Abstract
Background
For ruminants reared on grazing systems, gastrointestinal nematode (GIN) parasite infections represent the class of diseases with the greatest impact on animal health and productivity. Among the many possible strategies for controlling GIN infection, the enhancement of host resistance through the selection of resistant animals has been suggested by many authors. Because of the difficulty of routinely collecting phenotypic indicators of parasite resistance, information derived from molecular markers may be used to improve the efficiency of classical genetic breeding.
Methods
A total of 181 microsatellite markers evenly distributed along the 26 sheep autosomes were used in a genome scan analysis performed in a commercial population of Spanish Churra sheep to detect chromosomal regions associated with parasite resistance. Following a daughter design, we analysed 322 ewes distributed in eight half-sib families. The phenotypes studied included two faecal egg counts (LFEC0 and LFEC1), anti-Teladorsagia circumcincta LIV IgA levels (IgA) and serum pepsinogen levels (Peps).
Results
The regression analysis revealed one QTL at the 5% genome-wise significance level on chromosome 6 for LFEC1 within the marker interval BM4621-CSN3. This QTL was found to be segregating in three out of the eight families analysed. Four other QTL were identified at the 5% chromosome-wise level on chromosomes 1, 10 and 14. Three of these QTL influenced faecal egg count, and the other one had an effect on IgA levels.
Conclusion
This study has successfully identified segregating QTL for parasite resistance traits in a commercial population. For some of the QTL detected, we have identified interesting coincidences with QTL previously reported in sheep, although most of those studies have been focused on young animals. Some of these coincidences might indicate that some common underlying loci affect parasite resistance traits in different sheep breeds. The identification of new QTL may suggest the existence of complex host-parasite relationships that have unique features depending on the host-parasite combination, perhaps due to the different mechanisms underlying resistance in adult sheep (hypersensitivity reactions) and lambs (immunity). The most significant QTL identified on chromosome 6 for LFEC1 may be the target for future fine-mapping research efforts.
Background
For ruminants reared on grazing systems, gastrointestinal nematode parasite infections represent the class of diseases with the greatest impact on animal health and productivity [1]. Due to the growing incidence of anthelmintic resistance among most parasite species, there is a need for a sustainable control of gastrointestinal nematode (GIN) parasites. Among the potential strategies, enhancement of host resistance through the selection of resistant animals has been suggested by many researchers. Because of the difficulty of routine collection of phenotypic indicators of parasite resistance, information based on molecular markers can be used to improve the efficiency of classical genetic breeding.
Most studies on the detection of QTL for parasite resistance in sheep have been carried out in sheep populations specialised for meat and/or wool production [2,3], and particularly in young animals [4-7]. However, the variety of sheep breeds and nematode species considered in these studies has resulted in little consensus among the results reported.
In the present study, we carried out a genome scan based on a daughter design in a commercial population of Spanish Churra sheep, an indigenous dairy breed from the region of Castilla y León where the traditional breeding system is based on autochthonous grazing breeds. Even when gastrointestinal parasite infections in Churra sheep are moderate, Strongylid nematode parasites are known to cause substantial production losses in the Churra flocks due to subclinical infection and reduction of the general immune response [8]. In addition, the infection of young replacement females turned out to pasture for the first time may lead to clinical signs of disease such as diarrhoea and even death in some cases [8].
Previously, we quantified the proportion of the phenotypic variation of four parasite resistance traits that are under genetic control [9]. The occurrence of heritable variation has been observed for the four parasite traits studied, which suggests that genetic improvement is possible for these traits. However, the low heritability estimates obtained for the studied indicators of parasite resistance (ranging from 0.09 to 0.21), together with the difficulty of routinely collecting these phenotypes, suggests that the use of marker assisted selection might be of special interest for enhancing the response to selection of these traits. Based on this, and taking advantage of the genotypic information generated in a previous genome screening program undertaken in Churra sheep, we performed an initial QTL scan for four parasite traits measured in eight half-sib families of the Selection Nucleus of ANCHE (National Association of Spanish Churra sheep Breeders).
Methods
Sampled Animals and Measurements
The experimental design used in the present study is the daughter design described by Soller and Genizi [10]. We analysed a total of 322 ewes belonging to eight half-sib families, with an average family size of 40.25 daughters per sire (range: 19-84). Samples from these animals were collected from seven flocks included in the Selection Nucleus of ANCHE (National Association of Spanish Churra sheep Breeders).
As indicators of parasite resistance following natural infection, we used the phenotypes studied by Gutiérrez-Gil et al. [9] for the estimation of genetic parameters of parasite resistance traits in a larger population of Churra sheep (928 ewes). From the 928 animals sampled for parasite resistance traits, those animals belonging to half-family groups with at least about 20 ewes were selected for the present QTL detection experiment, avoiding the analysis of very small families. Hence a total of 322 animals were included in the genome scan analysis reported here. The methodology and techniques used to determine these phenotypes have been described in detail by Gutiérrez-Gil et al. [9]. Below is a brief description of the four phenotypes analysed, followed by a brief comment on the aspect of parasite resistance to which each trait is related:
(i) FEC0: Faecal egg count per gram (epg) at day 0 of the experiment, when all sampled animals received anthelmintic treatment. A modified McMaster technique was used to determine faecal egg counts. After the anthelmintic treatment the animals were exposed to natural infection in the fields following the normal management used by Churra sheep breeders. After a period of about 60 days, the following three measures were performed.
