Summary
Catenins of the p120 subclass display an array of intracellular localizations and functions. Although the genetic knockout of mouse δ-catenin results in mild cognitive dysfunction, we found severe effects of its depletion in Xenopus. δ-catenin in Xenopus is transcribed as a full-length mRNA, or as three (or more) alternatively spliced isoforms designated A, B and C. Further structural and functional complexity is suggested by three predicted and alternative translation initiation sites. Transcript analysis suggests that each splice isoform is expressed during embryogenesis, with the B and C transcript levels varying according to developmental stage. Unlike the primarily neural expression of δ-catenin reported in mammals, δ-catenin is detectable in most adult Xenopus tissues, although it is enriched in neural structures. δ-catenin associates with classical cadherins, with crude embryo fractionations further revealing non-plasma-membrane pools that might be involved in cytoplasmic and/or nuclear functions. Depletion of δ-catenin caused gastrulation defects, phenotypes that were further enhanced by co-depletion of the related p120-catenin. Depletion was significantly rescued by titrated p120-catenin expression, suggesting that these catenins have shared roles. Biochemical assays indicated that δ-catenin depletion results in reduced cadherin levels and cell adhesion, as well as perturbation of RhoA and Rac1. Titrated doses of C-cadherin, dominant-negative RhoA or constitutively active Rac1 significantly rescued δ-catenin depletion. Collectively, our experiments indicate that δ-catenin has an essential role in amphibian development, and has functional links to cadherins and Rho-family GTPases.
Keywords: Gastrulation, Rho, Rac
Introduction
Vertebrate catenins are related to Drosophila Armadillo (with the exception of α-catenin), and associate with the cytoplasmic regions of cadherins localized to cell-cell contacts, while also transducing cellular and developmental signals (Gumbiner, 2005; Lien et al., 2006). δ-catenin (NPRAP, also known as CTNND2) is a member of the p120-catenin subfamily that includes ARVCF (armadillo repeat protein deleted in velo-cardio-facial syndrome), p0071 (PKP4) and plakophilins PKP1-PKP3 (Hatzfeld, 2005; Kosik et al., 2005; McCrea and Park, 2007). Various characteristics distinguish the p120-catenin from the β-catenin subfamily; for example, members of the former contain 9 as opposed to 12 central armadillo repeats, bind to proximal as opposed to distal membrane tail regions of cadherins, and exhibit the capacity to modulate small GTPases (Anastasiadis, 2007; Anastasiadis and Reynolds, 2001; Choi and Weis, 2005).
Mammalian δ-catenin was identified in a search for proteins homologous to plakophilin-1 (Paffenholz and Franke, 1997), and in yeast two-hybrid screens for binding partners of the Alzheimer's pathogenic protein presenilin-1 (PSEN1) (Levesque et al., 1999; Tanahashi and Tabira, 1999; Zhou et al., 1997). Characterization of δ-catenin in mice and humans revealed predominant expression in neural tissues such as brain, whereas hemizygous deletion of the human chromosomal region containing δ-catenin is associated with mental retardation of Cri-du-chat syndrome (Medina et al., 2000). Direct evidence for neural functions of δ-catenin was indicated by gene targeting studies in mice. Gene knockout resulted in severe impairments in cognitive functions and abnormalities in synaptic plasticity, but otherwise few observed effects (Israely et al., 2004). Increased δ-catenin levels were recently identified in human prostatic adenocarcinomas (Burger et al., 2002; Kim, K. et al., 2008; Lu et al., 2005; Lu et al., 2008; Wang et al., 2008), with additional studies also pointing to potential roles in carcinogenesis (Westbrook et al., 2005).
Similarly to the prototypic p120-catenin (Reynolds, 2007), δ-catenin has been reported to have diverse functions in different cellular compartments. For instance, δ-catenin colocalizes with classic cadherins at cell borders (Lu et al., 1999), where together with fellow subfamily members it is thought to modulate cadherin turnover and clustering, thereby affecting cadherin-catenin-mediated adhesive and motility functions (Xiao et al., 2007). Upon growth factor stimulation of epithelial cells, δ-catenin promotes scattering and enhances cell outgrowth (Lu et al., 2002; Lu et al., 1999). In primary hippocampal neurons, overexpression promotes dendritic branching and the protrusion of spines (Arikkath et al., 2008); similarly, in NIH3T3 fibroblasts, δ-catenin induces cytoskeletal reorganization and process extension (Kim et al., 2002). In common with other p120 subfamily members, such effects are believed to occur mainly through the direct or indirect association of δ-catenin with small GTPases (RhoA and Rac1), and consequent downstream effectors (Abu-Elneel et al., 2008; Kim et al., 2008a; Kim et al., 2008b; Martinez et al., 2003). Finally, δ-catenin co-precipitates with Kaiso (Rodova et al., 2004), a POZ zinc-finger transcription factor that acts in various ways at gene promoters (Daniel, 2007; Ioka et al., 2009; Ruzov et al., 2009a; Ruzov et al., 2009b; van Roy and McCrea, 2005). Dynamic relocalization of δ-catenin within membrane, cytosolic and nuclear compartments probably reflects the varied roles of δ-catenin in cell adhesion, motility and nuclear transcription.
To further explore the physiological functions of δ-catenin in a distinct vertebrate system, we used the amphibian Xenopus laevis. Recognized experimental advantages of this system include rapid external development, large embryos that facilitate microinjections and the isolation of explants for phenotypic analyses, and the ability to test mechanistic hypotheses using rescue assays (Sive et al., 2000). Here, we report the isolation and characterization of δ-catenin; our results indicate that it is required in amphibian development and that its in vivo functions are linked to those of cadherins and small GTPases.
Results
Xenopus laevis δ-catenin cDNA isolation
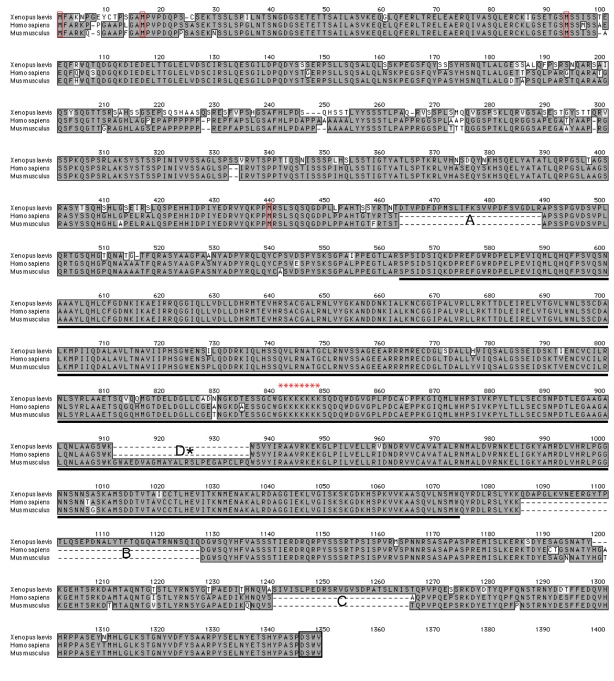
To isolate Xenopus laevis δ-catenin, we used cDNA from adult Xenopus laevis brain in conjunction with 5′ RACE and available Xenopus cDNA sequence information (Ensembl, Xenbase and NIBB XDB). PCR products migrating at the predicted size of full-length δ-catenin were excised from agarose gels, purified and ligated into the Topo vector (Invitrogen). Approximately 30 potential δ-catenin clones were sequenced. The longest isoform isolated was encoded by 3942 base pairs, corresponding to 1314 amino acids with a predicted protein molecular mass of 144 kDa. Xenopus δ-catenin shows strong sequence identity (89.9%) with human or mouse δ-catenin (Fig. 1 and supplementary material Fig. S1A). In common with p120-catenin (Aho et al., 2002; Keirsebilck et al., 1998), we identified four conserved methionines upstream of the central armadillo domain, which might serve as alternative translation initiation sites. Furthermore, alignment of all sequenced cDNAs revealed three sequence elements (A, B and C) present in Xenopus δ-catenin that have not been reported in mammals. Conversely, mouse δ-catenin contains a splicing variant (D) that we have not yet identified in Xenopus (Kawamura et al., 1999). The A, B and C variants are likely to arise from alternative splicing, because these sequences occur precisely within predicted exon junctions (see also below). In keeping with other p120 subfamily members, the multiple predicted translation initiation sites observed in Xenopus δ-catenin, as well as alternative splicing events, would be expected to result in several protein isoforms (major potential isoforms are illustrated in supplementary material Fig. S2B along with their calculated molecular masses), possibly having distinct functional attributes (see also Discussion).
Fig. 1.
δ-catenin primary sequence alignment. Xenopus δ-catenin exhibits strong homology with human and mouse δ-catenin (amino acid identity ∼90%). Alternative splicing events identified in Xenopus are labeled A-C (with D reported in mouse). The central armadillo domain is underlined; a black box outlines the extreme C-terminal PDZ-binding motif (DSWV). Red boxes indicate the four conserved methionines that potentially serve as alternative translation initiation sites. The conserved putative nuclear localization sequence (NLS) is labeled with asterisks.
