30.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polashoch J J, Hillman B I. Proc Natl Acad Sci USA. 1994;91:8680–8684. doi: 10.1073/pnas.91.18.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;347:166. (abstr.). [Google Scholar]
- 33.Wickner R B. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo H, Uyeda I, Shikata E. J Gen Virol. 1991;72:2857–2866. doi: 10.1099/0022-1317-72-12-2857. [DOI] [PubMed] [Google Scholar]
- 35.Xu Z, Anzola J, Nalin C M, Nuss D. Virology. 1989;170:511–522. doi: 10.1016/0042-6822(89)90443-1. [DOI] [PubMed] [Google Scholar]
- 36.Koonin E V, Dolja V V. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 37.Koonin E V. Semin Virol. 1992;3:327–339. [Google Scholar]
- 38.Bruenn J A. Nucleic Acids Res. 1993;21:5667–5669. doi: 10.1093/nar/21.24.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duncan K, Edwards R M, Coggins J R. Biochem J. 1987;246:375–386. doi: 10.1042/bj2460375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalghatgi K K, Nambudiri A M D, Bhat J V, Rao S P V. Indian J Biochem Biophys. 1974;11:116–118. [PubMed] [Google Scholar]
- 41.Wat C K, Towers G H N. In: Recent Advances in Phytochemistry: Biochemistry of Plant Phenolics. Swain T, Harvorne J B, Van Sumere C F, editors. New York: Plenum; 1977. pp. 371–432. [Google Scholar]
- 42.Frank J A, Francis S K. Can J Bot. 1976;54:2536–2540. [Google Scholar]
- 43.Hawkins A R, Lamb H K, Moore J D, Charles I G, Roberts C F. J Gen Microbiol. 1993;139:2891–2899. doi: 10.1099/00221287-139-12-2891. [DOI] [PubMed] [Google Scholar]
- 44.Lamb H K, van der Holmbergh J P T W, Newton G H, Moore J D, Roberts C F, Hawkins A R. Biochem J. 1992;284:181–187. doi: 10.1042/bj2840181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huiet L. Proc Natl Acad Sci USA. 1984;81:1174–1178. doi: 10.1073/pnas.81.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]