(ii) FEC1: Faecal egg count per gram (epg) at approximately day 60 after beginning the experiment. A modified McMaster technique was used to determine faecal egg counts.
(iii) IgA: The IgA (IgA) levels against a somatic extract of the fourth stage larvae (LIV) from Teladorsagia circumcincta were measured using an ELISA test based on the technique described by Martínez-Valladares et al. [11].
(iv) Peps: The concentration of serum pepsinogen, as measured by fluorometric determination in a 96-well microtitre plate using a technique adapted from Edwards et al. [12].
The number of eggs per gram of faeces (epg) is a measure of eggs produced by adult female parasites within the host animal and is thought to be a good indicator of the parasite infection status of the host [13]. In addition, the serum anti-Teladorsagia circumcincta LIV level (IgA) is an indicator of a specific immune reaction to the fourth stage larvae of T. circumcinta, the most important parasite in Churra sheep. The serum pepsinogen level is an indicator of gastric damage associated with the progression of larvae to adult stages [14]. In Churra sheep, the increase in serum pepsinogen has been found to be triggered by the action of the LIV and early non-egg-laying adults [11]. Hence, the traits studied are likely to represent different aspects related to the host-parasite interaction during infection.
Age of the animals was sorted in six different levels according to the lambing number (from 1 to 5 years, and 6 or more than 6), and their physiological status varied among four different states (dairy, pregnant, dry-not pregnant or peripartum), as we have previously reported [9].
Basic statistics for the four measured traits are given in Additional File 1. Regarding the faecal egg count, the most prevalent genera encountered was Teladorsagia (65.5%), followed by Trichostrongylus spp. (30.5%), Nematodirus spp. (3.1%) and some less frequent genera (1% Chabertia spp. and Oesophagostomum spp.).
Prior to further analysis, the distribution values for FEC0 and FEC1, which were positively skewed, were transformed using a logarithmic transformation [LFEC0 = ln (FEC0+1); LFEC1 = ln (FEC1+1)]. IgA and Peps did not require any transformation. The influence of fixed factors and the estimation of genetic parameters for the studied traits have been reported elsewhere [9].
Data Analysis
Genotyping and Linkage maps
A total of 322 ewes from the complete set of animals sampled for parasite resistance traits (928 ewes) were included in a genome scan analysis carried out in Churra sheep to detect QTL for dairy traits [15-17]. Taking advantage of the genotypic information generated in that genome scan and the linkage maps built for the Churra sheep population, we performed a QTL analysis for the four traits related to parasite resistance considered in this experiment. In this genome scan a total of 182 markers (181 microsatellites and 1 SNP) distributed along the 26 ovine autosomes were genotyped across 1,421 animals belonging to 11 half-sib families. The procedures used for the genotyping of the 182 markers have been described in detail elsewhere [16,17]. The linkage map used in the current work was that generated for the most complete Churra sheep population genotyped (1.421 ewes), which has been reported by Gutiérrez-Gil et al. [18]. This map, which was built with the CRI-MAP 2.4 software [19], showed an average marker interval of 17.86 cM [18] and an information content (IC) for QTL detection of about 0.6 [17]. The use of this map for the parasite resistance genome scan allowed a more accurate estimation of the phase of the paternal sires, yielding therefore more reliable QTL results.
QTL Analysis
Mapping of quantitative trait loci was performed by the multimarker regression method described by Knott et al. [20] for half-sib designs implemented with the HSQM software [21]. Response variables used in the QTL analysis were the Yield Deviations (YD) [22], which are the records expressed as deviations from the population mean and corrected for the corresponding environmental effects. For each trait, the effects included in the YD calculation were those considered in the estimation of the genetic parameters, which had been shown to have a significant influence on the trait (Flock-Year-Season (5 levels), Lambing Number or age (6 levels), and Permanent Environmental effects for the four traits; the sampling interval was also considered for LFEC1). When evidence for a significant effect was found in the across-family analysis, the position with the greatest F-value was considered as the most likely location of the QTL, and the within-family analysis was examined to identify the segregating families and to estimate the QTL size effect.
Chromosome-wise significance thresholds were obtained for each trait-chromosome combination by performing 10,000 random permutations of the phenotypic data [23]. QTL effects were considered significant if they exceeded the 5% chromosome-wise significance threshold (pc-value < 0.05). Genome-wide p-values were obtained by applying the following Bonferroni correction: Pgenomewise = 1-(1-Pchromosomewise)(1/r), where r indicates the contribution of the chromosome to the total genome length [24]. The r parameter was calculated based on the last update of the Australian sheep linkage map [25] (consulted September 2008). The results of the within-family analyses were used to identify the families segregating for each of the QTL identified at the whole population level (those with a within-family pc < 0.05, as determined through permutation testing). Correction for multiple traits was not performed due to the preliminary nature of the genome scan so that we could compare our results with other studies [24]. Empirical 95% confidence intervals (95% CI) were calculated by the bootstrapping method [26].