Xenopus δ-catenin temporal profile
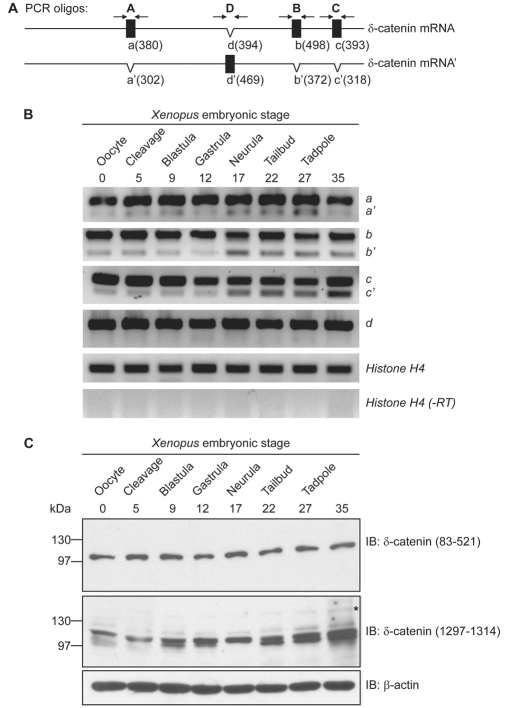
To assess the temporal expression pattern of δ-catenin, we performed semi-quantitative RT-PCR of total RNA isolated from Xenopus embryos at varying developmental stages. Xenopus δ-catenin mRNA transcripts were deposited maternally and expressed throughout early embryonic development (Fig. 2A and 2B). Both the long and short splicing variants of A, B, and C were observed; the short forms of B and C showed increased expression following neurulation, suggesting the possibility of distinct functions later in development. To characterize Xenopus δ-catenin at the protein level, we generated a polyclonal antibody directed against its N-terminal domain (amino acids 83-521). The affinity-purified antibody recognized a δ-catenin isoform migrating at approximately 100 kDa. Using immune-depletion and cadherin co-immunoprecipitation strategies, this band was demonstrated to be authentic δ-catenin (results not shown). We further tested a commercial δ-catenin antibody directed against the C-terminus of mouse δ-catenin (amino acids 1229-1247 that correspond to 1297-1314 of Xenopus δ-catenin). This antibody principally resolves a doublet that likewise migrates at approximately 100 kDa, with additional reactivity sometimes appearing at 130 kDa and 160 kDa (Fig. 2C and results not shown). Consistent with the temporal profile of δ-catenin transcript expression, δ-catenin protein was evident throughout Xenopus embryogenesis, suggesting that δ-catenin might function throughout amphibian development.
Fig. 2.
Xenopus δ-catenin temporal expression profiles. (A) Schematic diagram of δ-catenin alternative splicing events, and the PCR primers used to resolve them. The nucleotide lengths of PCR products are indicated in parentheses. (B) RT-PCR analyses indicate δ-catenin transcripts are deposited maternally and expressed throughout early embryonic stages. Both long (a,b,c) and short (a′,b′,c′) splicing variants were detected, with b′ and c′ having increased expression following neurulation. (C) Immunoblotting confirms δ-catenin protein expression throughout Xenopus embryogenesis. Antibodies directed against Xenopus amino acids 83-521 (N-terminal domain) recognize a δ-catenin isoform migrating at approximately 100 kDa; antibodies directed against Xenopus amino acids 1297-1314 (C-terminal domain) react mainly with a 100 kDa doublet, with reactivity additionally appearing at 130 kDa (marked with an asterisk) and 160 kDa (not shown, but see Fig. 3C). The 130 and 160 kDa bands are most evident following immunoprecipitation (results not shown).
Xenopus δ-catenin spatial characterization
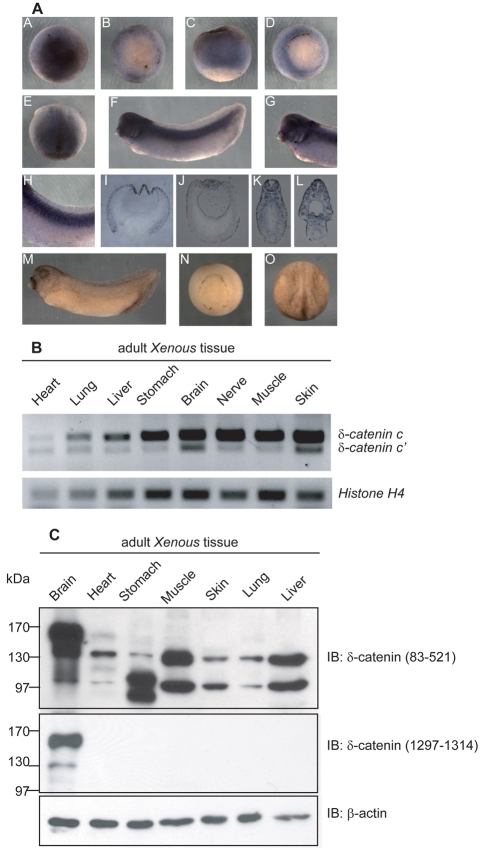
We next examined the spatial expression of δ-catenin by whole-mount in situ RNA hybridization. A dioxigenin-labeled antisense δ-catenin RNA probe was used to detect transcripts concentrated in the ectodermal regions of blastula and gastrula embryos. At neurulation, the anterior and dorsal neural regions displayed δ-catenin signals, with tadpole-stage embryos showing staining in neural or neural crest tissues, such as brain, eye vesicle, ear vesicle, branchial arches and spinal cord, as well as the somites. As a negative control, embryos were hybridized and processed in parallel using the sense probe, wherein no significant signal was detected (Fig. 3A). The wider expression of δ-catenin in Xenopus embryos relative to the near brain-exclusive pattern reported in mammals prompted us to verify our findings using other approaches. All adult Xenopus tissues tested by semi-quantitative RT-PCR reproducibly showed the presence of δ-catenin transcripts (splicing variants c and c′). Tissues showing stronger expression included brain, nerve, muscle and skin, the latter two being derived from mesoderm and non-neural ectoderm sources, respectively (Fig. 3B).
Fig. 3.
Spatial expression of Xenopus δ-catenin. (A) As viewed in animal versus vegetal regions, whole-mount in situ RNA hybridization detects δ-catenin mRNA signals in the ectoderm regions of blastula (subpanels A-C) and gastrula (subpanel D) embryos. At neurulation (subpanel E), the anterior and dorsal neural regions displayed the most apparent signals. Embryos at tadpole stages (subpanel F) showed a distinctive staining pattern in tissues of neural derivation such as brain, eye vesicle, ear vesicle, branchial arches (higher magnification in subpanel G) and spinal cord as well as somites (higher magnification in subpanel H). Subpanels I-L are cross-section views of paraffin-fixed embryos from corresponding stages. Sense probe hybridization was processed in parallel as negative controls (subpanels M-O). (B) RT-PCR analyses detect δ-catenin transcripts in all adult Xenopus tissues examined, with stronger expression in brain, nerve, muscle and skin. (C) Immunoblotting using an N-terminus-directed antibody detected three δ-catenin isoforms migrating at approximately 160, 130 and 100 kDa. The 130 and 100 kDa isoforms are ubiquitously present, whereas the 160 kDa appears to be brain specific. An antibody directed against the δ-catenin C-terminus reacts with the 160 kDa and 130 kDa isoforms in brain.
We further explored the spatial profile of δ-catenin isoform expression using immunoblot analysis of adult Xenopus tissue extracts. Intriguingly, the δ-catenin N-terminal antibody (directed against Xenopus amino acids 83-521), detected three isoforms migrating at approximately 160, 130 and 100 kDa. The 130 and 100 kDa isoforms were ubiquitous, whereas the 160 kDa form appeared to be brain-specific. The δ-catenin C-terminal antibody (directed against Xenopus amino acids 1297-1314), however, detected only the 160 and 130 kDa isoforms in brain using standard film exposures. At longer exposures, the 100 kDa isoform in brain and the 130 kDa isoform in muscle and liver were also observed (Fig. 3C and results not shown). The differing patterns seen upon use of N-terminus versus C-terminus directed δ-catenin antibodies presumably result from distinct immunoreactivities that will require future investigation. In addition to possible post-translational modifications that might differ among isoforms and affect antibody recognition, the N- and C-terminal (and armadillo) domains each undergo alternative splicing events, with the N-terminus likely to undergo further alternative translational initiation. Whatever the underlying basis, our results indicate that Xenopus δ-catenin is expressed in most tissues at the mRNA and protein levels, and that it is later most evident in tissues or organs of neural derivation.
Xenopus δ-catenin subcellular localization
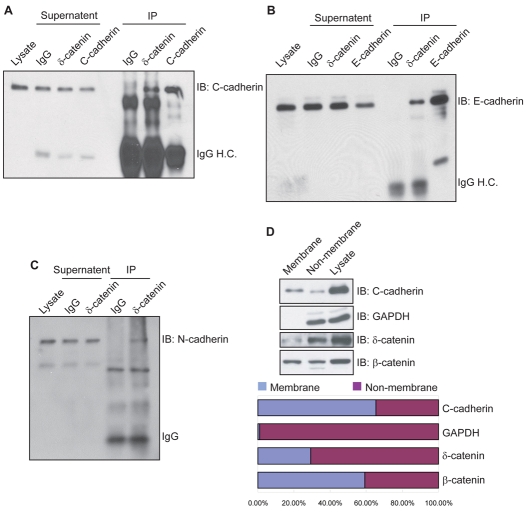
Previous studies of p120-catenin subfamily members indicated diverse functions in differing subcellular compartments (Hatzfeld, 2005; McCrea and Park, 2007; Reynolds, 2007). This includes binding to the membrane-proximal region of cadherin cytoplasmic tails at cell-cell junctions, interaction with small GTPases in cytoplasm, and modulation of gene activity in nucleus. To test for the association of endogenous δ-catenin with classical cadherins, we performed co-immunoprecipitations from gastrula embryo lysates. Pull-downs of δ-catenin included C-cadherin, which is principally expressed at early embryonic cleavage stages (Fig. 4A). Since δ-catenin is expressed more widely across Xenopus tissues than in mammals, we then tested for its interaction with epithelial enriched E-cadherin, as well as neural concentrated N-cadherin. As assessed from neurula and tailbud lysates, both of these cadherins were associated with δ-catenin (Fig. 4B and 4C), further suggesting that Xenopus δ-catenin is present within a number of cadherin complexes existing within cleaving blastomeres, epithelial layers and neural tissues.
Fig. 4.
Xenopus δ-catenin associates with classical cadherins and displays a significant non-membrane-associated fraction. Endogenous δ-catenin complexes were immunoprecipitated using a C-terminus-directed antibody (1297-1314), and immunoblotted using antibodies direct against cadherins. Positive co-immunoprecipitation results suggest an association of δ-catenin with C-cadherin in gastrulating embryos (A), and with E-cadherin (B) and N-cadherin (C) in neurulation stage embryos. Immunoglobulin heavy chain (IgG H.C.) bands are included to reflect the specific versus negative control antibody input. (D) Crude membrane fractionations of gastrula embryos followed by immunoblot analyses indicate the predominant localization (∼70%) of endogenous δ-catenin within non-plasma-membrane pools. As expected, GAPDH is almost exclusively evident in the non-plasma-membrane pools. C-cadherin predominantly resides within the plasma-membrane pool (65%), with the remaining fraction likely to reflect associations with non-sedimenting vesicular stores, endoplasmic reticulum or Golgi.