Results
The regression analysis revealed five significant QTL at the 5% chromosome-wise level on chromosomes 1, 6, 10 and 14, and the QTL on chromosome 6 exceeded the 5% genome-wise significance level. Details regarding the QTL position, significance level and 95% CI calculated for all the QTL identified by the across-family regression analysis are given in Table 1, along with the position and estimated effect for each of the segregating families identified in the within-family analysis.
Table 1.
Characterisation of QTL influencing parasite resistance traits that exceed the 5% chromosome-wise significance threshold in the commercial population of Spanish Churra sheep analysed in this study
| ACROSS-FAMILY ANALYSIS | WITHIIN-FAMILY ANALYSIS | |||||
|---|---|---|---|---|---|---|
| Chr.1 | Trait
Position2 [95% CI]3 |
Flanking interval4 | Pc5 (Pg)6 | Segregating families Pc7 |
Position8 Flanking interval9 | Size effect10 (SD units) |
| 1 |
IgA 35 cM [1-320]. |
BMS835-ILSTS044 | 0.038 | Family 1 0.005 |
38 cM BMS835-ILSTS044 |
0.111 (1.70 SD) |
|
LFEC1 152 cM [122-374] |
INRA006-BMS574 | 0.016 | Family 8 0.013 |
134 cM INRA006-BMS574 |
0.129 (1.31 SD) |
|
| 6 |
LFEC1 84 cM [49-140] |
BM4621-CSN3 | 0.002 (0.041) |
Family 1 0.002 Family 2 0.041 Family 7 0.049 |
79 cM BM4621-CSN3 113 cM CSRD2158-MCM214 105 cM CSN3-CSRD2158 |
0.160 (1.63 SD) 0.082 (0.83 SD) 0.117 (1.19 SD) |
| 10 |
LFEC0 59-60 cM [1-95] |
BM4621-CSN3 | 0.018 | Family 7 0.014 |
65 cM BMS975-TGLA441 |
0.324 (2.53 SD) |
| 14 |
LFEC0 1-2 cM [1-125] |
TGLA357-CSRD247 | 0.018 | Family 1 0.029 Family 6 0.015 |
2 cM TGLA357-CSRD247 1-2 cM TGLA357-CSRD247 |
0.137 (1.07 SD) 0.136 (1.06 SD) |
1 Chromosome number
2,8 Position (cM Haldane) of the chromosome where the maximum F-statistic value was obtained in the across- and within-family analyses, respectively
3 The 95% confidence interval obtained by bootstrapping analysis [26] is shown in square brackets (cM Haldane)
4,9 Markers flanking the position of the maximum F-statistic in the across-family and within- analysis, respectively. Markers in bold caps are < 1 cM from the maximum F-statistic
5,7 Pc = chromosome-wide p-value obtained by permutation test for that position [23]
6 Pg = genome-wide p-value for that position obtained by applying the following Bonferroni correction: Pgenomewide = 1-(1-Pchromosomewise)(1/r), where r indicates the contribution of the chromosome to the total genome length [24]; only indicated for Pg < 0.05
10 Magnitude of the allelic substitution effect calculated for each segregating family, expressed in units of the trait (egg count/g faeces for LFEC0 and LFEC1, D.O ratio for IgA and mUTyr Peps) and in phenotypic SD units of the analysed YDs (value in brackets)
Significant QTL were found for three out of the four traits investigated. Four of the significant linkage associations identified influenced the faecal egg count, and one chromosomal region was associated with the IgA serum indicator. No QTL were observed for Peps. The statistical profiles for the four parasite resistance traits obtained along the four chromosomes where the significant QTL were detected are represented in Figures 1 and 2.
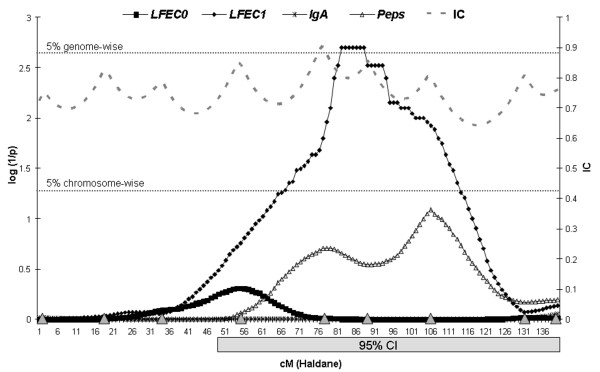
Figure 1.
Across-family statistical profiles obtained on chromosome 6 for the four parasite resistance traits analysed in the present study. The x-axis indicates the relative position on the linkage map (cM Haldane); the y-axis represents the log (1/pg-value); the horizontal lines indicate the 5% genome-wise and 5% chromosome-wise significance thresholds. Information content (IC) obtained along the linkage map is represented at the right, on the y-axis; beginning at the centromeric end, the triangles on the x-axis indicate the relative positions of the markers analysed on this chromosome, which were INRA133, MCM53, MCMA14, BM143, BM4621, CSN3, CSRD2158, MCM214 and BL1038; confidence interval (95% CI), calculated by bootstrapping analysis of the LFEC1 QTL, is shown as a grey box at the bottom of the figure.
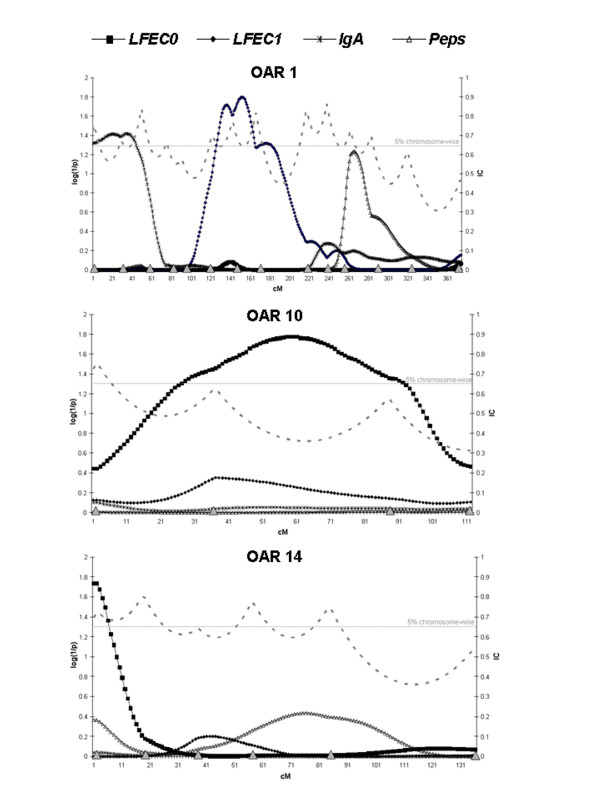
Figure 2.
Across-family statistical profiles obtained on chromosomes 1, 10 and 14 for the four parasite resistance traits analysed in the present study. The x-axis indicates the relative position on the linkage map (cM Haldane); the y-axis represents the log (1/pg-value); information content (IC) obtained along the linkage map of each chromosome is represented at the right, on the y-axis; the horizontal lines indicate the 5% chromosome-wise significance threshold; beginning at the centromeric end, the triangles on the x-axis indicate the relative positions of the markers analysed on each chromosome; see Gutiérrez-Gil et al. [18] for details about marker names and genetic distances.
The most significant QTL was located on the second half of chromosome 6, within the marker interval BM4621-CSN3, and influenced LFEC1 (Figure 1). This QTL reached genome-wise significance (pg = 0.041) and was found to segregate in three out of the eight analysed half-sib groups (Families 1, 2 and 7). For Family 1, which showed the highest significance level, the QTL position suggested by the within-family analysis was coincident with the results of the across-family analysis. For the two other segregating families, the QTL were localised within the first and second downstream marker intervals with regard to the QTL across-family position. Here, it should be noted that the estimation of the across-family QTL position may be biased towards the marker with the highest informativeness in the region, microsatellite marker BM4621, for which all the sires included in the study were heterozygous. This discrepancy regarding the within-family QTL positions may explain the large 95% CI obtained for this QTL, which spanned 91 cM of the chromosome length. However, the possibility that the effect detected at the across-family level can be due to different QTL segregating in the different families can not be ruled out. The magnitude of the allelic substitution effect for this QTL in the segregating families ranged from 0.83 (Family 2) to 1.63 (Family 1) phenotypic SD units (Table 1).
On chromosome 1, a QTL associated with LFEC1 was also found at the 5% chromosome-wise significance level. This QTL was found in the central region of the chromosome (152 cM). Close to the proximal end of the same chromosome, there was evidence for an additional 5% chromosome-wise significant QTL influencing IgA. The other two significant QTL identified by the across-family analysis were found on chromosomes 10 and 14 and showed effects on LFEC0. The QTL on chromosome 10 was located approximately in the middle of the chromosome, whereas the QTL on chromosome 14 was found at the proximal end, close to the first marker analysed on this chromosome.
For each of the QTL identified at the 5% chromosome-wise level, only one family within the population was found to be segregating. The exception to this was the QTL identified on chromosome 14, where the within-family analysis indicates that two of the eight sires are likely to be heterozygous for this QTL (Families 1 and 6). The QTL position suggested by the within-family analysis for these two families was coincident with that estimated in the across-family analysis. The magnitude of the estimated allelic substitution effects for the QTL identified at the 5% chromosome-wise level ranged from 0.83 (chromosome 6 QTL for LFEC1, Family 2) to 2.53 (chromosome 10 QTL for LFEC0, Family 7) phenotypic SD units.