To characterize δ-catenin further, we examined its subcellular localization in Xenopus early embryos by membrane fractionation. Lysates of gastrula embryos were separated into plasma-membrane versus non-plasma-membrane fractions, and subjected to immunoblot analysis. Endogenous δ-catenin was localized preferably within the non-plasma-membrane pool that includes cytosolic and nuclear components (Fig. 4D). Indeed, relative to β-catenin, a larger proportion of δ-catenin was not associated with the membrane fraction. Our results are therefore consistent with the possibility that Xenopus δ-catenin participates in cytosolic and/or nuclear processes, as well as plasma-membrane- or cadherin-dependent events.
Knockdown of endogenous δ-catenin using antisense morpholinos
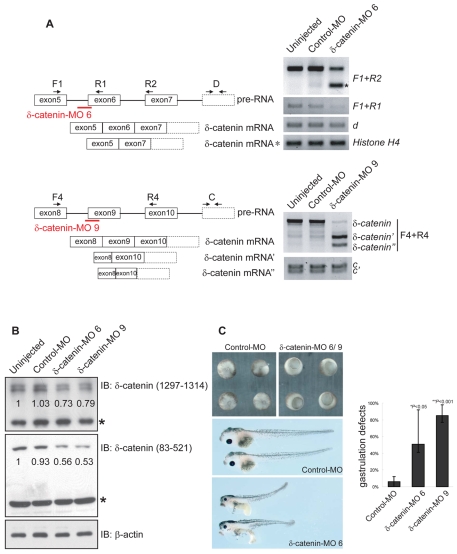
The expression profile of δ-catenin suggests that it might be required during amphibian embryogenesis. To address the developmental functions of δ-catenin, we used an antisense morpholino strategy to disrupt expression of the endogenous protein. As Xenopus δ-catenin is putatively translated from up to four distinct initiation codons, we largely avoided use of morpholinos directed to block any single such initiation. Instead, we used splice junction morpholinos that interfere with pre-RNA splicing, thereby disrupting the protein-coding frame (Draper et al., 2001). Exon 6 of δ-catenin was targeted based on predicted frame shifts that would generate the smallest possible protein product(s). We acquired the sequence of Xenopus δ-catenin exon 6 using PCR amplification of tadpole-stage genomic DNA. δ-catenin morpholino 6 (MO 6) was designed to disrupt splicing between intron 5 and exon 6 (directed against the intron-exon junction). MO 6 was injected into early cleavage stage embryos, and its efficacy was assayed using RT-PCR. Starting at blastula stages, injected embryos displayed a small PCR product (Fig. 5A upper right), which was consistent with the expected alteration in pre-RNA splicing (skipping of exon 6) and was confirmed by DNA sequencing (results not shown). We then used oligos associating with more downstream sequences (splicing variant d), and found no sign of alterations in δ-catenin transcription and RNA stability. Using a similar strategy, we designed another splice junction morpholino (MO 9), targeting the junction of intron 8 with exon 9, and thus predicted to have an impact upon protein products originating from all four putative translational initiation sites. δ-catenin MO 9 activated cryptic splicing sites present in exon 8 and 10, causing exon 9 skipping as well as partial skipping of exon 8(′) and 10(″) (Fig. 5A lower right). At the protein level, we observed that both MO 6 and MO 9 reproducibly reduced the intensity of δ-catenin in immunoblots, with no obvious effects on nonspecific (crossreacting) or control proteins (Fig. 5B).
Fig. 5.
Antisense morpholino depletion of endogenous δ-catenin results in developmental defects. (A) Schematic diagram of the morpholino-based strategy, including the relative positions of PCR diagnostic primers. MO 6 targets δ-catenin RNA at the splice junction between intron 5 and exon 6, whereas MO 9 targets the junction between intron 8 and exon 9. MO 6 produces exon 6 elimination, with predicted codon frame-shift and early polypeptide termination. In addition to the normal transcript, RT-PCR confirmed (via DNA sequencing) the expected alteration in pre-RNA splicing (using oligos F1+R2; marked with an asterisk). Likewise, MO 9 results in alternative splicing, exon elimination and translational termination (PCR product marked with a single apostrophe was confirmed to be skipping of exon 9 and part of exon 8. An additional partial exon 10 deletion is indicated with a double apostrophe). δ-catenin transcription and mRNA stability did not appear to be significantly altered (oligos D and C, see also Fig. 2A), with histone H4 serving as an internal control. (B) Immunoblotting of gastrula stage embryo extracts confirmed the reduction of δ-catenin protein following morpholino injection. Non-specific bands (labeled with asterisks) and β-actin act as loading controls. Numbers indicate relative band intensities normalized to the uninjected control. (C) δ-catenin knockdown results in developmental phenotypes, including significant delays in blastopore closures and gastrulation defects (upper right panel). Although most embryos outwardly appear to recover from these effects and complete blastopore closure, the majority of MO-9-injected embryos were developmentally arrested during early tailbud or tadpole stages and subsequently died. For surviving embryos, abnormalities were again outwardly evident, particularly at tadpole stages, including shortened anterior-posterior axes, smaller craniofacial skeletons and eyes, malformed gut and edema (lower panel). Control embryos injected with standard morpholino displayed no obvious phenotypes (upper left and middle panel). P-values indicate statistical significance.
δ-catenin depletion results in developmental phenotypes, including gastrulation defects
To evaluate the effects of δ-catenin loss-of-function, we injected Xenopus embryos with the morpholinos described earlier. Such embryos underwent early cleavage and blastula stages without noticeable phenotypes. However, at gastrulation, embryos displayed severe delays in blastopore closure with occasional protrusions of endoderm. Although based upon external observation, many embryos appeared to recover and complete gastrulation, most MO-9-injected embryos were arrested and died during early tadpole stages. We then used biochemical assays to assess the effect of δ-catenin depletion upon cell proliferation (phospho-histone H3) or apoptosis (active caspase-3) but our results were inconclusive (data not shown). It is possible that the effect of δ-catenin knockdown upon cell death is below our detection methods when applied at early developmental stages, but is ongoing and ultimately results in observable late-stage tissue necrosis. Such effects might arise from as yet unexamined processes, for example, a poorly understood nuclear function of δ-catenin. Tadpoles that survived displayed abnormalities including shortened anterior-posterior axes and gut malformations, which might have resulted from underlying earlier gastrulation defects. As in p120-catenin depletions (Ciesiolka et al., 2004), these embryos showed reductions in craniofacial skeletal components, suggestive of developmental defects in neural crest populations, accompanied by smaller eyes and reduced eye pigment (Fig. 5C and results not shown). Embryos injected with a standard control morpholino and manipulated similarly displayed no obvious phenotypes.
Since δ-catenin depletion affected blastopore closure, which is a morphogenetic process dependent on the proper orientation of cell intercalations (Keller, 2005; Keller et al., 2003), we investigated whether knockdown of δ-catenin altered convergence-extension movements. For this purpose, we excised explants of dorsal mesoderm that are able to recapitulate ex vivo the movements observed in vivo. Intriguingly, δ-catenin-depleted explants did not show significant defects in elongation compared with controls (supplementary material Fig. S3A). Among other possibilities, δ-catenin knockdown might alter the directed cell rearrangements of more medial or ventral tissues (not examined), or alternatively, the larger coordination of such cell or tissue movements. We then scored δ-catenin-knockdown embryos for their convergence-extension in vivo. δ-catenin MO 6 together with an Alexa Fluor 488 fluorescent tracer were injected into the equatorial region of one dorsal blastomere of four-cell-stage embryos. At mid-gastrula stages, the length-to-width ratios (`aspect ratios') of fluorescence intensity within the dorsal marginal zones (DMZs) were assessed. We did not observe significant differences between δ-catenin-depleted and control embryos (supplementary material Fig. S3B). Collectively, our results indicate that although δ-catenin is required for normal blastopore closure and later developmental events at neurula stages, its depletion does not significantly affect convergent-extension morphogenesis in the dorsal mesoderm compartment of gastrulating embryos.
δ-catenin-knockdown phenotypes are rescued with either δ-catenin or p120-catenin
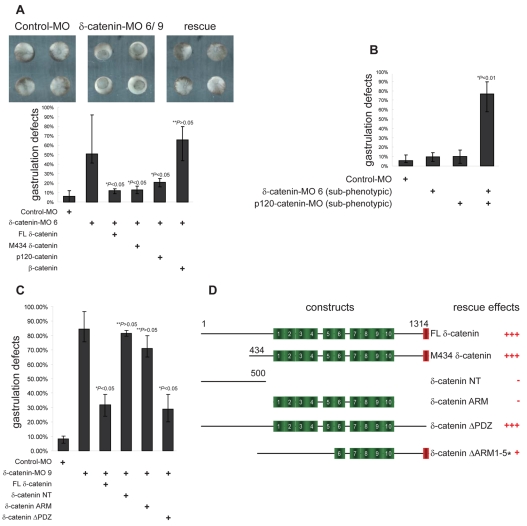
To verify the specificity of observed phenotypes and to test the functional relationship of δ-catenin with selected proteins, we used a rescue strategy based on scoring for blastopore closure effects. Co-injection of a carefully titrated dose (250 pg, see also supplementary material Fig. S4A,B) of full-length δ-catenin RNA (insensitive to splice junction morpholinos) effectively rescued blastopore closure defects, confirming the specificity of the morpholinos. Since p120 subfamily proteins share certain attributes, such as modulation of cadherin and small GTPase functions (Hatzfeld, 2005; McCrea and Park, 2007), we further investigated whether p120-catenin could rescue δ-catenin depletion. Indeed, p120-catenin reproducibly displayed significant rescuing activity, whereas the more distantly related β-catenin did not (Fig. 6A). Moreover, when morpholinos were injected at subphenotypic doses, the combined depletion of δ-catenin and p120-catenin generated much more pronounced effects than either morpholino alone (Fig. 6B). These experiments suggest that δ-catenin and p120-catenin share some functions during Xenopus gastrulation, although importantly, the endogenous level of either protein is not sufficient to compensate for depletion of the other.
Fig. 6.
δ-catenin-knockdown phenotypes are rescued by overexpression of either δ-catenin or p120-catenin. (A) The specificity of δ-catenin-depletion phenotypes were verified through rescue with select constructs. Exogenous and titrated FL (full-length) δ-catenin (see also supplementary material Fig. 4A,B) as well as M434 δ-catenin (initiated with the fourth potential translation start site, see also Fig. 1) largely rescued blastopore closure defects arising from endogenous δ-catenin depletion. Titrated levels of p120-catenin, but not β-catenin, also reproducibly displayed significant rescuing activity. (B) Co-injection of δ-catenin and p120-catenin morpholino, each at subphenotypic doses, produces enhanced phenotypic effects.(C) Use of a depletion-rescue strategy suggests that the PDZ-binding motif of δ-catenin is dispensable for the rescuing capacity of δ-catenin in blastopore closure. By contrast, the δ-catenin N-terminus or armadillo domain in isolation failed to rescue δ-catenin depletion. (D) Schematic presentation of various rescue constructs with a summary of their rescuing effects (asterisk indicates data from Fig. 7D). For all panels P-values indicate statistical significances.
Similarly, a δ-catenin construct beginning at the most downstream putative translation initiation site (M434), or a δ-catenin mutant lacking the C-terminal PDZ-binding motif (δ-catenin ΔPDZ), partially rescued blastopore closure defects (Fig. 6A,C). These results suggest that the δ-catenin depletion phenotypes observed during gastrulation are more likely to be due to reduced functions normally provided by its armadillo and/or C-terminal regions, rather than its N-terminal domain (1-433) or its PDZ motif functions or interactions. In support of this, the N-terminal domain (NT construct; amino-acids 1-500) displayed no rescuing effects. Intriguingly, the isolated δ-catenin armadillo domain (δ-catenin ARM) also failed to produce statistically significant rescues, suggesting it is necessary, but not sufficient for rescuing activity (Fig. 6C). As was the case for the depletion of δ-catenin, its overexpression likewise perturbed development in a dose-dependent manner (supplementary material Fig. S4A). Thus, similarly to other p120-catenin subfamily members (Ciesiolka et al., 2004; Fang et al., 2004; Geis et al., 1998; Paulson et al., 1999), it appears that δ-catenin levels must be maintained within a defined range for embryogenesis to proceed normally.
δ-catenin depletion leads to reduced levels of cadherins and cell adhesion
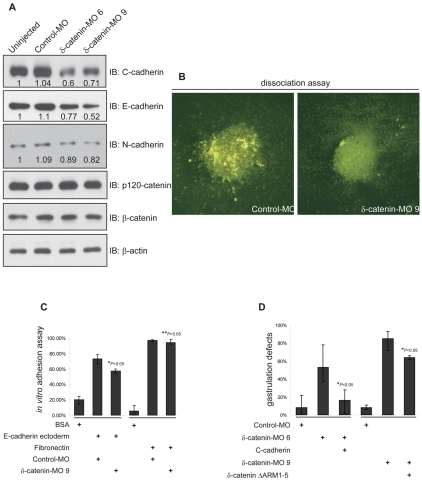
To investigate mechanisms underlying δ-catenin knockdown phenotypes, we assessed cadherin levels and cell adhesive functions. Immunoblotting reproducibly showed considerable to modest reductions in C-, E- and N-cadherin levels following depletion of δ-catenin (Fig. 7A). Other catenin proteins, including p120 and β-catenin, were not significantly altered. Next we evaluated cell adhesive functions using standard animal cap (naive ectoderm) dissociation and re-aggregation assays. In MO-9-injected embryos, cell-cell adhesion was reduced when evaluated using either of these tests (Fig. 7B and results not shown). To further evaluate the effect of δ-catenin depletion upon cadherin-mediated adhesion, we performed in vitro experiments wherein dissociated naive animal cap cells were incubated with glass coverslips coated either with the extracellular domain of mouse E-cadherin, or with the extracellular matrix component fibronectin (negative control). Here, the percentage of cells remaining after washing is taken as a rough measure of adhesive strength. Although we observed modest reductions in cadherin heteromeric interactions as a consequence of δ-catenin depletion (endogenous C-cadherin with exogenous E-cadherin), changes were not observed in the relative levels of fibronectin association (presumably occurring via integrins) (Fig. 7C). Next, we addressed whether reduced cadherin-mediated adhesion might contribute to the observed developmental defects by scoring whether carefully titrated doses of C-cadherin could rescue δ-catenin-knockdown phenotypes. Indeed, C-cadherin displayed a significant if not complete capacity to rescue blastopore closure defects that would otherwise have followed δ-catenin depletion (Fig. 7D). Furthermore, we wished to address the relevance of interactions between δ-catenin and cadherin to the capacity of δ-catenin or C-cadherin to perform rescues. An E-cadherin triple point mutant (AAA) is reported to fail to interact with p120-catenin in mammalian cell lines (Thoreson et al., 2000). Yet in Xenopus embryos, our unpublished results indicate that the corresponding C-cadherin mutant retains partial association with ARVCF, a p120-catenin subfamily member. Therefore, the cadherin AAA mutant might not be an ideal rescue construct to address this question. Instead, we generated a δ-catenin deletion mutant (ΔARM1-5), which lacks armadillo repeats 1-5. ΔARM1-5 failed to interact with endogenous C-cadherin in Xenopus co-immunoprecipitation tests (supplementary material Fig. S5A). However, transfection of ΔARM1-5 in neuro-2a neuroblastoma cells enhanced the formation of neurite-like structures as expected and seen for full-length δ-catenin (supplementary material Fig. S5B). This suggested that although it lacks the ability to bind cadherin, ΔARM1-5 largely retains the capability to modulate small GTPase functions. Such effects were verified through Rho or Rac pull-down assays in HeLa cells, where, in common with full-length protein, δ-catenin ΔARM1-5 inhibits Rho while activating Rac (supplementary material Fig. S5C,D). Notably, δ-catenin ΔARM1-5 showed only weak rescue effects following δ-catenin depletion compared with the full-length construct (Fig. 7D and Fig. 6E). Our findings thus point to the significance of the δ-catenin-cadherin association in carrying out successful Xenopus gastrulation. In this regard, δ-catenin depletion appears to result in reduced cadherin levels and lowered cell-cell adhesion, leading to the observed developmental phenotypes.
Fig. 7.
δ-catenin depletion leads to reduced cadherin functions. (A) Immunoblotting shows that δ-catenin depletion reproducibly leads to reduced C-, E- and N-cadherin levels, whereas p120-catenin and β-catenin levels are not significantly altered.(B) Calcium-dependant adhesive functions were decreased in δ-catenin-depleted naive ectoderm cells. Following calcium removal from ectoderm explants, note the larger cell aggregates remaining after control MO injection. (C) Using an in vitro assay with the extracellular domain of E-cadherin tethered to a solid substrate (chamber glass), cadherin-mediated adhesion is decreased in naive ectoderm cells depleted of δ-catenin. By contrast, a similar assay that uses tethered fibronectin, did not resolve changes in cell attachment (presumably integrin mediated). (D) A titrated dose of exogenous C-cadherin significantly rescues blastopore closure defects induced by depletion of endogenous δ-catenin. By contrast, a δ-catenin mutant construct lacking armadillo repeats 1-5, and failing to co-immunoprecipitate with C-cadherin (supplementary material Fig. 5A) showed minimal rescuing effects. For all panels P-values indicate statistical significance.
δ-catenin depletion perturbs Rho-GTPase activity
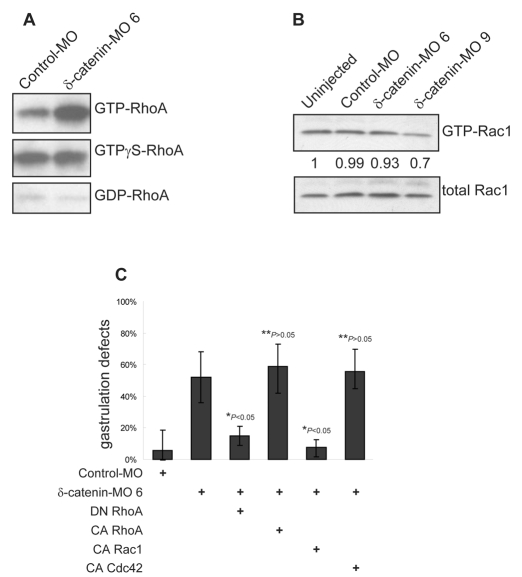
Rho-family small GTPases are critical regulators of actin dynamics (Hall, 1998) and probably interact with δ-catenin in mediating some downstream effects (Kim et al., 2007; Martinez et al., 2003). To test whether Rho activity responds to δ-catenin depletion in Xenopus embryos, we measured GTP-bound levels of RhoA relative to total RhoA levels. Remarkably, a significant activation of RhoA followed δ-catenin knockdown (Fig. 8A). To test whether such changes might be relevant to the developmental phenotypes observed following δ-catenin depletion, we applied a rescue strategy wherein a titrated dose of dominant-negative RhoA (RhoA N19) was co-injected with MO 6. Consistently, we observed significant rescue using the dominant-negative RhoA (RhoA N19) in conjunction with δ-catenin depletion, whereas as a negative control, rescue did not occur using constitutively active RhoA (RhoA V14) (Fig. 8C). Given multiple reports in the literature regarding the activation of Rac1 by p120 subfamily members, including δ-catenin (Ciesiolka et al., 2004; Elia et al., 2006; Fang et al., 2004; Grosheva et al., 2001; Hou et al., 2006; Wildenberg et al., 2006), we measured Rac1 activity following δ-catenin depletion using a similar pull-down approach and, as expected, resolved modest Rac1 inhibition (Fig. 8B). Supporting the functional relevance of such observations, constitutively active Rac1 (Rac1 V12) likewise rescued blastopore closure defects following δ-catenin knockdown, whereas constitutively active Cdc42 did not (Cdc42 V12; negative control) (Fig. 8C). Thus, consistent with cell line and in vivo studies, our results support models wherein knockdown of δ-catenin activates RhoA function, while repressing Rac1.
Fig. 8.