Discussion
Via a genome scan analysis, this study, based on the daughter design described by Soller and Genizi [10], has identified five QTL influencing parasite resistance traits on four sheep autosomes. Considering that two independent traits were analysed (according to a principal component analysis performed with the SAS® package [27]; results not shown), the numbers of tests in our experiment expected by chance alone to be significant at the 5% genome-wise and chromosome-wise level are 0.13 and 2.6, respectively. We identified one and four significant associations in our across-family analysis for these respective significance levels, providing evidence in favour of genuine segregating QTL for parasite resistance traits in the studied population of Churra sheep.
By adapting the method proposed by Weller et al. [28] to our experimental conditions (e.g., the number of ewes and families analysed, marker density and marker informativeness), we estimated that the power of this experiment to detect a QTL with two alleles that occur with equal frequency and influence a trait with a heritability of 0.20 varied between 16% (0.3 phenotypic SD units) and 42% (0.5 phenotypic SD units) according to the magnitude of the allelic substitution effect that we considered. This estimation was performed assuming a type I error rate of 0.05 and 10% recombination between a marker and the QTL. Hence, we should take into account the fact that the low number of animals analysed in the regression analysis had an important negative influence on the statistical power of the experiment, and that a substantial proportion of genuine segregating QTL, especially those with small effects, may not have been identified by the across-family regression analysis performed. Therefore, we suggest that some of the other regions that were identified at a lower significance level in the across-family analysis might represent genuine QTL segregating in individual families. Some of these weak associations, e.g., QTL identified at the 10% chromosome-wise significance level for Peps on chromosomes 1, 2 and 24, IgA on chromosomes 9 and 13, and LFEC1 on chromosome 26 (data not shown), might be confirmed if additional animals were to be included in the analyses.
The lack of coincidence among the QTL identified for the different traits analysed here supports our previously mentioned hypothesis that the traits studied may represent different aspects of the host-parasite interaction during infection. It is possible that the QTL detected for IgA and Peps could be related to the early response to incoming larvae (i.e., hypersensitivity reactions), whereas the QTL for faecal egg counts may be associated with the ability to avoid the development of adult parasites. This agrees with the observations reported by Davies et al. [7], who did not find any coincident QTL between parasitic traits and IgA activity. The lack of coincidence between the QTL influencing LFEC0 and LFEC1, although intriguing, agrees with certain differences observed regarding the correlations between these two traits and the serum indicator traits [9]. As suggested in that work, this could be related to the limited sample period between the faecal egg counts, which could indicate that LFEC1 is a better indicator of the initial immune response triggered by larvae at the beginning of infection.
On the other hand, the allelic substitution effects estimated for the QTL reported herein are likely to be overestimated as a result of the low power of the experiment at the sire-marker level. As shown by Lynch and Walsh [29], the lower the power, the more the effects of a detected QTL are overestimated. Hence, the genuine QTL effects are likely to be much smaller. This result would be in accordance with the work of Houle et al. [30], who suggested that parasite resistance is likely to be controlled by several loci and, therefore, may receive a strong mutation input, which generates genetic variation. This agrees with the complexity of the physiological processes that lead to nematode resistance [31].
In order to compare our QTL analysis results with chromosomal regions previously identified in sheep in relation to parasite resistance traits, we consulted the Sheep Quantitative Trait Loci (QTL) database [32] and other reports available in the literature. We found that some previously published QTL are coincident with the results reported herein. It is worth noting, however, that most of the QTL mapping studies targeting parasite resistance traits in sheep have typically used experimentally challenged animals, and that the parasite species considered vary between studies. In addition, most of the previously reported studies consider parasite resistance traits measured in young animals, mainly meat production lambs.
Marshall et al. [33] recently reported a QTL on chromosome 1 for Haemonchus contortus faecal egg count in 13-month-old Australian sheep. This QTL is close to the marker ADMST4, which maps within the flanking interval of the chromosome 1 QTL reported here for LFEC1. At the proximal end of the same chromosome, within the marker interval EPCDV010-ILSTS044, Díez-Tascón et al. [5] reported a within-family QTL for faecal strongyle egg count and an across-family significant QTL for adult T. columbriformis recovered from the gastric contents of outcrossed lambs at slaughter. These significant associations co-localise with the position of the chromosome 1 QTL influencing IgA that was identified in Churra sheep in our analysis.
On chromosome 6, Beh et al. [4] reported a genome-wise significant QTL for faecal T. columbriformis egg count in lambs after primary challenge. This QTL was confirmed to have a chromosome-wise significance following a secondary challenge and mapped to the interval between markers MCMA22 and MCM214. According to the latest version of the Australian Sheep Linkage Map (v 4.7) [25], the first of these two markers is 16 cM distal to CSN3 (male map), one of the markers flanking the genome-wise significant QTL identified by our across-family regression analysis.
On chromosome 14, Davies et al. [7] reported three QTL related to Nematodirus egg count in Scottish blackface lambs that were located in the last third of the chromosome, whereas the QTL for LFEC0 that we identified mapped to the centromeric end of chromosome 14.