δ-catenin depletion results in activation of RhoA and inhibition of Rac1. (A) Rhotekin pull-down assays reveal strong activation of RhoA following δ-catenin depletion. (B) δ-catenin knockdown modestly reduces active Rac1 levels as measured by PBD (PAK-binding domain) affinity pull-down experiments. (C) Consistent with our biochemical evidence, a titrated dose of dominant-negative (DN) RhoA, but not constitutively active (CA) RhoA, significantly rescues δ-catenin-depletion phenotypes. Likewise, constitutively active Rac1, but not constitutively active Cdc42, rescues blastopore closure defects in δ-catenin-depleted embryos. P-values indicate statistical significance.
Discussion
δ-catenin belongs to the p120-catenin subfamily of armadillo-domain proteins that are capable of modulating various activities, including those involving the cytoskeleton, cadherin complex and developmental signaling (Hatzfeld, 2005; Kosik et al., 2005; Lien et al., 2006; McCrea and Park, 2007; Reynolds, 2007). Similarly to the β-catenin subfamily, altered expression of p120 subfamily members has been correlated with the progression of certain human pathologies (Reynolds and Roczniak-Ferguson, 2004; van Hengel and van Roy, 2007), with causative links to disease now being examined.
δ-catenin associates with multiple proteins of the neural adherens or synaptic junction (Arikkath et al., 2008; Deguchi et al., 2000; Fujita et al., 2004; Ide et al., 1999; Izawa et al., 2002; Jones et al., 2002; Kim et al., 2006; Laura et al., 2002; Lu et al., 2002; Mackie and Aitken, 2005; Martinez et al., 2003; Munoz et al., 2007; Silverman et al., 2007). Most of these interactions occur through its central armadillo domain or its C-terminal PDZ (PSD-95/Discs-large/ZO-1) binding motif. Although the functions of most such associations are unclear, δ-catenin might contribute scaffolding functions and act as a sensor for synaptic activity in neurons (Arikkath et al., 2008; Arikkath et al., 2009; Kosik et al., 2005). Following the removal of δ-catenin in mice, N-cadherin levels are significantly reduced. Consequent effects upon small GTPases may also have affected synapses and additional cell-cell junctions, as well as neural cell shape and migratory behavior (Bamji, 2005). Given its reported association with the transcriptional regulator Kaiso (Rodova et al., 2004), δ-catenin might further modulate gene programs necessary for synaptic or other cell functions.
In contrast to the evident phenotypes that arise in mice following tissue-specific knockout of p120-catenin (Davis and Reynolds, 2006; Elia et al., 2006; Perez-Moreno et al., 2006; Perez-Moreno et al., 2008), even whole-animal targeting of δ-catenin did not generate dramatic developmental or morphogenetic defects. The targeting strategy used for the mouse δ-catenin knockout might have generated an N-terminal fragment that exhibited partial activity. Therefore, we examined δ-catenin in another vertebrate model, Xenopus laevis, using knockdown as opposed to genetic approaches.
Our isolation and characterization of δ-catenin in Xenopus resulted in the identification of three RNA splicing variants not previously identified. The shorter B and C variants are principally expressed in later development, whereas the A variant is more uniformly expressed over time. Using peptide sequences encoded by these elements, we failed to identify (BLAST) similar hits in the human or mouse genome, suggesting that these alternative splicing events may be restricted to amphibians. Element A is located towards the N-terminus of δ-catenin, and when analyzed in silico as an isolated entity (Jpred program), is predicted to form helical structure. Elements B and C are located towards its C-terminus and potentially encode sheet structures (supplementary material Fig. S2A). When we included in the analysis 50 amino acids residing on either side of the element, splicing variant A in particular is predicted to form an alpha-helix spanning nearly 20 amino acids (supplementary material Fig. S2B). In conjunction with the armadillo domain, these secondary motifs may contribute to its larger topology and protein associations.
In contrast to the pattern of mouse δ-catenin transcripts (Ho et al., 2000), we found that Xenopus δ-catenin mRNA and protein exists across all embryonic stages and in adulthood, presumably contributing to development and/or tissue maintenance or repair. Although whole-mount in situ analysis indicated enrichment of δ-catenin RNA in ectodermal and neural derivatives, further differences included the consistent detection of δ-catenin transcripts and protein isoforms in a wider variety of adult amphibian tissues. The largest δ-catenin isoform resolved (160 kDa), migrated at a position consistent with the molecular size predicted from our longest δ-catenin cDNA. Since this isoform showed brain-specific expression, it might possess unique functions in neural tissues. The principal two shorter protein isoforms (130 and 100 kDa), might originate from alternative splicing and/or translation initiation sites, possibly in combination with post-translational modifications, such as proteolytic cleavage or phosphorylation. These modifications and their corresponding functional consequences will require future exploration. Compared with adult tissues, we noticed in embryo extracts, that the longer 130 and 160 kDa δ-catenin isoforms were more difficult to detect. Their presence was ultimately confirmed, however, using combined immunoprecipitation and immunoblot approaches and a panel of available δ-catenin antibodies (supplementary material Table S1). Interestingly, antibodies directed the most N-terminal region failed to recognize the shortest δ-catenin isoform (100 kDa), indicating that it might arise from either a predicted downstream translational initiation site or from N-terminal proteolysis.
To study the in vivo functions of δ-catenin, we used antisense morpholinos designed to interfere with normal RNA splicing, causing frame-shifts (verified via DNA sequencing) and thus premature termination of translation. Using two distinct morpholinos, this approach resulted in consistent phenotypic outcomes that were further indicated to be specific based upon self-rescue analysis. MO 9, directed to the splice junction between intron 8 and exon 9, displayed the greatest effectiveness. In common with the other δ-catenin morpholinos, it altered blastopore closure. This process occurs during gastrulation wherein mesoderm involutes in response to multiple signaling events and mechanical driving forces (such as convergent-extension) (Keller, 2005; Wallingford et al., 2002). Our use of Keller open-face explants, as well as fluorescent tracer analysis within dorsal tissues, failed to reveal an obvious reduction of convergence-extension movements in δ-catenin-depleted embryos. It is plausible that the assays we applied largely reflect superficial cell intercalations, as opposed to deeper cell movements where δ-catenin has a more obvious role. Indeed, in support of this view, dorsal head mesoderm of bisected embryos depleted of δ-catenin displayed a reduced association with the blastocoel roof, perhaps contributing to aberrant tissue movements and blastopore closure (results not shown). Alternatively, δ-catenin depletion might affect more lateral or ventral convergent-extension, or the interrelation between these processes. Additionally, given the complexity of this developmental stage, disruptions other than those upon convergence-extension might produce effects appearing outwardly similar to those observed.
To begin to address biochemical mechanisms by which δ-catenin depletion leads to perturbed gastrulation, we used a δ-catenin ΔARM1-5 construct that fails to interact with endogenous cadherin yet retains considerable capacity to modulate small GTPase functions. Given that ΔARM1-5 exhibited little ability to rescue δ-catenin depletion relative to full-length δ-catenin, the knockdown phenotypes might have arisen largely from altered cadherin-dependent adhesion and/or downstream signaling. As the appropriate forms of exogenous RhoA or Rac1 could rescue δ-catenin-knockdown phenotypes, small GTPases are further likely to be involved in producing the depletion effects. As others have indicated, one possibility is that these small GTPases reside downstream of cadherin (Charrasse et al., 2007; Charrasse et al., 2002; Fukuyama et al., 2006; Goodwin et al., 2003; Johnson et al., 2004; Lampugnani et al., 2002; Nakagawa et al., 2001; Nelson and Chen, 2003; Semina et al., 2009; Yap and Kovacs, 2003), such that their reintroduction restores needed downstream signals, possibly even allowing for cadherin function to be likewise rescued (positive feedback) (Braga, 2002; Braga and Yap, 2005). In all cases, the gross effects we observe following δ-catenin depletion in Xenopus point to its essential role in embryogenesis.
The requirement for δ-catenin in amphibian development, however, is in contrast to the mild effects seen with the knockout mouse. One possible explanation is that during mammalian evolution, other catenins assumed roles that are maintained by δ-catenin in amphibians. In this case, the transition would have occurred while δ-catenin remained the primordial member of the p120 subfamily, displaying the highest primary sequence homology to the single subfamily member present in Drosophila or Caenorhabditis elegans. Interestingly, this invertebrate gene product appears to be dispensable when removed in isolation (Myster et al., 2003; Pettitt et al., 2003), although one study has indicated otherwise (Magie et al., 2002). A second possibility is that the mouse knockout did not produce a complete null; the knockout strategy used is expected to generate an N-terminal fragment that might possess partial native activity. Indeed, a truncated δ-catenin fragment migrating at approximately 50 kDa was seen upon immunoblotting of mouse brain lysates. In Xenopus, our use of MO 9 is likewise predicted to generate an N-terminal δ-catenin fragment. However, in contrast to the mouse knockout, the N-terminal fragment was not detected in immunoblots probed with antibodies that readily detect similar protein fragments expressed in vitro (results not shown). In Xenopus, therefore, such a protein fragment may be targeted for rapid proteolysis. Although the morpholinos we used were effective and specific, Xenopus knockdowns are generally incomplete, as we observed for δ-catenin. Thus, although the depletion phenotypes we observed may be distinct from those seen in a knockout, our results clearly point to a number of key in vivo functions of δ-catenin, and its essential developmental role.
In mice, the expression of δ-catenin is largely restricted to brain, with gene targeting across all tissues producing cognitive deficits and altered synaptic functions, but no obvious additional phenotypes. In our Xenopus studies, δ-catenin depletion interfered with early development. Such distinctions could well result from its wider temporal and spatial expression in amphibians. Indeed, our preliminary results suggest that targeted depletion of δ-catenin in the prospective neural ectoderm inhibits neural crest migration and results in craniofacial cartilage malformations, whereas morpholino delivery to the ventral vegetal region results in kidney defects (results not shown). A final possibility arises from differences in the experimental means by which the loss of δ-catenin function was accomplished in mice versus Xenopus. For example, one could imagine that an acute knockdown might have more profound consequences than the corresponding constitutive knockout carried out at an earlier developmental stage, should the latter provide a more opportune environment for functional compensation to arise.
In summary, our findings are the first to identify an essential role for δ-catenin in amphibian development, as well as providing in vivo evidence of a functional relationship with cadherins and small GTPases. Perturbation of other known or unknown δ-catenin interactions might contribute further to the observed depletion phenotypes, including its putative, but still largely uncharacterized, roles in gene regulation.
Materials and Methods
5′ RACE and cDNA cloning
5′ rapid amplification of cDNA ends (RACE) was performed to isolate Xenopus laevis δ-catenin 5′UTR according to the manufacturer's protocol (5′/3′ RACE kit, Roche). Xenopus δ-catenin cDNA was directly isolated using a high-fidelity PCR system (Roche) and a Xenopus brain cDNA pool reverse transcribed using oligo-dT adaptors (Invitrogen).
RNA isolation and RT-PCR
Total RNA was extracted from Xenopus embryos or adult tissues using Trizol according to the manufacturer's instructions (Invitrogen). Approximately 1 μg total RNA was reverse-transcribed into cDNA pools using oligo-dT or random hexamer oligos (Roche). cDNA was then used as template for PCR amplification. Negative controls for RNA purification were performed by omitting reverse transcriptase in the reverse transcription reaction. PCR primers are as follows: splicing variant A forward, 5′-GATCGGGTGTATCAGAAGCCAC-3′ and reverse, 5′-CCTTCTGGTGGGATAGCTGGT-3′; B forward, 5′-GTAGTAAAGGCAGCGTCTCAG-3′ and reverse, 5′-AGGGGTACCATAGGAATTCC-3′; C forward, 5′-GAACACACGTCTAGGAAAG-3′ and reverse, 5′-AAGTTCACTATAGGGACGAGCAG-3′; D forward, 5′-CAGATCCACCAAAAGGAATA-3′ and reverse, 5′-ATGGCAGTAACAGTGTCATC-3′; histone H4 forward, 5′-CGGGATAACATTCAGGGTA-3′ and reverse, 5′-TCCATGGCGGTAACTGTC-3′; F1, 5′-ATGGCCAAAAAGACATAGAGGATG-3′; R1, 5′-TTGATGAATACTGGAAAGACC-3′; R2, 5′-CCTCACAATCACATGCTGCCTCCCAGTC-3′; F4, 5′-GTCCCTGATTTTTAAGAGTG-3′; R4, 5′-AAGGGAACTGATGCTGTAAC-3′.
Whole-mount in situ RNA hybridization
Procedures used for whole-mount in situ RNA hybridization were as published (Sive et al., 2000). In brief, dioxigenin-labeled sense and antisense RNA probes were generated through in vitro transcription (DIG RNA Labeling Kit, Roche), while NBT/BCIP (Roche) was used as the substrate for alkaline phosphatase reactions. To reveal internal signals, embryos were paraffin embedded and sectioned (10 μm slices) using a Leica RM2245 semi-motorized rotary microtome.
Antibodies, membrane fractionation and immunoblotting
Antibodies generated against a recombinant (6× histidine-tagged) N-terminal domain of Xenopus δ-catenin (amino acids 83-521), were affinity-purified from rabbit crude serum. δ-catenin C-terminal antibody was purchased from either Abcam or Sigma (directed against amino acids 1229-1247 of mouse δ-catenin). Other antibodies were purchased from commercial suppliers (β-actin, Sigma A2066; GAPDH, Santa Cruz sc-25778; E-cadherin, BD Transduction Laboratories 610405; N-cadherin, Calbiochem 205605; RhoA, Santa Cruz Biotechnology sc-179; Rac1, Millipore MAB3735), or are described elsewhere (Fang et al., 2004). Membrane fractionation for Xenopus embryo lysates was performed according to the protocol described by Fagotto and Gumbiner (Fagotto and Gumbiner, 1994). Immunoblotting was performed following standard procedures. Immunoblot band density was quantified using ImageJ 1.38x.
Plasmid constructs and in vitro transcription
To generate plasmids harboring cDNAs for the subsequent production of in vitro transcribed RNAs, high-fidelity PCR approaches were used to place restriction sites into the 5′ and 3′ ends of Xenopus δ-catenin constructs. Inserts were then subcloned into pCS2 vectors, fusing an HA- or Myc-epitope tag to their N- or C-termini. P120-catenin, β-catenin, C-cadherin, RhoA N19 (dominant-negative), RhoA V14 (constitutively active), Rac1 V12 (constitutively active) and Cdc42 V12 (constitutively active) constructs were described previously (Fang et al., 2004; Park et al., 2006). Capped RNAs were synthesized in vitro from NotI-linearized plasmids using the mMESSAGE SP6 kit (Ambion). The integrity of RNA was checked via agarose gel electrophoresis.
Antisense morpholinos
Xenopus δ-catenin intron 5, exon 6, intron 8 and exon 9 sequences were obtained using the Roche high-fidelity PCR system and genomic DNA isolated from tadpole stage embryos. δ-catenin splice junction morpholinos (δ-catenin MO 6 5′-GTACTTGTCCACTTACTTGACTGTA-3′ and δ-catenin MO 9 5′-GCTACGACAGGAAAGTAGGGACAAA-3′), and a standard morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were obtained from Gene Tools. P120-catenin and Kaiso morpholinos were reported previously (Fang et al., 2004; Kim et al., 2004).
Embryos and microinjections
Xenopus embryos were obtained and microinjected according to standard protocols (Sive et al., 2000). Unless indicated, morpholinos and RNA constructs were injected into animal hemisphere(s) of one-, two- or four-cell cleavage stage embryos. Injected embryos were cultured in 0.1× MMR containing 50 μg/ml gentamycin until desired stages. The injection volume for morpholinos or mRNAs was 20 nl at the one-cell stage, 10 nl per blastomere at the two-cell stage and beyond with the doses as follows: 80 ng for δ-catenin and standard morpholinos; 20 ng for subphenotypic dose of δ-catenin morpholinos; 40 ng for p120-catenin morpholino; 10 ng for subphenotypic dose of p120-catenin morpholino; 250 pg for FL δ-catenin, M434 δ-catenin, ΔPDZ and ΔARM1-5 RNA constructs; 500 pg for δ-catenin NT and armadillo constructs; 100 pg for p120-catenin and C-cadherin RNAs; 200 pg for β-catenin RNA, 50 pg for RhoA N19 and RhoA V14 RNAs, 5 pg for Rac1 V12 and Cdc42 V12 RNAs. Embryonic phenotypes were observed and evaluated using a standard binocular dissecting microscope (Zeiss Stemi DV4).
Explants and adhesion assays
Control or δ-catenin morpholino oligonucleotides were injected equatorially into the dorsal blastomeres of four-cell-stage embryos; dorsal marginal zones were excised from injected embryos at the mid-gastrula stage and cultured as open-face Keller explants, as previously described (Kim et al., 2004). Explants were cultured in vitro until control embryonic stage 15. For cell dissociation and re-aggregation assays, animal caps (naive ectoderm) from late blastula or dorsal marginal zones from mid-gastrula embryos were isolated and cultured in calcium free buffer until the cells dissociated. Dissociated cell cultures were then treated with 1 mM each of calcium and magnesium. Dissociation and re-aggregation were monitored to evaluate cell-cell adhesion in control versus δ-catenin-depleted embryos. In vitro cadherin and integrin adhesion assays were based upon published procedures (Ogata and Cho, 2007), with modifications. In brief, Lab-Tek Chamber Slides were coated with either mouse E-cadherin extracellular domain (Sigma E2153) or fibronectin (Sigma F4759), and blocked with BSA (bovine serum albumin). Disassociated blastomeres (typically triplicates of two animal caps per condition) were allowed to adhere for 1 hour at 16°C. Chambers were then inverted and washed in 0.1× MMR for 5 minutes on a rotary shaker. Cell numbers before and after washing were photographically recorded and quantified using Adobe Photoshop CS3.
Rho and Rac activation assay
RhoA activities were measured according to the manufacturer's instructions (Rho Activation Assay Kit, Upstate). In brief, Xenopus embryo or mammalian cell lysates were prepared using the buffer provided, incubated with agarose-coated Rhotekin RBD, pelleted by centrifugation and washed gently for further immunoblot detection. GDP and non-hydrolysable GTPγS were pre-incubated with embryo lysates before Rhotekin RBD binding for negative and positive controls, respectively. Relative active Rac1 levels were determined using a similar experimental design, except that PBD (Pak binding domain) was used for precipitation and total Rac1 as loading control.
Biostatistics
Statistical significance for phenotypic, adhesion or rescue experiments was calculated using SigmaPlot and error bars represent s.d. P<0.05 was deemed significant.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/22/4049/DC1
The authors wish to thank Warren S.-L. Liao, Lei Li, Malgorzata Kloc and Milan Jamrich for constructive comments, as well as laboratory members Jon P. Lyons, Jae-Il Park, Ji-Yeon Hong, Rachel K. Miller, William A. Munoz and Moon-Sup Lee for helpful discussions. P.D.M. was funded through an NIH RO1 (GM52112), a Texas ARP Grant, and an M. D. Anderson Cancer Center Institutional Research Grant. Assistance with DNA sequencing and other core facilities was provided from a National Cancer Institute Core Grant (CA-16672) to M. D. Anderson Cancer Center. Q.L. was supported through an NIH RO1 (CA111891) and a DOD Grant (PC040569). Deposited in PMC for release after 12 months.