Considering the low resolution of the preliminary genome scans that have been reported thus far regarding QTL position, some of these coincidences might indicate common underlying loci affecting parasite resistance traits. However, this possibility should be confirmed with further studies. Taking into account the high degree of variation between different experiments due to factors such as the type of parasite exposure (natural or artificial challenge), the parasite species, the phenotypic indicators and the breeds of sheep studied, the identification of non-coincident QTL in different experiments may suggest the existence of complex host-parasite relationships that have unique features that depend on the host-parasite combination.
Curiously, our analysis did not find any significant association within two of the regions for which consensus has been found in different studies. These are the regions close to IFNG on chromosome 3 [7,34] and the histocompatibility complex (MHC) region on chromosome 20 [7,35,36]. This discrepancy may be explained by the fact that the studies that found significant associations in these two regions were focused on lambs, whereas our study considered adult ewes. Marshall et al. [33] reported an important age and/or immune status specificity of the QTL for resistance to Haemonchus contortus that they identified in Australian sheep. This specificity is based on the low overlapping levels observed for the QTL that influenced the faecal egg counts measured in animals 6 and 13 months of age. This kind of age-specific mode of action could apply to most parasite infections, which would provide support at the genetic level for the hypothesis suggested by Stear et al. [37] that describes the different mechanisms controlling GIN parasite infections in lambs (antibody response) and adult sheep (hypersensitivity reaction). Also, Balic et al. [38] suggested that the genes that control key mechanisms preventing the establishment of worms in primary infections are different from those involved in subsequent infections. This idea is based on the different pathways that are involved in innate and acquired resistance. However, this hypothesis is challenged by the fact that overall immunity has been successfully achieved through selection for acquired resistance rather than via resistance to primary exposure to worms [31]. All these observations highlight the complexity of parasite resistance and the difficulty of completely understanding the genetic architecture of the physiological mechanisms underlying resistance as well as resilience. As mentioned by Dominik [31], consistency in protocols, experimental materials and analysis approaches would facilitate the generation of phenotypic information that would help to increase our knowledge on this topic.
Conclusion
In conclusion, we present evidence for a significant number of QTL that influence parasite resistance indicator traits in adult dairy sheep. Some of these linkage associations appear to confirm and support the presence of previously published QTL for parasite resistance in lambs, which could indicate that common genes underlie these traits throughout an animal's life. This study represents a starting point for a better understanding of the genetic architecture of parasite resistance in Churra dairy sheep. Further fine-mapping research efforts focused on the most promising regions, e.g., the genome-wise significant QTL identified on chromosome 6, might be simplified as sheep SNP chips become affordable.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BG-G coordinated the genotyping experiments, performed error-checking on genotype data, contributed to interpretation of results and drafted the manuscript. JP, AM and MMV obtained the parasite resistance phenotypic data by collection and analysis of the corresponding samples. LA and YB performed microsatellite genotyping. LFdlF participated in the design and coordination of the study, performed the analyses of genetic parameters and helped to draft the manuscript. FSP selected the animals to be sampled and compiled genealogical information. FARV supervised the collection of phenotypic data and revised the manuscript. JJA conceived of the study, selected the initial marker panel, performed QTL analyses and participated in drafting the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Descriptive statistics of phenotypes analysed in this study. The data provided represents basis statistic of parasite resistance traits including: the total number of observations analysed, mean, range, percentage of 0-values and SD for each studied trait.
Contributor Information
Beatriz Gutiérrez-Gil, Email: beatriz.gutierrez@unileon.es.
Jorge Pérez, Email: dsajpo@unileon.es.
Lorena Álvarez, Email: lalvg@unileon.es.
Maria Martínez-Valladares, Email: mmarva@unileon.es.
Luis-Fernando de la Fuente, Email: f.fuente@unileon.es.
Yolanda Bayón, Email: yolanda.bayon@unileon.es.
Aranzazu Meana, Email: meana@vet.ucm.es.
Fermin San Primitivo, Email: fsant@unileon.es.
Francisco-Antonio Rojo-Vázquez, Email: francisco.rojo@unileon.es.
Juan-José Arranz, Email: jjarrs@unileon.es.
Acknowledgements
This work was supported by the Spanish Ministry of Education and Science (Projects 1FD97-0225 and 1FD97-0427) and by the European Union through the project genesheepsafety (QLK5-2000-00656). Financial support from the Castilla and León regional government (Junta de Castilla y León) by a grant for research groups of excellence (Project GR43) is acknowledged. Beatriz Gutiérrez-Gil is funded by the "Juan de la Cierva Program" of the Spanish Ministry of Education and Science.
References
- Perry BD, Randolph RF, McDermott JJ, Sones KR, Thornton PK. In: Investing in animal health research to alleviate poverty. Perry BD, Randolph, TF, McDermott JJ, Sones KR, Thornton PK, editor. International Livestock Research Institute (ILRI), Nairobi, Kenya; 2002. p. 148.http://www.ilri.org/InfoServ/Webpub/Fulldocs/InvestAnim/index.htm [Google Scholar]
- Morris CA, Bisset SA, Vlassoff A, West CJ, Wheeler M. Genetic parameters for Nematodirus spp. egg counts in Romney lambs in New Zealand. Anim Sci. 2004;79:33–39. [Google Scholar]
- Bishop SC, Jackson F, Coop RL, Stear MJ. Genetic parameters for resistance to nematode infections in Texel lambs and their utility in breeding programmes. Anim Sci. 2004;78:185–194. [Google Scholar]
- Beh KJ, Hulme DJ, Callaghan MJ, Leish Z, Lenane I, Windon RG, Maddox JF. A genome scan for quantitative trait loci affecting resistance to Trichostrongylus colubriformis in sheep. Anim Genet. 2002;33:97–106. doi: 10.1046/j.1365-2052.2002.00829.x. [DOI] [PubMed] [Google Scholar]
- Diez-Tascon C, Macdonald PA, Dodds KG, McEwan JC, Crawford AM. A screen of chromosome 1 for QTL affecting nematode resistance in an ovine outcross population. Proceedings of the 7th World Congress on Genetics Applied to Livestock Production: 19-23 August 2002; Montpellier. 2002. pp. 13–37.
- Crawford AM, Paterson KA, Dodds KG, Diez Tascon C, Williamson PA, Roberts Thomson M, Bisset SA, Beattie AE, Greer GJ, Green RS, Wheeler R, Shaw RJ, Knowler K, McEwan JC. Discovery of quantitative trait loci for resistance to parasitic nematode infection in sheep: I. Analysis of outcross pedigrees. BMC Genomics. 2006;18(7):178. doi: 10.1186/1471-2164-7-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Stear MJ, Benothman M, Abuagob O, Kerr A, Mitchell S, Bishop SC. Quantitative trait loci associated with parasitic infection in Scottish blackface sheep. Heredity. 2006;96:252–258. doi: 10.1038/sj.hdy.6800788. [DOI] [PubMed] [Google Scholar]
- Rojo-Vázquez FA. In: Proceedings of the IXth European Multicolloquium of Parasitology: 18-23 July, Valencia. Mas-Coma S, Bargues MD, Esteban JG, Valero MA, editor. 2004. The present status of helminth infections in small ruminants in Spain: prevalence, anthelmintic resistance and some considerationson their control; pp. 227–231. [Google Scholar]
- Gutiérrez-Gil B, Pérez J, de la Fuente LF, Meana A, Martínez-Valladares M, San Primitivo F, Rojo-Vázquez FA, Arranz JJ. Genetic parameters for resistance to trichostrongylidinfection in dairy sheep. Animal. in press . [DOI] [PubMed]
- Soller MA, Genizi A. The efficiency of experimental designs for the detection of linkage between a marker locus and a locus affecting a quantitative trait in segregating populations. Biometrics. 1978;34:47–55. doi: 10.2307/2529587. [DOI] [Google Scholar]
- Martínez-Valladares M, Vara-Del Río MP, Cruz-Rojo MA, Rojo-Vázquez FA. Genetic resistance to the infection by Teladorsagia circumcincta: IgA and parameters measured at slaughter in Churra sheep. Parasite Immunol. 2005;27:213–218. doi: 10.1111/j.1365-3024.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- Edwards K, Jepson RP, Wood KF. Value of plasma pepsinogen estimation. Br Med J. 1960;2:30–31. doi: 10.1136/bmj.1.5165.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear MJ, Bairden K, Duncan JL, Gettinby G, McKellar QA, Murray M, Wallace DS. The distribution of faecal nematode egg counts in Scottish Blackface lambs following natural, predominantly Ostertagia circumcincta infection. Parasitology. 1995;110:573–581. doi: 10.1017/S003118200006529X. [DOI] [PubMed] [Google Scholar]
- Murray M, Jennings FW, Armour J. Bovine ostertagiasis: structure, function and mode of differentiation of the bovine gastric mucosa and kinetics of the worm loss. Res Vet Sci. 1970;11:417–427. [PubMed] [Google Scholar]
- Gutiérrez-Gil B, El-Zarei MF, Bayón Y, de la Fuente LF, San Primitivo F, Arranz JJ. Genome Scan analysis for detection of QTL influencing somatic cell score in dairy sheep. J Dairy Sci. 2007;90:422–426. doi: 10.3168/jds.s0022-0302(07)72643-7. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Gil B, El-Zarei MF, Alvarez L, Bayón Y, de la Fuente LF, San Primitivo F, Arranz JJ. Quantitative Trait Loci underlying udder morphology traits in dairy sheep. J Dairy Sci. 2008;91:3672–3681. doi: 10.3168/jds.2008-1111. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Gil B, El-Zarei MF, Alvarez L, Bayón Y, de la Fuente LF, San Primitivo F, Arranz JJ. Quantitative trait loci underlying milk production traits in sheep. Anim Genet. 2009;40:423–434. doi: 10.1111/j.1365-2052.2009.01856.x. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Gil B, Arranz JJ, El-Zarei MF, Álvarez L, Pedrosa S, San Primitivo F, Bayón Y. A male linkage map constructed for QTL mapping in Spanish Churra sheep. J Anim Breed Genet. 2008;125:201–204. doi: 10.1111/j.1439-0388.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Documentation for CRI-MAP, version 2.4. Washington University School of Medicine, St Louis, USA; 1990. [Google Scholar]