References
- Abu-Elneel, K., Ochiishi, T., Medina, M., Remedi, M., Gastaldi, L., Caceres, A. and Kosik, K. S. (2008). A delta-catenin signaling pathway leading to dendritic protrusions. J. Biol. Chem. 283, 32781-32791. [DOI] [PubMed] [Google Scholar]
- Aho, S., Levansuo, L., Montonen, O., Kari, C., Rodeck, U. and Uitto, J. (2002). Specific sequences in p120ctn determine subcellular distribution of its multiple isoforms involved in cellular adhesion of normal and malignant epithelial cells. J. Cell Sci. 115, 1391-1402. [DOI] [PubMed] [Google Scholar]
- Anastasiadis, P. Z. (2007). p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta. 1773, 34-46. [DOI] [PubMed] [Google Scholar]
- Anastasiadis, P. Z. and Reynolds, A. B. (2001). Regulation of Rho GTPases by p120-catenin. Curr. Opin. Cell Biol. 13, 604-610. [DOI] [PubMed] [Google Scholar]
- Arikkath, J., Israely, I., Tao, Y., Mei, L., Liu, X. and Reichardt, L. F. (2008). Erbin controls dendritic morphogenesis by regulating localization of delta-catenin. J. Neurosci. 28, 7047-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikkath, J., Peng, I. F., Ng, Y. G., Israely, I., Liu, X., Ullian, E. M. and Reichardt, L. F. (2009). Delta-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J. Neurosci. 29, 5435-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamji, S. X. (2005). Cadherins: actin with the cytoskeleton to form synapses. Neuron 47, 175-178. [DOI] [PubMed] [Google Scholar]
- Braga, V. M. (2002). Cell-cell adhesion and signalling. Curr. Opin. Cell Biol. 14, 546-556. [DOI] [PubMed] [Google Scholar]
- Braga, V. M. and Yap, A. S. (2005). The challenges of abundance: epithelial junctions and small GTPase signalling. Curr. Opin. Cell Biol. 17, 466-474. [DOI] [PubMed] [Google Scholar]
- Burger, M. J., Tebay, M. A., Keith, P. A., Samaratunga, H. M., Clements, J., Lavin, M. F. and Gardiner, R. A. (2002). Expression analysis of delta-catenin and prostate-specific membrane antigen: their potential as diagnostic markers for prostate cancer. Int. J. Cancer 100, 228-237. [DOI] [PubMed] [Google Scholar]
- Charrasse, S., Meriane, M., Comunale, F., Blangy, A. and Gauthier-Rouviere, C. (2002). N-cadherin-dependent cell-cell contact regulates Rho GTPases and beta-catenin localization in mouse C2C12 myoblasts. J. Cell Biol. 158, 953-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse, S., Comunale, F., Fortier, M., Portales-Casamar, E., Debant, A. and Gauthier-Rouviere, C. (2007). M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol. Biol. Cell 18, 1734-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H. J. and Weis, W. I. (2005). Structure of the armadillo repeat domain of plakophilin 1. J. Mol. Biol. 346, 367-376. [DOI] [PubMed] [Google Scholar]
- Ciesiolka, M., Delvaeye, M., Van Imschoot, G., Verschuere, V., McCrea, P., van Roy, F. and Vleminckx, K. (2004). p120 catenin is required for morphogenetic movements involved in the formation of the eyes and the craniofacial skeleton in Xenopus. J. Cell Sci. 117, 4325-4339. [DOI] [PubMed] [Google Scholar]
- Daniel, J. M. (2007). Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim. Biophys. Acta. 1773, 59-68. [DOI] [PubMed] [Google Scholar]
- Davis, M. A. and Reynolds, A. B. (2006). Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell 10, 21-31. [DOI] [PubMed] [Google Scholar]
- Deguchi, M., Iizuka, T., Hata, Y., Nishimura, W., Hirao, K., Yao, I., Kawabe, H. and Takai, Y. (2000). PAPIN. A novel multiple PSD-95/Dlg-A/ZO-1 protein interacting with neural plakophilin-related armadillo repeat protein/delta-catenin and p0071. J. Biol. Chem. 275, 29875-29880. [DOI] [PubMed] [Google Scholar]
- Draper, B. W., Morcos, P. A. and Kimmel, C. B. (2001). Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis 30, 154-156. [DOI] [PubMed] [Google Scholar]
- Elia, L. P., Yamamoto, M., Zang, K. and Reichardt, L. F. (2006). p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron 51, 43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto, F. and Gumbiner, B. M. (1994). Beta-catenin localization during Xenopus embryogenesis: accumulation at tissue and somite boundaries. Development 120, 3667-3679. [DOI] [PubMed] [Google Scholar]
- Fang, X., Ji, H., Kim, S. W., Park, J. I., Vaught, T. G., Anastasiadis, P. Z., Ciesiolka, M. and McCrea, P. D. (2004). Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. J. Cell Biol. 165, 87-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, T., Okada, T., Hayashi, S., Jahangeer, S., Miwa, N. and Nakamura, S. (2004). Delta-catenin/NPRAP (neural plakophilin-related armadillo repeat protein) interacts with and activates sphingosine kinase 1. Biochem. J. 382, 717-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama, T., Ogita, H., Kawakatsu, T., Inagaki, M. and Takai, Y. (2006). Activation of Rac by cadherin through the c-Src-Rap1-phosphatidylinositol 3-kinase-Vav2 pathway. Oncogene 25, 8-19. [DOI] [PubMed] [Google Scholar]
- Geis, K., Aberle, H., Kuhl, M., Kemler, R. and Wedlich, D. (1998). Expression of the Armadillo family member p120cas1B in Xenopus embryos affects head differentiation but not axis formation. Dev. Genes Evol. 207, 471-481. [DOI] [PubMed] [Google Scholar]
- Goodwin, M., Kovacs, E. M., Thoreson, M. A., Reynolds, A. B. and Yap, A. S. (2003). Minimal mutation of the cytoplasmic tail inhibits the ability of E-cadherin to activate Rac but not phosphatidylinositol 3-kinase: direct evidence of a role for cadherin-activated Rac signaling in adhesion and contact formation. J. Biol. Chem. 278, 20533-20539. [DOI] [PubMed] [Google Scholar]
- Grosheva, I., Shtutman, M., Elbaum, M. and Bershadsky, A. D. (2001). p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J. Cell Sci. 114, 695-707. [DOI] [PubMed] [Google Scholar]
- Gumbiner, B. M. (2005). Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell. Biol. 6, 622-634. [DOI] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, M. (2005). The p120 family of cell adhesion molecules. Eur. J. Cell Biol. 84, 205-214. [DOI] [PubMed] [Google Scholar]
- Ho, C., Zhou, J., Medina, M., Goto, T., Jacobson, M., Bhide, P. G. and Kosik, K. S. (2000). delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 420, 261-276. [PubMed] [Google Scholar]
- Hou, J. C., Shigematsu, S., Crawford, H. C., Anastasiadis, P. Z. and Pessin, J. E. (2006). Dual regulation of Rho and Rac by p120 catenin controls adipocyte plasma membrane trafficking. J. Biol. Chem. 281, 23307-23312. [DOI] [PubMed] [Google Scholar]
- Ide, N., Hata, Y., Deguchi, M., Hirao, K., Yao, I. and Takai, Y. (1999). Interaction of S-SCAM with neural plakophilin-related Armadillo-repeat protein/delta-catenin. Biochem. Biophys. Res. Commun. 256, 456-461. [DOI] [PubMed] [Google Scholar]
- Ioka, H., Doerner, S. K. and Tamai, K. (2009). Kaiso is a bimodal modulator for Wnt/beta-catenin signaling. FEBS Lett. 583, 627-632. [DOI] [PubMed] [Google Scholar]
- Israely, I., Costa, R. M., Xie, C. W., Silva, A. J., Kosik, K. S. and Liu, X. (2004). Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 14, 1657-1663. [DOI] [PubMed] [Google Scholar]
- Izawa, I., Nishizawa, M., Ohtakara, K. and Inagaki, M. (2002). Densin-180 interacts with delta-catenin/neural plakophilin-related armadillo repeat protein at synapses. J. Biol. Chem. 277, 5345-5350. [DOI] [PubMed] [Google Scholar]
- Johnson, E., Theisen, C. S., Johnson, K. R. and Wheelock, M. J. (2004). R-cadherin influences cell motility via Rho family GTPases. J. Biol. Chem. 279, 31041-31049. [DOI] [PubMed] [Google Scholar]
- Jones, S. B., Lanford, G. W., Chen, Y. H., Morabito, M., Kim, K. and Lu, Q. (2002). Glutamate-induced delta-catenin redistribution and dissociation from postsynaptic receptor complexes. Neuroscience 115, 1009-1021. [DOI] [PubMed] [Google Scholar]
- Kawamura, Y., Fan, Q. W., Hayashi, H., Michikawa, M., Yanagisawa, K. and Komano, H. (1999). Expression of the mRNA for two isoforms of neural plakophilin-related arm-repeat protein/delta-catenin in rodent neurons and glial cells. Neurosci. Lett. 277, 185-188. [DOI] [PubMed] [Google Scholar]
- Keirsebilck, A., Bonne, S., Staes, K., van Hengel, J., Nollet, F., Reynolds, A. and van Roy, F. (1998). Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics 50, 129-146. [DOI] [PubMed] [Google Scholar]
- Keller, R. (2005). Cell migration during gastrulation. Curr. Opin. Cell Biol. 17, 533-541. [DOI] [PubMed] [Google Scholar]
- Keller, R., Davidson, L. A. and Shook, D. R. (2003). How we are shaped: the biomechanics of gastrulation. Differentiation 71, 171-205. [DOI] [PubMed] [Google Scholar]
- Kim, H., Han, J. R., Park, J., Oh, M., James, S. E., Chang, S., Lu, Q., Lee, K. Y., Ki, H., Song, W. J. et al. (2008a). Delta-catenin-induced dendritic morphogenesis. An essential role of p190RhoGEF interaction through Akt1-mediated phosphorylation. J. Biol. Chem. 283, 977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Oh, M., Lu, Q. and Kim, K. (2008b). E-Cadherin negatively modulates delta-catenin-induced morphological changes and RhoA activity reduction by competing with p190RhoGEF for delta-catenin. Biochem. Biophys. Res. Commun. 377, 636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S., Bareiss, S., Kim, K. K., Tatum, R., Han, J. R., Jin, Y. H., Kim, H., Lu, Q. and Kim, K. (2006). Presenilin-1 inhibits delta-catenin-induced cellular branching and promotes delta-catenin processing and turnover. Biochem. Biophys. Res. Commun. 351, 903-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., Sirota, A., Chen, Yh, Y. H., Jones, S. B., Dudek, R., Lanford, G. W., Thakore, C. and Lu, Q. (2002). Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp. Cell Res. 275, 171-184. [DOI] [PubMed] [Google Scholar]
- Kim, K., Oh, M., Ki, H., Wang, T., Bareiss, S., Fini, M. E., Li, D. and Lu, Q. (2008). Identification of E2F1 as a positive transcriptional regulator for delta-catenin. Biochem. Biophys. Res. Commun. 369, 414-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. W., Park, J. I., Spring, C. M., Sater, A. K., Ji, H., Otchere, A. A., Daniel, J. M. and McCrea, P. D. (2004). Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat. Cell Biol. 6, 1212-1220. [DOI] [PubMed] [Google Scholar]
- Kosik, K. S., Donahue, C. P., Israely, I., Liu, X. and Ochiishi, T. (2005). Delta-catenin at the synaptic-adherens junction. Trends Cell Biol. 15, 172-178. [DOI] [PubMed] [Google Scholar]
- Lampugnani, M. G., Zanetti, A., Breviario, F., Balconi, G., Orsenigo, F., Corada, M., Spagnuolo, R., Betson, M., Braga, V. and Dejana, E. (2002). VE-cadherin regulates endothelial actin activating Rac and increasing membrane association of Tiam. Mol. Biol. Cell 13, 1175-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laura, R. P., Witt, A. S., Held, H. A., Gerstner, R., Deshayes, K., Koehler, M. F., Kosik, K. S., Sidhu, S. S. and Lasky, L. A. (2002). The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 277, 12906-12914. [DOI] [PubMed] [Google Scholar]
- Levesque, G., Yu, G., Nishimura, M., Zhang, D. M., Levesque, L., Yu, H., Xu, D., Liang, Y., Rogaeva, E., Ikeda, M. et al. (1999). Presenilins interact with armadillo proteins including neural-specific plakophilin-related protein and beta-catenin. J. Neurochem. 72, 999-1008. [DOI] [PubMed] [Google Scholar]
- Lien, W. H., Klezovitch, O. and Vasioukhin, V. (2006). Cadherin-catenin proteins in vertebrate development. Curr. Opin. Cell Biol. 18, 499-506. [DOI] [PubMed] [Google Scholar]
- Lu, Q., Paredes, M., Medina, M., Zhou, J., Cavallo, R., Peifer, M., Orecchio, L. and Kosik, K. S. (1999). delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144, 519-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., Mukhopadhyay, N. K., Griffin, J. D., Paredes, M., Medina, M. and Kosik, K. S. (2002). Brain armadillo protein delta-catenin interacts with Abl tyrosine kinase and modulates cellular morphogenesis in response to growth factors. J. Neurosci. Res. 67, 618-624. [DOI] [PubMed] [Google Scholar]
- Lu, Q., Dobbs, L. J., Gregory, C. W., Lanford, G. W., Revelo, M. P., Shappell, S. and Chen, Y. H. (2005). Increased expression of delta-catenin/neural plakophilin-related armadillo protein is associated with the down-regulation and redistribution of E-cadherin and p120ctn in human prostate cancer. Hum. Pathol. 36, 1037-1048. [DOI] [PubMed] [Google Scholar]
- Lu, Q., Zhang, J., Allison, R., Gay, H., Yang, W. X., Bhowmick, N. A., Frelix, G., Shappell, S. and Chen, Y. H. (2008). Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate 69, 411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie, S. and Aitken, A. (2005). Novel brain 14-3-3 interacting proteins involved in neurodegenerative disease. FEBS J. 272, 4202-4210. [DOI] [PubMed] [Google Scholar]
- Magie, C. R., Pinto-Santini, D. and Parkhurst, S. M. (2002). Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development 129, 3771-3782. [DOI] [PubMed] [Google Scholar]
- Martinez, M. C., Ochiishi, T., Majewski, M. and Kosik, K. S. (2003). Dual regulation of neuronal morphogenesis by a delta-catenin-cortactin complex and Rho. J. Cell Biol. 162, 99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea, P. D. and Park, J. I. (2007). Developmental functions of the P120-catenin sub-family. Biochim. Biophys. Acta. 1773, 17-33. [DOI] [PubMed] [Google Scholar]
- Medina, M., Marinescu, R. C., Overhauser, J. and Kosik, K. S. (2000). Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics 63, 157-164. [DOI] [PubMed] [Google Scholar]
- Munoz, J. P., Huichalaf, C. H., Orellana, D. and Maccioni, R. B. (2007). cdk5 modulates beta- and delta-catenin/Pin1 interactions in neuronal cells. J. Cell Biochem. 100, 738-749. [DOI] [PubMed] [Google Scholar]
- Myster, S. H., Cavallo, R., Anderson, C. T., Fox, D. T. and Peifer, M. (2003). Drosophila p120catenin plays a supporting role in cell adhesion but is not an essential adherens junction component. J. Cell Biol. 160, 433-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, M., Fukata, M., Yamaga, M., Itoh, N. and Kaibuchi, K. (2001). Recruitment and activation of Rac1 by the formation of E-cadherin-mediated cell-cell adhesion sites. J. Cell Sci. 114, 1829-1838. [DOI] [PubMed] [Google Scholar]
- Nelson, C. M. and Chen, C. S. (2003). VE-cadherin simultaneously stimulates and inhibits cell proliferation by altering cytoskeletal structure and tension. J. Cell Sci. 116, 3571-3581. [DOI] [PubMed] [Google Scholar]
- Ogata, S. and Cho, K. W. (2007). Dissection of organizer and animal pole explants from Xenopus laevis embryos and assembly of a cell adhesion assay. J. Vis. Exp. p. 187. [DOI] [PMC free article] [PubMed]
- Paffenholz, R. and Franke, W. W. (1997). Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation 61, 293-304. [DOI] [PubMed] [Google Scholar]
- Park, J. I., Ji, H., Jun, S., Gu, D., Hikasa, H., Li, L., Sokol, S. Y. and McCrea, P. D. (2006). Frodo links Dishevelled to the p120-catenin/Kaiso pathway: distinct catenin subfamilies promote Wnt signals. Dev. Cell 11, 683-695. [DOI] [PubMed] [Google Scholar]
- Paulson, A. F., Fang, X., Ji, H., Reynolds, A. B. and McCrea, P. D. (1999). Misexpression of the catenin p120(ctn)1A perturbs Xenopus gastrulation but does not elicit Wnt-directed axis specification. Dev. Biol. 207, 350-363. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno, M., Davis, M. A., Wong, E., Pasolli, H. A., Reynolds, A. B. and Fuchs, E. (2006). p120-catenin mediates inflammatory responses in the skin. Cell 124, 631-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno, M., Song, W., Pasolli, H. A., Williams, S. E. and Fuchs, E. (2008). Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc. Natl. Acad. Sci. USA 105, 15399-15404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt, J., Cox, E. A., Broadbent, I. D., Flett, A. and Hardin, J. (2003). The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J. Cell Biol. 162, 15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, A. B. (2007). p120-catenin: Past and present. Biochim. Biophys. Acta. 1773, 2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, A. B. and Roczniak-Ferguson, A. (2004). Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene 23, 7947-7956. [DOI] [PubMed] [Google Scholar]
- Rodova, M., Kelly, K. F., VanSaun, M., Daniel, J. M. and Werle, M. J. (2004). Regulation of the rapsyn promoter by kaiso and delta-catenin. Mol. Cell. Biol. 24, 7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov, A., Hackett, J. A., Prokhortchouk, A., Reddington, J. P., Madej, M. J., Dunican, D. S., Prokhortchouk, E., Pennings, S., and Meehan, R. R. (2009a). The interaction of xKaiso with xTcf3: a revised model for integration of epigenetic and Wnt signalling pathways. Development 136, 723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzov, A., Savitskaya, E., Hackett, J. A., Reddington, J. P., Prokhortchouk, A., Madej, M. J., Chekanov, N., Li, M., Dunican, D. S., Prokhortchouk, E., Pennings, S. and Meehan, R. R. (2009b). The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development 136, 729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina, E. V., Rubina, K. A., Rutkevich, P. N., Voyno-Yasenetskaya, T. A., Parfyonova, Y. V. and Tkachuk, V. A. (2009). T-cadherin activates Rac1 and Cdc42 and changes endothelial permeability. Biochemistry. Mosc. 74, 362-370. [DOI] [PubMed] [Google Scholar]
- Silverman, J. B., Restituito, S., Lu, W., Lee-Edwards, L., Khatri, L. and Ziff, E. B. (2007). Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J. Neurosci. 27, 8505-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive, H. L., Grainger, R. M. and Harland, R. M. (2000). Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press.
- Tanahashi, H. and Tabira, T. (1999). Isolation of human delta-catenin and its binding specificity with presenilin 1. NeuroReport 10, 563-568. [DOI] [PubMed] [Google Scholar]
- Thoreson, M. A., Anastasiadis, P. Z., Daniel, J. M., Ireton, R. C., Wheelock, M. J., Johnson, K. R., Hummingbird, D. K. and Reynolds, A. B. (2000). Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 148, 189-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel, J. and van Roy, F. (2007). Diverse functions of p120ctn in tumors. Biochim. Biophys. Acta. 1773, 78-88. [DOI] [PubMed] [Google Scholar]
- van Roy, F. M. and McCrea, P. D. (2005). A role for Kaiso-p120ctn complexes in cancer? Nat. Rev. Cancer 5, 956-964. [DOI] [PubMed] [Google Scholar]
- Wallingford, J. B., Fraser, S. E. and Harland, R. M. (2002). Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev. Cell 2, 695-706. [DOI] [PubMed] [Google Scholar]
- Wang, T., Chen, Y. H., Hong, H., Zeng, Y., Zhang, J., Lu, J. P., Jeansonne, B. and Lu, Q. (2008). Increased nucleotide polymorphic changes in the 5′-untranslated region of delta-catenin (CTNND2) gene in prostate cancer. Oncogene 28, 555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook, T. F., Martin, E. S., Schlabach, M. R., Leng, Y., Liang, A. C., Feng, B., Zhao, J. J., Roberts, T. M., Mandel, G., Hannon, G. J. et al. (2005). A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837-848. [DOI] [PubMed] [Google Scholar]
- Wildenberg, G. A., Dohn, M. R., Carnahan, R. H., Davis, M. A., Lobdell, N. A., Settleman, J. and Reynolds, A. B. (2006). p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell 127, 1027-1039. [DOI] [PubMed] [Google Scholar]
- Xiao, K., Oas, R. G., Chiasson, C. M. and Kowalczyk, A. P. (2007). Role of p120-catenin in cadherin trafficking. Biochim. Biophys. Acta. 1773, 8-16. [DOI] [PubMed] [Google Scholar]
- Yap, A. S. and Kovacs, E. M. (2003). Direct cadherin-activated cell signaling: a view from the plasma membrane. J. Cell Biol. 160, 11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Liyanage, U., Medina, M., Ho, C., Simmons, A. D., Lovett, M. and Kosik, K. S. (1997). Presenilin 1 interaction in the brain with a novel member of the Armadillo family. NeuroReport 8, 2085-2090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.