- Knott SA, Elsen JM, Haley CS. Methods for multiple-marker mapping of quantitative trait loci in half-sib populations. Theor Appl Genet. 1996;93:71–80. doi: 10.1007/BF00225729. [DOI] [PubMed] [Google Scholar]
- Coppieters W, Kvasz A, Farnir F, Arranz JJ, Grisart B, Mackinnon M, Georges M. A rank-based nonparametric method for mapping quantitative trait loci in outbred half-sib pedigrees: application to milk production in a granddaughter design. Genetics. 1998;149:1547–1555. doi: 10.1093/genetics/149.3.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanraden PM, Wiggans GR. Derivation, calculation, and use of national animal model information. J Dairy Sci. 1991;74:2737–2746. doi: 10.3168/jds.S0022-0302(91)78453-1. [DOI] [PubMed] [Google Scholar]
- Churchill G, Doerge R. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning DJ, Janss LL, Rattink AP, van Oers PA, de Vries BJ, Groenen MA, Piel JJ van der, de Groot PN, Brascamp EW, van Arendonk JA. Detection of quantitative trait loci for backfat thickness and intramuscular fat content in pigs (Sus scrofa) Genetics. 1999;152:1679–1690. doi: 10.1093/genetics/152.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australian Sheep Gene Mapping Web Site. http://rubens.its.unimelb.edu.au/~jillm/jill.htm
- Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS® Institute Inc. SAS/STAT® User's Guide, Version 6.12. Cary, NC, USA. 1999.
- Weller JI, Kashi Y, Soller M. Power of daughter and granddaughter designs for determining linkage between marker loci and quantitative trait loci in diary cattle. J Dairy Sci. 1990;73:2525–2537. doi: 10.3168/jds.S0022-0302(90)78938-2. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh JB. Genetics and Analysis of QuantitativeTraits. Sinauer Associates, Inc.; 1998. p. 475. [Google Scholar]
- Houle D, Morikawa B, Lynch M. Comparing mutational variabilities. Genetics. 1996;143:1467–1483. doi: 10.1093/genetics/143.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominik S. Quantitative trait loci for internal nematode resistance in sheep: a review. Genet Sel Evol. 2005;37(Suppl 1):S83–96. doi: 10.1186/1297-9686-37-S1-S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheep Quantitative Trait Loci (QTL) database (SheepQTLdb) http://sphinx.vet.unimelb.edu.au/QTLdb/
- Marshall K, Maddox JF, Lee SH, Zhang Y, Kahn L, Graser HU, Gondro C, Walkden-Brown SW, Werf JH van der. Genetic mapping of quantitative trait loci for resistance to Haemonchus contortus in sheep. Anim Genet. 2009;40:262–272. doi: 10.1111/j.1365-2052.2008.01836.x. [DOI] [PubMed] [Google Scholar]
- Coltman DW, Wilson K, Pilkington JG, Stear MJ, Pemberton JM. A microsatellite polymorphism in the gamma interferon gene is associated with resistance to gastrointestinal nematodes in a naturally-parasitized population of Soay sheep. Parasitology. 2001;122:571–582. doi: 10.1017/S0031182001007570. [DOI] [PubMed] [Google Scholar]
- Schwaiger FW, Gostomski D, Stear MJ, Duncan JL, McKellar QA, Epplen JT. et al. An ovine Major Histocompatibilty Complex DRB1 allele is associated with low faecal egg counts following natural, predominantly Ostertagia circumcincta infection. Int J Parasitol. 1995;25:815–822. doi: 10.1016/0020-7519(94)00216-B. [DOI] [PubMed] [Google Scholar]
- Janssen M, Weimann C, Gauly M, Erhardt G. Associations between infections with haemonchus contortus and genetic markers on ovine chromosome 20. Proceedings of the 7th World Congress on Genetics Applied to Livestock Production: 19-23 August 2002; Montpellier. 2002. pp. 13–11.
- Stear MJ, Strain S, Bishop SC. Mechanisms underlying resistance to nematode infection. Int J Parasitol. 1999;29:51–56. doi: 10.1016/S0020-7519(98)00179-9. [DOI] [PubMed] [Google Scholar]
- Balic A, Bowles VM, Liu YS, Meeusen EN. Local immune responses in sensitized sheep following challenge infection with Teladorsagia circumcincta. Parasite Immunol. 2003;25:375–381. doi: 10.1046/j.1365-3024.2003.00646.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Descriptive statistics of phenotypes analysed in this study. The data provided represents basis statistic of parasite resistance traits including: the total number of observations analysed, mean, range, percentage of 0-values and SD for each studied trait.